Abstract
Current literature accepts 17 species in Penicillium section Sclerotiora. Several produce colonies in bright yellow to orange colours and have monoverticillate conidiophores, apart from P. herquei, P. malachiteum and P. nodositatum, which are biverticillate. The focus of this paper is to refine the concepts of the species currently accepted in the section and introduce five new species, named after the Dutch Royal family as P. vanoranjei, P. maximae, P. amaliae, P. alexiae and P. arianeae. Penicillium vanoranjei produces orange (Dutch = oranje) colonies in culture, and is named after Willem-Alexander Claus George Ferdinand, ‘Zijne Koninklijke Hoogheid de Prins van Oranje’ (translated from Dutch as: ‘His Royal Highness the Prince of Orange’) and his family, to coincide with his coronation. We review the current taxonomic positions of P. lilacinoechinulatum and P. nodositatum, both currently considered to be synonyms of P. bilaiae. Sequence data generated in this study show that both species are phylogenetically distinct. Penicillium lilacinoechinulatum is closely related to P. amaliae sp. nov., whereas P. nodositatum does not belong to Penicillium sensu stricto. All species were compared morphologically and phylogenetically, based on β-tubulin and calmodulin DNA data. A table summarising the morphological characters of all species is included, together with photomicrographs and recommended DNA markers for identification.
Keywords: arthropod vectoring, beta-tubulin, internal transcribed spacer region (ITS)
INTRODUCTION
In anticipation of recent changes in the International Code of Nomenclature for algae, fungi and plants (ICN, previously known as the International Code of Botanical Nomenclature, ICBN), which ended dual nomenclature in fungi (McNeill et al. 2012), the family Trichocomaceae was revised using a four-gene phylogeny by Houbraken & Samson (2011). In that paper, Penicillium subgenus Biverticillium was separated from Penicillium sensu stricto (s.str.) and its species were combined into Talaromyces in a subsequent paper (Samson et al. 2011). Species of Penicillium s.str. were divided among two subgenera, Aspergilloides and Penicillium, and 25 sections (Houbraken & Samson 2011). The section Sclerotiora represents one of these. Seventeen species were included in section Sclerotiora by Houbraken & Samson (2011), most with monoverticillate conidiophores. Some exceptions are P. herquei, P. malachiteum and P. nodositatum (= P. bilaiae fide Houbraken & Samson 2011), which produce biverticillate conidiophores. In general, colonies have bright yellow or orange pigments, which may occur in mycelia, sclerotia, ascocarps, soluble pigments or colony reverse pigmentation (Pitt 1979, Savard et al. 1994, Houbraken & Samson 2011, Rivera & Seifert 2011). Also, species often produce loosely funiculose colony textures and conidiophores with short stipes, as seen in P. bilaiae, P. hirayamae and P. viticola (Nonaka et al. 2011).
Section Sclerotiora is phylogenetically well defined, with several recent studies introducing new species (Peterson 2000, Peterson et al. 2003, 2004, Nonaka et al. 2011, Rivera & Seifert 2011, Rivera et al. 2012). Rivera & Seifert (2011) addressed the problematic P. sclerotiorum complex using a five-gene phylogeny combined with morphological studies, introducing three new species and a phylogenetically accurate description for P. sclerotiorum. In a subsequent paper, Rivera et al. (2012) introduced two additional species to the complex.
In this paper, we describe five new species isolated from soil. Although this group is phylogenetically well studied, several species descriptions have not been updated using modern, standardized techniques. Therefore, we summarise the most important morphological characters in table format and provide photomicrographs for the remaining species of section Sclerotiora not treated by Rivera & Seifert (2011) or Rivera et al. (2012).
Several names associated with section Sclerotiora, including putative synonyms of some species, are also considered in this paper. Based on sequence data, P. nodositatum was tentatively placed in synonymy with P. bilaiae by Houbraken & Samson (2011). The uncertainty reflected the fact that the latter species is strictly monoverticillate, whereas P. nodositatum was described as biverticillate (Valla et al. 1989). The synonymy of P. nodositatum with P. bilaiae would thus be surprising, and is reconsidered below. Penicillium multicolor was previously mentioned as a problematic taxon (Pitt 1979, Houbraken & Samson 2011, Rivera & Seifert 2011). The descriptions by Grigorieva-Manoilova & Poradielova (1915) and Raper & Thom (1949) are at odds, and Pitt (1979) considered P. multicolor a dubious name. Two different strains have been considered to represent ex-type cultures, namely (CBS 501.73), closely related to P. fellutanum (Houbraken & Samson 2011, Rivera & Seifert 2011), and Raper & Thom’s strain (NRRL 2060 = CBS 134565), considered the ex-type strain by Peterson & Horn (2009). The latter has unique sequences in section Sclerotiora (Peterson & Horn 2009, Houbraken & Samson 2011) and we formally describe it as P. maximae below. Finally, reconsideration of P. lilacinoechinulatum, previously considered a synonym of P. bilaiae (Pitt 1979, Pitt et al. 2000), shows it to be a distinct species in section Sclerotiora.
MATERIALS AND METHODS
Strains
Reference and ex-type strains (summarised in Table 1) were obtained from the public collection of the CBS-KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands (CBS), with additional strains retrieved from the working culture collection of the Applied and Industrial Mycology department (DTO) at the same institution. Some strains were originally received and deposited into the collection from J.I. Pitt (Australia), C. Silva Pereira (Portugal), C.M. Visagie and K. Jacobs (South Africa). Many of the South African strains are also deposited in the Canadian Collection of Fungal Cultures associated with the herbarium DAOM, Agriculture & Agri-Food Canada, Ottawa. Raper & Thom’s P. multicolor strain (NRRL 2060) was provided by the United States Department of Agriculture, Agricultural Research Service (USDA-ARS), Peoria, United States.
Table 1.
Strains used for phylogenetic analyses of Penicillium section Sclerotiora.
| Species | Strains | Substrate and locality | GenBank accession no. |
||
|---|---|---|---|---|---|
| ITS | Btub | CMD | |||
| P. adametzii | CBS 209.28T = ATCC 10407 = IMI 039751 = NRRL 737 | Soil under conifers, Poznan, Poland | JN714929 | JN625957 | KC773796 |
| DTO 190A8 | Soil, Spanderswoud, Netherlands | KC773822 | KC773772 | KC773797 | |
| P. adametzioides | CBS 313.59T = ATCC 18306 = IMI 068227 = NRRL 3405 | Soil, Japan | JN686433 | JN799642 | JN686387X |
| DTO 78A7 = IBT 23667 | Unknown | KC773825 | KC773775 | KC773800 | |
| DTO 78A9 = IBT 27906 | Unknown | KC773826 | KC773776 | KC773801 | |
| DTO 78F2 = IBT 10870 | Unknown | KC773827 | KC773777 | KC773802 | |
| P. alexiae | CBS 134558T = DTO 118H8 | Quercus suber forest soil, Tunisia | KC790400 | KC773778 | KC773803 |
| P. amaliae | CBS 134209T = DTO 183F3 = DAOM 241034 = CV1875 | Protea repens infructescence, Struisbaai, South Africa | JX091443 | JX091563 | JX141557 |
| CBS 134211 = DTO 181C6 = CV 204 | Protea repens infructescence, Stellenbosch, South Africa | JX091444 | JX091560 | JX141554 | |
| CBS 134212 = DTO 181F7 = DAOM 241031 = CV 401 | Protea repens infructescence, Stellenbosch, South Africa | JX091440 | JX091558 | JX141556 | |
| CBS 134555 = DTO 181A5 = DAOM 241032 = CV 112 | Protea repens infructescence, Stellenbosch, South Africa | JX091441 | JX091559 | JX141553 | |
| CBS 134556 = DTO 181C9 = DAOM 241033 = CV 227 | Protea repens infructescence, Stellenbosch, South Africa | JX091445 | JX091561 | JX141555 | |
| CBS 134557 = DTO 57A8 | Insect larva, New South Wales, Australia | KC790401 | KC773788 | KC779542 | |
| P. angulare | CBS 130293T = IBT 27051 = NRRL 28157 | Polypore on dead conifer stump, New Mexico, USA | AY313613 | KC773779 | KC773804 |
| DTO 190B8 | Soil, Spanderswoud, Netherlands | KC773829 | KC773780 | KC773805 | |
| DTO 41A2 | Soil, Poland | KC773830 | KC773781 | KC773806 | |
| DTO 41E6 | Soil, Poland | KC773831 | KC773782 | KC773807 | |
| DTO 42A9 | Soil, Poland | KC773832 | KC773783 | KC773808 | |
| NRRL 35630 | Cork bark, Alentejo, Portugal | EF200087 | EF198554 | EF198582 | |
| NRRL 35633 | Cork bark, Alentejo, Portugal | EF200088 | EF198555 | EF198583 | |
| P. arianeae | CBS 134559T = DTO 20B8 | Soil, Spanderswoud, Netherlands | KC773833 | KC773784 | KC773811 |
| P. bilaiae | CBS 221.66T = ATCC 22348 = IMI 113677 = NRRL 3391 | Soil, Kiev, Ukraine | JN714937 | JN625966 | JN626009 |
| CBS 330.90 | Unknown | KC773834 | KC773785 | KC773812 | |
| DTO 181D8 = CV 255 | Mite inside Protea repens infructescence, Stellenbosch, South Africa | JX091437 | JX091565 | JX141560 | |
| P. brocae | CBS 116113T = IBT 26293 = NRRL 31472 | Coffee berry borer faeces, Tapachula, Chiapas, Mexico | KC773835 | KC773787 | KC773814 |
| P. cainii | DAOM 239914T | Nuts of Juglans nigra, Niagara, Canada | JN686435 | JN686366 | JN686389X |
| DAOM 239915 | Nuts of Carya ovate, Niagara, Canada | JN686436 | JN686367 | JN686390 | |
| P. guanacastense | DAOM 239912 T | Gut of the caterpillar Eutelia sp. reared on leaves of Spondias mombin, Santa Rosa, Costa Rica | JN626098 | JN625967 | JN626010 |
| DAOM 239913 | Gut of the caterpillar Eutelia sp. reared on leaves of Spondias mombin, Santa Rosa, Costa Rica | JN626099 | JN625968 | JN626011 | |
| P. herquei | CBS 336.48T = ATCC 10118 = IMI 28809 = NRRL 1040 | Leaf of Agauria pirifolia, France | JN626101 | JN625970 | JN626013 |
| CBS 136.22 = NRRL 2113 | Unknown, France | JN626100 | JN625969 | JN626012 | |
| P. herquei | CBS 347.51 = ATCC 18237 = IMI 107651 = NRRL 3450 | Ex-type of P. luteocoeruleum, Wakamoto corn and rice cake, Nehira, Japan | JN617703 | JN625971 | JN626014 |
| P. hirayamae | CBS 229.60T = ATCC 18312 = IMI 078255 = NRRL 143 | Milled rice, Thailand | JN626095 | JN625955 | JN626003 |
| CBS 134207 = DTO 182B9 = DAOM 241115 = CV 887 | Fynbos soil, Riverlands Nature Reserve, Malmesbury, South Africa | JX091453 | JX091572 | JX141568 | |
| CBS 134208 = DTO 182D2 = DAOM 241116 = CV 916 | Fynbos soil, Riverlands Nature Reserve, Malmesbury, South Africa | JX091454 | JX091573 | JX141569 | |
| CBS 238.65 | Corn meal, South Africa | JN626096 | JN625956 | JN626004 | |
| P. jacksonii | DAOM 239937T | Forest soil, Queensland, Australia | JN686437 | JN686368 | JN686391 |
| DAOM 239938 | Forest soil, Queensland, Australia | JN686438 | JN686369 | JN686392 | |
| P. johnkrugii | DAOM 239943T | Forest soil, Langkawi, Kedah, Malaysia | JN686447 | JN686378 | JN686401 |
| DAOM 239939 | Rainforest soil, Langkawi, Kedah, Malaysia | JN686443 | JN686374 | JN686397 | |
| DAOM 239940 | Forest soil, Langkawi, Kedah, Malaysia | JN686444 | JN686375 | JN686398 | |
| DAOM 239941 | Forest soil, Langkawi, Kedah, Malaysia | JN686445 | JN686376 | JN686399 | |
| DAOM 239942 | Forest soil, Langkawi, Kedah, Malaysia | JN686446 | JN686377 | JN686400 | |
| DAOM 239944 | Forest soil, Langkawi, Kedah, Malaysia | JN686448 | JN686379 | JN686402 | |
| DAOM 239945 | Forest soil, Langkawi, Kedah, Malaysia | JN686449 | JN686380X | JN686403 | |
| DAOM 239946 | Forest soil, Langkawi, Kedah, Malaysia | JN686450 | JN686381X | JN686404 | |
| P. jugoslavicum | CBS 192.87T = IMI 314508 | Seed of Helianthus annuus, Yugoslavia | KC773836 | KC773789 | KC773815 |
| P. levitum | CBS 345.48T = ATCC 10464 = IMI 039735 = NRRL 705 | Modelling clay, USA | GU981607 | JN714938 | JN714939 |
| P. lilacinoechinulatum | CBS 454.93T = ATCC 18309 = IMI 068211 | Soil, Japan | KC773837 | KC773790 | KC773816 |
| CBS 134563 = DTO 17E2 | Soil, Spanderswoud, Netherlands | KC773791 | KC773817 | ||
| CBS 134564 = DTO 17I3 | Soil, Spanderswoud, Netherlands | KC773792 | KC773818 | ||
| CBS 134560 = DTO 42A2 | Soil, Poland | KC773793 | KC773819 | ||
| P. malachiteum | CBS 647.95 = IBT 17515 | Soil, Japan | KC773838 | KC773794 | KC773820 |
| P. mallochii | DAOM 239917T | Caterpillar on Spondias mombin, Santa Rosa, Costa Rica | JN626104 | JN625973 | JN626016 |
| DAOM 239919 | Midgut of the caterpillar Citheronia lobesis feeding on Spondias mombin, Santa Rosa, Costa Rica | JN626106 | JN625975 | JN626018 | |
| DAOM 239922 | Hindgut of the caterpillar Rothschildia lebeau reared on leaves of Spondias mombin, Santa Rosa, Costa Rica | JN626109 | JN625978 | JN626021 | |
| DAOM 239925 | Guts of the caterpillar Citheronia lobesis reared on leaves of Cochlospermum vitifolium, Santa Rosa, Costa Rica | JN626112 | JN625980 | JN626023 | |
| DAOM 239926 | Frass of the caterpillar Rothschildia lebeau reared on leaves of Spondias mombin, Santa Rosa, Costa Rica | JN626111 | JN625981 | JN626024 | |
| DAOM 239927 | Gut of the caterpillar Rothschildia lebeau reared on leaves of Spondias mombin, Santa Rosa, Costa Rica | JN626113 | JN625982 | JN626025 | |
| P. maximae | CBS 134565T = NRRL 2060 | Weathering treated cellophane, Florida, USA | EU427298 | KC773795 | KC773821 |
| P. multicolor | CBS 501.73T = ATCC 24723 = IMI 174716 | Soil, Russia | KC790402 | JN799645 | JN799646 |
| P. sclerotiorum | CBS 287.36T = ATCC 10494 = IMI 040569 = NRRL 2074 | Air, Java, Indonesia | JN626132 | JN626001 | JN626044 |
| CBS 118889 | Soil, Korea | JN686454 | JN686385X | JN686408 | |
| CBS 128.65 | Forest litter, Leopoldville, Zaire | JN686452 | JN686383X | JN686406 | |
| CBS 258.55 | Culture contaminant, Istanbul, Turkey | JN686453 | JN686384X | JN686407 | |
| DAOM 239930 | Forest soil, Hua Hin, Thailand | JN626129 | JN625998 | JN626041 | |
| DAOM 239931 | Forest soil, Barron Falls, Queensland, Australia | JN626130 | JN625999 | JN626042 | |
| DAOM 239932 | Forest soil, Barron Falls, Queensland, Australia | JN626131 | JN626000 | JN626043 | |
| NRRL 32583 | Coffee seeding crown, Kuauai, Hawari, USA | JN626133 | JN626002 | JN626045 | |
| P. vanoranjei | CBS 134406T = DTO99H6 | Quercus suber forest soil, Tunisia | KC695696 | KC695686 | KC695691 |
| CBS 134404 = DTO99F3 | Quercus suber forest soil, Tunisia | KC695694 | KC695684 | KC695689 | |
| CBS 134405 = DTO99G1 | Quercus suber forest soil, Tunisia | KC695695 | KC695685 | KC695690 | |
| CBS 134407 = DTO119G8 | Quercus suber forest soil, Tunisia | KC695692 | KC695682 | KC695687 | |
| CBS 134408 = DTO120C8 | Quercus suber forest soil, Tunisia | KC695693 | KC695683 | KC695688 | |
| P. viticola | DAOM 239933 | Forest soil, Barron Falls, Queensland, Australia | JN686439 | JN686370 | JN686393 |
| DAOM 239934 | Forest soil, Atherton, Queensland, Australia | JN686440 | JN686371 | JN686394 | |
| DAOM 239935 | Rainforest soil, Atherton, Queensland, Australia | JN686441 | JN686372 | JN686395 | |
| DAOM 239936 | Rainforest soil, Atherton, Queensland, Australia | JN686442 | JN686373 | JN686396 | |
x In Rivera et al. (2011) some GenBank accession numbers were listed incorrectly and are corrected here. Those published as JN686779–686788 are correctly JN686379–686388
Morphology
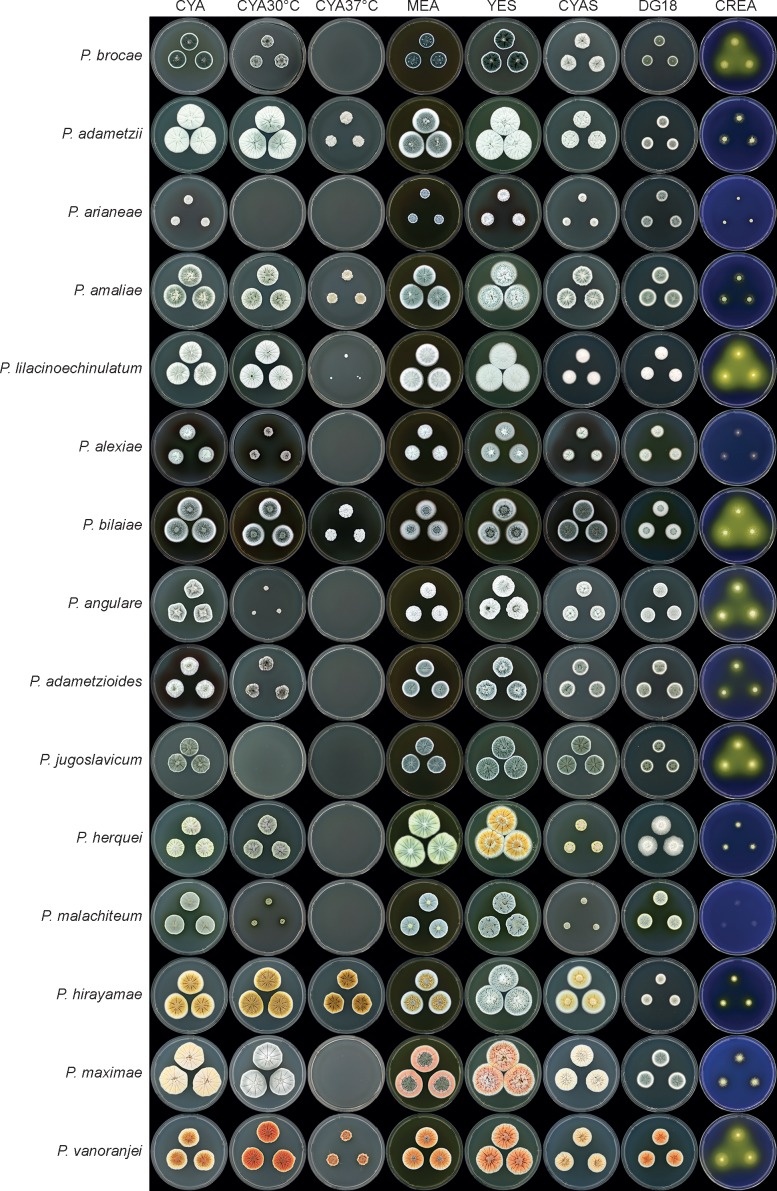
Great care was taken to use standardised conditions for culture incubation (Okuda et al. 2000). Media were dispensed in 90 mm Petri dishes with a volume of 20 mL. Strains were plated onto media from spore suspensions in a three-point pattern using a micropipette and inocula of 0.5–1 μL. Cultures were incubated for 7 d in the dark (Okuda et al. 2000) as described below. Each species was characterised using colony characters on Czapek yeast autolysate agar (CYA), malt extract agar (Oxoid) (MEA), yeast extract sucrose agar (YES), dichloran 18 % glycerol agar (DG18), CYA supplemented with 5 % NaCl (CYAS) and creatine sucrose agar (CREA) grown at 25 °C, with additional CYA plates incubated at 30 and 37 °C. After incubation, strains were described using the models of Pitt (1979) and Frisvad & Samson (2004). All colour names and codes refer to the Methuen Handbook of Colour (Kornerup & Wanscher 1967).
For microscopy, slides were prepared from cultures grown on MEA, with 60 % lactic acid used as mounting fluid. In species producing abundant conidia, conidiophores were washed with 70 % ethanol, then mounted in lactic acid to release air bubbles and wash away excess spores. Microscopic examinations were made using a stereo- (Olympus SZX12) and light-microscope (Olympus BX50 and Zeiss Axioskop 2 Plus). Pictures were taken using an Evolution MP digital microscope camera and ImagePro v. 6.0 software. Conidiophore structures were measured with ImagePro v. 6.0 and Nikon NIS-elements D v. 4.0. In species descriptions, average measurements and standard deviations are provided between square brackets, generally based on 30–50 measurements. Plates of photomicrographs were assembled using Adobe® Photoshop® Creative Suite v. 6. The healing brush tool was used to clean up some images for aesthetic reasons, without altering parts of the images of scientific relevance. Colony textures were captured using multiple focal planes and assembled with Helicon Focus v. 4.2 Z-stacking software.
Low-temperature scanning electron microscopy (SEM)
Strains of P. vanoranjei were grown for 2–3 wk on MEA. By this time, micro-colonies with typical sclerotia and conidiophores had developed next to the three major colonies. Colonies were selected using a dissecting microscope (10–50× magnification, Nikon SMZ 1500), and agar blocks (5 × 5 mm) were cut out with a surgical blade and carefully transferred into a copper cup (diam 10 mm, height 8 mm). To prevent dislodging during freezing, agar blocks were glued to the copper cup with frozen tissue medium (KP-Cryoblock, Klinipath, Duiven, the Netherlands) mixed with one part colloidal graphite (Agar Scientific, Stansted, UK). The copper cup was placed onto wet agar to maintain humid conditions and prevent drying of the sample. The sample was snap-frozen in nitrogen slush and immediately transferred to a JEOL 5600LV scanning electron microscope (JEOL, Tokyo, Japan) equipped with an Oxford CT1500 Cryostation for cryo-electron microscopy (cryoSEM). The sample was sputter-coated by means of a gold target for three times 90 s holding the sample at different angles for an optimal coating. Electron micrographs were acquired with the F4 scan at 4 kV and contrast levels digitally enhanced in Adobe® Photoshop® Creative Suite v. 6.
DNA extraction, sequencing and phylogenetic analysis
Representative strains for each species were grown on MEA and DNA extracted using the UltracleanTM Microbial DNA isolation Kit (MoBio, Solana Beach, USA). DNA preps were stored at −20 °C until used for PCR.
Amplification of target genes employed Kapa ReadyMix (Kapa Biosystems, Woburn, USA). Reactions had a final volume of 25 μL, consisting of 12.5 μL ReadyMix, 10.5 μL MilliQ H2O, 1 μL DNA and 0.25 μM of both forward and reverse primers. Primer pairs used for amplification and sequencing included ITS1–ITS4 (White et al. 1990) for internal transcribed spacer regions of the nrDNA operon (ITS), Bt2a–Bt2b (Glass & Donaldson 1995) for partial β-tubulin and CMD5–CMD6 (Hong et al. 2006) for partial calmodulin. The thermocycling conditions for amplifications had an initial denaturing step of 94 °C for 5 min, 36 cycles of 94 °C for 45 s, 56 °C for 45 s, 72 °C for 60 s, followed by a final elongation step at 72 °C for 10 min. For some strains, annealing temperatures were dropped to 52 °C to enable better amplification. Sequence reactions were set up using the BigDye Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems, CA). The thermocycling profile had an initial denaturing step at 94 °C for 5 min and 25 cycles at 94 °C for 10 sec, 50 °C for 10 sec, 60 °C for 4 min, with sequences determined on an ABI PRISM 3730xl genetic analyser.
Sequence contigs were assembled and edited using CodonCode Aligner v. 4.0.1 (CodonCode Corporation, Centerville, USA). A sequence database was established using newly generated sequences and those previously published in GenBank. Strains used for sequence comparisons and their GenBank accession numbers are summarised in Table 1. Alignments were done using MAFFT v. 6.850b (Katoh et al. 2009) using the G-INS-I option. NEXUS files were analysed in PAUP v. 4.0b10 (Swofford 2002) using the BioNJ option (Gascuel 1997) and MEGA v. 5.1 using Maximum Likelihood with the Tamurai-Nei model and Nearest-Neighbour-Interchange option selected. Confidence levels in nodes were determined using bootstrap analyses of 1 000 replicates. Alignment matrices and tree files were deposited in TreeBASE (www.treebase.org) with submission ID 13796. Newly generated sequences were submitted to GenBank, and their accession numbers are included in Table 1.
RESULTS
Molecular markers, phylogeny and morphology
Phylogenetic and morphological results show that section Sclerotiora contains 22 species, including the five that are newly described here. ITS barcodes were generated for these new species and are listed in Table 1. Analysis of the ITS, β-tubulin and calmodulin gene regions resulted in coherent, monophyletic clades (Fig. 1 & 2). The aligned datasets for ITS, β-tubulin and calmodulin were respectively 513, 355 and 478 bp long, including alignment gaps. Tree topologies were identical for both Maximum Likelihood and Neighbour-Joining analysis. ITS barcodes were suitable identification markers, with all species having unique sequences, although some have only two or three base pair differences, i.e. P. malachiteum and P. herquei, as well as P. amaliae and P. lilacinoechinulatum (Fig. 1). Therefore, as is normal in Penicillium, a secondary identification marker is recommended for accurate and robust identification of sibling species. ITS barcodes and proposed markers for all species accepted in the section are summarised in the taxonomy section below and in Table 1. Infraspecific sequence variations seem to be the norm for section Sclerotiora, as also noted by Rivera & Seifert (2011). This is especially true among strains of P. hirayamae, P. johnkrugii, P. mallochii, P. angulare, P. amaliae and P. sclerotiorum. However, morphological examination revealed no consistent morphological or phylogenetic characters that could separate putative species within these clades.
Fig. 1.

Neighbour-Joining tree based on ITS nucleotide sequences, showing the relationship of species in the section Sclerotiora. Penicillium levitum (CBS 345.48T) was chosen as outgroup. Bootstrap values above 80 % for Maximum Likelihood and Neighbour-Joining are presented at nodes (ML-bs/NJ-bs), with a hyphen (−) indicating no support. (T = ex-type). Coloured names indicate strains that belong to the new species.
Fig. 2.
Neighbour-Joining trees based on β-tubulin and calmodulin nucleotide sequences, showing the relationship of species in the section Sclerotiora. Penicillium levitum (CBS 345.48T) and P. multicolor (CBS 501.73T) were chosen as outgroups for the β-tubulin and calmodulin phylogenies. Bootstrap values above 80 % Maximum Likelihood and Neighbour-Joining are presented at nodes (ML-bs/NJ-bs), with a hyphen (−) indicating no support. (T = ex-type). Coloured names indicate strains that belong to the new species.
The phylogenies consistently resolved strains into three main clades (Fig. 2). These include the P. sclerotiorum complex (clade 1), species closely related to P. adametzii (clade 2) and the P. herquei complex (clade 3). Assignment of specific morphological characters to individual clades is not straightforward. Characters are summarised in Table 2 and in Fig. 3, 4, 5. The P. herquei complex contains two species that produce biverticillate conidiophores, in contrast with species assigned to clades 1 and 2, which are monoverticillate. Clades 1 and 2 are more difficult to define. Generally, species in clade 1 have colonies in orange colours and lack the strongly coloured, soluble pigments such as those generally seen in species of clade 2. However, P. johnkrugii (clade 1) lacks orange colours in colonies, because the sclerotia remain white or grey on media other than MEA. Micromorphology also cannot be used to characterise clades 1 and 2. Species of both clades are monoverticillate, typically have vesiculated stipes and produce conidia in a variety of shapes and ornamentations.
Table 2.
Summary of morphological features for the identification of species of Penicillium section Sclerotiora.
| Growth rates (in mm) |
Colony characters |
Conidiophores |
Conidia |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | CYA | CYA 30 °C | CYA 37 °C | MEA | YES | DG18 | CYAS | Acid on CREA | Sclerotia | Sclerotia colour | Soluble pigments CYA | Reverse colour on CYA | Branching | Stipe roughening | Roughening | Shape |
| P. cainii*1 | 23–29 | n.a. | n.a. | 28–35 | 31–34 | n.a. | n.a. | strong | absent | n.a. | absent | golden to brownish yellow | monoverticillate/single metula rare | rough | finely rough | spheroid |
| P. guanacastense*1 | 25–33 | n.a. | n.a. | 29–33 | n.a. | n.a. | n.a. | strong | absent | n.a. | absent/yellow | orange | monoverticillate/single metula rare | finely rough | finely rough | spheroid |
| P. hirayamae1 | 25–33 | 30–33 | 25–33 | 25–30 | 29–35 | 8–15 | 29–33 | weak | present | orange | absent | orange | monoverticillate/single metula rare | smooth | smooth | spheroid to subspheroid |
| P. jacksonii*1 | 30–33 | n.a. | n.a. | 31–37 | 30–32 | n.a. | n.a. | strong | absent | n.a. | absent | yellow | monoverticillate/single metula rare | smooth to rough | finely rough | spheroid |
| P. johnkrugii*1 | 30–38 | n.a. | n.a. | 26–36 | 28–38 | n.a. | n.a. | strong | present | white/grey | absent | yellow/orange | monoverticillate | smooth to finely roughened | finely rough | spheroid to subspheroid |
| P. mallochii*1 | 29–39 | n.a. | n.a. | 19–35 | n.a. | n.a. | n.a. | strong | absent | n.a. | absent/orange | yellow/orange | monoverticillate/single metula rare | smooth to finely roughened | finely rough | spheroid to subspheroid |
| P. maximae1 | 34–37 | 32–37 | no growth | 34–37 | 40–43 | 15–22 | 27–30 | absent | absent | n.a. | absent | reddish brown | monoverticillate/single metula rare | smooth | smooth | ellipsoid |
| P. sclerotiorum*1 | 18–40 | n.a. | n.a. | 15–32 | 20–44 | n.a. | n.a. | strong | present | orange | absent/orange | orange/reddish yellow/brown | monoverticillate | smooth to finely roughened | finely rough | subspheroid to ellipsoid |
| P. vanoranjei1 | 21–28 | 25–30 | 12–18 | 22–30 | 30–35 | 15–25 | 17–23 | moderate to strong | present | bright orange | orange (incon-spicuous) | brownish orange/reddish brown | monoverticillate | smooth | smooth to finely rough | spheroid to subspheroid |
| P. viticola*1 | 26–36 | n.a. | n.a. | 25–40 | 29–33 | n.a. | n.a. | strong | absent | n.a. | absent | yellow / orange | monoverticillate/single metula rare | rough | smooth | spheroid |
| P. adametzii2 | 27–35 | 27–37 | 10–16 | 28–35 | 27–36 | 9–16 | 18–21 | absent | absent | n.a. | absent | pale to yellow brown | monoverticillate | smooth | smooth to finely rough | spheroid to subspheroid |
| P. adametzioides2 | 20–26 | 15–20 | no growth | 20–26 | 20–30 | 17–21 | 15–24 | weak | absent | n.a. | reddish brown | deep brown | monoverticillate | smooth | smooth to finely rough | subspheroid |
| P. alexiae2 | 18–21 | 8–15 | no growth | 18–21 | 21–26 | 15–17 | 15–18 | absent | present | white | orange brown | brown | monoverticillate | smooth | smooth | subspheroid |
| P. amaliae2 | 20–30 | 22–30 | 3–11 | 27–33 | 30–36 | 14–24 | 24–27 | weak | absent | n.a. | absent | light to dull yellow | monoverticillate | smooth | rough | spheroid to subspheroid |
| P. angulare2 | 13–25 | 2–5 | no growth | 10–20 | 16–28 | 12–20 | 10–20 | weak to moderate | absent | n.a. | absent | pale yellow | monoverticillate | smooth | smooth | subspheroid to ellipsoid |
| P. arianeae2 | 9–12 | no growth | no growth | 10–14 | 14–16 | 11–15 | 9–10 | absent | absent | n.a. | orange brown | pale orange/greyish yellow | monoverticillate | smooth | thick rough walled | spheroid |
| P. bilaiae2 | 25–33 | 25–30 | 13–15 | 25–28 | 29–31 | 20–22 | 25–29 | strong | absent | n.a. | orange brown | deep brown | monoverticillate/single metula rare | smooth | rough | spheroid to subspheroid |
| P. brocae2 | 17–23 | 15–18 | no growth | 14–22 | 25–32 | 14–16 | 16–20 | moderate | absent | n.a. | absent | yellow | monoverticillate | smooth | finely rough | spheroid |
| P. jugoslavicum2 | 20–22 | no growth | no growth | 21–25 | 27–30 | 15–17 | 24–25 | moderate | absent | n.a. | absent | pale yellow | monoverticillate | smooth | finely rough to rough | ellipsoid |
| P. lilacinoechinulatum2 | 25–30 | 30–33 | 2–4 | 27–32 | 32–35 | 17–20 | 22–25 | moderate | absent | n.a. | absent | light to dull yellow | monoverticillate | smooth | rough | spheroid to subspheroid |
| P. herquei3 | 25–35 | 25–34 | no growth | 30–40 | 30–40 | 15–25 | 5–15 | absent | absent / sometimes present | cream | yellow | olive brown/brown | biverticillate | smooth | smooth to rough | ellipsoid |
| P. malachiteum3 | 20–25 | 5–10 | no growth | 28–32 | 28–32 | 17–21 | 10–17 | absent | present | cream | yellow | olive brown/brown | biverticillate | smooth | smooth to rough | ellipsoid |
* Data from Rivera & Seifert (2011) and Rivera et al. (2012).
1/2/3 Indicative of clade (from Fig. 2) to which species belong.
n.a. Data not available.
Fig. 3.
Overview of colony characters in Penicillium section Sclerotiora species treated in this paper.
Fig. 4.
Overview of conidiophores (a–af) and conidia (ag–an) in Penicillium section Sclerotiora species treated in this paper: a–d, ag. P. brocae; e–h, ah. P. adametzii; i–l, ai. P. arianeae; m–p, aj. P. amaliae; q–t, ak. P. lilacinoechinulatum; u–x, al. P. alexiae; y–ab, am. P. bilaiae; ac–af, an. P. angulare. — Scale bar in an = 10 μm, applies to a–an.
Fig. 5.
Overview of conidiophores (a–ab) and conidia (ac–aj) in Penicillium section Sclerotiora species treated in this paper: a–d, ac. P. adametzioides; e–h, ad. P. jugoslavicum; i–n, ae, af. P. herquei; o–q, ag. P. malachiteum; r–t, ah. P. hirayamae; u–x, ai. P. maximae; y–ab, aj. P. vanoranjei. — Scale bar in an = 10 μm, applies to a–aj.
Our phylogenetic analysis confirmed the status of NRRL 2060 (CBS 134565) as a unique taxon, as suggested by Peterson & Horn (2009) and Houbraken & Samson (2011), described as P. maximae below. The ex-type strain of P. lilacinoechinulatum (CBS 454.93) resolved separately from P. bilaiae, along with several strains previously deposited in the DTO collection, and as a sister taxon to P. amaliae, newly described below. Morphologically, P. lilacinoechinulatum has weaker sporulation on most media, and slower growth on CYAS (Fig. 3). More noticeable, however, is the absence of brown or yellow exudates consistently produced in P. bilaiae on CYA and its slower growth at 37 °C. Compared to P. amaliae, P. lilacinoechinulatum has stronger acid production on CREA and more restricted growth at 37 °C. Newly generated sequences of the ITS (KC790403) and β-tubulin (KC790399) gene regions of P. nodositatum (CBS 333.90, ex-type) show that it is closely related to P. kabunicum and does not belong in Penicillium s.str. (Houbraken & Samson 2011). This corresponds with morphological features of its conidiophores, which are symmetrically biverticillate. The phialides are not penicillium-like, but are broad and taper into very fine and long necks, rather similar to those of some Talaromyces species, e.g. T. verruculosus.
The phylogenies revealed that several strains with unique morphological characters are grouped in distinct clades. These are considered new species and described in the taxonomy section as P. vanoranjei, P. maximae, P. amaliae, P. alexiae and P. arianeae. Following these descriptions, morphological characters of the remaining section Sclerotiora species not treated by Rivera & Seifert (2011) are summarised in Table 2 and illustrated in Fig. 3, 4, 5.
Scanning electron microscopy of P. vanoranjei
Characters observed from samples prepared from a well developed colony and a young microcolony were similar (Fig. 7). Sclerotia had a highly complex and well organised structure. Cell walls were distinctly and consistently roughened and cells appeared to be linked by some kind of extracellular matrix (Fig. 7e). On the majority of the sclerotia, characteristic sheet-like structures were observed. Conidial roughening and size develop with age (Fig. 7h–j).
Fig. 7.
Scanning electron microscope pictures showing characteristic features of P. vanoranjei (CBS 134406T). a, b. Young colony showing development of sclerotia together with conidiophores; c–e. sclerotia produced on MEA, showing the sheets of dried-out exudates covering sclerotia; f–j. conidiophores and conidia; SEM pictures clearly show that conidial roughness develops as conidia become older. Also, connectives between conidia visible in (j) makes it possible for this species to support very long chains of conidia. — Scale bars: a = 100 μm; b, f = 50 μm; c, d = 20 μm; e, g = 10 μm; h, i = 5 μm; j = 2 μm.
TAXONOMY
Penicillium vanoranjei Visagie, Houbraken, Samson, sp. nov. — MycoBank MB803782; Fig. 3, 5, 6, 7
Fig. 6.
Penicillium vanoranjei (CBS 134406T). a. Colonies on CYA, MEA and YES from left to right (top = obverse, bottom = reverse); b. texture on CYA; c, d. texture on MEA; e. sclerotia; f–k. conidiophores; l. conidia. — Scale bar in e = 100 μm; in k = 10 μm, applies to f–l.
ITS barcode. KC695696.
Alternative markers. KC695686 (β-tubulin), KC695691 (calmodulin).
Etymology. Latin, vanoranjei: named, in reference to the orange (Dutch = oranje) coloured colonies produced by this species, after Willem-Alexander Claus George Ferdinand, ‘Zijne Koninklijke Hoogheid de Prins van Oranje’ (translated from Dutch as: ‘His Royal Highness the Prince of Orange’) and the new King of the Netherlands upon the retirement of Queen Beatrix on 30 April 2013.
Diagnosis — Bright orange sclerotia dominate colony appearance on most media. Conidiophores monoverticillate; stipes smooth walled, vesiculate; phialides 8.5–12.5 × 3–3.5 μm; conidia smooth, spheroidal to subspheroidal, 2.5–3.5 × 2.5–3 μm.
Colony morphology — Colony diam, 7 d, in mm: CYA 21–28; CYA 30 °C 25–30; CYA 37 °C 12–18; MEA 22–30; YES 30–35; DG18 15–25; CYAS 17–23; CREA 10–20.
CYA, 25 °C, 7 d: Colonies slightly raised at centre, radially sulcate, abundant bright orange sclerotia produced; margins low, narrow, entire; mycelia white near margin, orange elsewhere; texture floccose; sporulation absent to sparse, conidial colour en masse cannot be determined; exudate bright orange, soluble pigment inconspicuously orange, reverse pigmentation brownish orange to Burnt Sienna (6C7–7D8). CYA, 30 °C, 7 d: Colonies showing no differences from those grown on CYA at 25 °C. MEA, 25 °C, 7 d: Colonies slightly raised at centre, radially sulcate, abundant bright orange sclerotia produced; margins low, narrow, entire; mycelia white near margin, orange elsewhere; texture floccose; sporulation sparse and only at colony centre, conidial colour en masse difficult to determine precisely, greyish green; exudate bright orange, soluble pigment absent, reverse pigmentation brownish orange (6A8–6C8). YES, 25 °C, 7 d: Colonies moderately deep, raised at centre, radially and concentrically sulcate; margins low, narrow (2 mm), entire; mycelia white near margin, orange elsewhere; texture floccose; sporulation absent; exudate absent, soluble pigment absent, reverse pigmentation brownish orange to Burnt Sienna (6C7–7D8). CREA, 25 °C, 7 d: Moderate to strong acid production.
Conidiophores strictly monoverticillate; stipes smooth walled, 65–220 × 2.5–3 μm, vesicle 4.5–6 μm (5.3 ± 0.6); phialides ampulliform, 10–20 per stipe, 8.5–12.5 × 3–3.5 μm (10.6 ± 0.9 × 3.3 ± 0.2); conidia smooth to slightly rough walled, connectives visible, spheroidal to subspheroidal, 2.5–3.5 × 2.5–3 μm (3.0 ± 0.2 × 2.9 ± 0.2), average width/length = 0.95 ± 0.04, n = 39; sclerotia bright orange, 85–190 × 70–150 μm.
Specimen examined. Tunisia, Tabarka, Quercus suber forest soil, 2 Feb. 2009, collected by C. Silva Pereira, CBS H-21145, holotype, culture ex-type CBS 134406 (= DTO 99H6 = AHS3SF_13).
Additional strains examined. Tunisia, Tabarka, Quercus suber forest soil, 2 Feb. 2009, collected by C. Silva Pereira, cultures CBS 134404 (= DTO 99F3 = AHS3SF_2); CBS 134405 (= DTO 99G1 = AHS3SF_10); CBS 134407 (= DTO 119G8); CBS 134408 (= DTO 120C8).
Notes — Penicillium vanoranjei is characterised by colonies dominated by bright orange sclerotia. Orange sclerotia also occur in P. sclerotiorum and P. johnkrugii in the P. sclerotiorum species-complex (clade 1), as recently revised by Rivera & Seifert (2011). Penicillium sclerotiorum was distinguished from P. johnkrugii based on more abundant conidiogenesis on CYA, orange sclerotia on most media and subspheroidal to ellipsoidal conidia in the former species, in contrast to orange sclerotia produced only on MEA and spheroidal to subspheroidal conidia in the latter. The new species is easily distinguished from P. sclerotiorum by the absence of conidiogenesis on CYA and spheroidal to subspheroidal conidia. The orange sclerotia of P. vanoranjei, produced on most media, distinguish it from P. johnkrugii. Also, the new species has more restricted growth on CYA. It must be noted that roughness on conidia of P. vanoranjei, although easily visible on SEM, was inconspicuous with light microscopy.
Penicillium maximae Visagie, Houbraken, Samson, sp. nov. — MycoBank MB803783; Fig. 3, 5, 8
Fig. 8.
Penicillium maximae (CBS 134565T) a. Colonies on CYA, MEA and YES from left to right (top = obverse, bottom = reverse); b. texture on CYA; c, d. texture on MEA; e–l. conidiophores; m. conidia. — Scale bars = 10 μm; l applies to f–m.
ITS barcode. EU427298.
Alternative markers. KC773795 (β-tubulin), KC773821 (calmodulin).
Etymology. Latin, maximae: named after ‘Hare Koninklijke Hoogheid Prinses Máxima der Nederlanden’ (translated from Dutch as: ‘Her Royal Highness Princess Máxima of the Netherlands’), wife of Prince Willem-Alexander of the Netherlands.
Diagnosis — A ring of orange mycelia masks sporulation on MEA. Conidiophores mostly monoverticillate, a minor proportion biverticillate; stipes smooth walled; phialides 6.5–10 × 2.5–3 μm; conidia smooth and ellipsoidal, 3–3.5 × 2.5–3 μm. Sclerotia are not produced.
Colony morphology — Colony diam, 7 d, in mm: CYA 34–37; CYA 30 °C 32–37; CYA 37 °C no growth; MEA 34–37; YES 40–43; DG18 15–22; CYAS 27–30; CREA 12–15.
CYA, 25 °C, 7 d: Colonies angular in outline, radially sulcate, moderately raised, with an orange pink colour; margins low, narrow, entire; mycelia white and pinkish orange; texture floccose; sporulation sparse, conidial colour en masse cannot be determined; exudate orange, soluble pigment orange, reverse pigmentation reddish brown (8E8) at centre fading to orange (6B7). CYA, 30 °C, 7 d: Colonies similar to CYA at 25 °C, except for denser sporulation, conidia en masse greyish green (25D5). MEA, 25 °C, 7 d: Colonies moderately deep, lightly radially sulcate, fluffy pinkish orange mycelia dominate margins and masks conidiogenesis; margins low, narrow, entire; mycelia white and pinkish orange elsewhere; texture floccose to somewhat velutinous in some areas; sporulation dense at centre, conidial colour en masse dark green (26F5); exudate clear, soluble pigment absent, reverse pigmentation brown to reddish brown (7E8–8E8). YES, 25 °C, 7 d: Colonies moderately deep, randomly sulcate; margins low, narrow (2 mm), entire; mycelia white and pinkish orange; texture floccose; sporulation sparse, conidial colour en masse cannot be determined; exudate absent, soluble pigment absent, reverse pigmentation reddish brown (8E8) at centre fading to orange (6B7). CREA, 25 °C, 7 d: Acid not produced.
Conidiophores monoverticillate, with a low proportion biverticillate; stipes smooth walled, 45–150 × 2–3 μm, vesicle 3–6 μm (4.5 ± 0.7); branches when present only two, 11–38 μm; phialides ampulliform, 8–16 per stipe, 6.5–10 × 2.5–3 μm (8.4 ± 0.2 × 2.8 ± 0.8); conidia smooth walled, ellipsoidal, 3–3.5 × 2.5–3 μm (3.0 ± 0.2 × 2.5 ± 0.04), average width/length = 0.8 ± 0.04, n = 38; sclerotia not produced.
Specimen examined. USA, Florida, weathering treated cellophane, 1945, collected by L. White, CBS H-21144, holotype, culture ex-type CBS 134565 (= NRRL 2060 = DTO 244C7).
Notes — Penicillium maximae typically produces fast growing colonies with pinkish orange mycelia that mask sporulation underneath. This colour is especially striking on MEA. The species is basal to the P. sclerotiorum species complex, as is P. hirayamae. Morphologically it most closely resembles P. sclerotiorum. However, the absence of sclerotia, lack of acid production on CREA and lack of sporulation on CYA distinguish P. maximae from P. sclerotiorum.
Penicillium amaliae Visagie, Houbraken & K. Jacobs, sp. nov. — Mycobank MB803784; Fig. 3, 4, 9
Fig. 9.
Penicillium amaliae (CBS 134209T). a. Colonies on CYA, MEA and YES from left to right (top = obverse, bottom = reverse); b. texture on CYA; c. texture on MEA; d–k. conidiophores; l. conidia. — Scale bar in e = 100 μm; k = 10 μm, applies to e–l.
ITS barcode. JX091443.
Alternative markers. JX091563 (β-tubulin), JX141557 (calmodulin).
Etymology. Latin, amaliae: Named after Catharina-Amalia Beatrix Carmen Victoria ‘Prinses der Nederlanden, Prinses van Oranje-Nassau’ (translated from Dutch as: ‘Princess of the Netherlands, Princess of Orange-Nassau’), first daughter of Princess Máxima of the Netherlands and Prince Willem-Alexander of the Netherlands.
Diagnosis — Conidial colour on all media pale green. Colonies on CYA at 37 °C 3–11 mm, no acid produced on CREA. Conidiophores strictly monoverticillate; stipes smooth walled and ending in a swollen vesicle; phialides 6.5–9 × 2.5–3.5 μm; conidia rough walled, spheroidal to subspheroidal, 2–3 × 2–2.5 μm. Sclerotia are not produced.
Colony morphology — Colony diam, 7 d, in mm: CYA 20–30; CYA 30 °C 22–30; CYA 37 °C 3–11; MEA 27–33; YES 30–36; DG18 14–24; CYAS 24–27; CREA 10–18.
CYA, 25 °C, 7 d: Colonies moderately deep, radially and concentrically sulcate; margins low, narrow (2 mm), entire; mycelia white; texture velutinous; sporulation moderately dense, conidial colour en masse dull green to greyish green (27D4–27D5), areas greenish grey to greyish green (27B2–27B4) especially near margin; exudate clear to almost yellowish brown, soluble pigment absent, reverse pigmentation pale to pale yellow (2A3–2A4), in some isolates a darker dull yellow (3B4). CYA, 30 °C, 7 d: Colonies showing no differences from those grown on CYA at 25 °C. MEA, 25 °C, 7 d: Colonies low, planar; margins low, narrow (2 mm), entire; mycelia white; texture velutinous; sporulation moderately dense, conidial colour en masse greyish green (27D5–27E5–27E6–27D6); exudate absent, soluble pigment absent, reverse pigmentation pale yellow (2A3) at centre, greyish yellow to greyish green (1B3–1C3) elsewhere. YES, 25 °C, 7 d: Colonies moderately deep, radially and concentrically sulcate; margins low, narrow (2 mm), entire; mycelia white; texture velutinous; sporulation moderately dense, conidial colour en masse green to greyish green (27D4–27D5), when sporulation denser, dull to greyish green (27E4–27D7); exudate absent, soluble pigment absent, reverse pigmentation greyish to olive yellow (3B6–3C6) at centre, pale to pastel yellow (2A3–2A4) near margin. CREA, 25 °C, 7 d: Weak acid production.
Conidiophores strictly monoverticillate; stipes smooth walled, 20–95 × 2–3 μm, vesicle 4.5–7(–9) μm (6.3 ± 0.8); phialides ampulliform, 12–24 per stipe, 6.5–9 × 2.5–3.5 μm (7.7 ± 0.5 × 2.9 ± 0.2); conidia rough walled, connectives visible, spheroidal to subspheroidal, 2–3 × 2–2.5 μm (2.5 ± 0.1 × 2.4 ± 0.1), average width/length = 0.93 ± 0.04, n = 127; sclerotia not produced.
Specimen examined. South Africa, Western Cape Province, Struisbaai, Protea repens infructescence, 1 Aug. 2009, collected by C.M. Visagie, CBS H-21141, holotype, culture ex-type CBS 134209 (= CV 1875 = DTO 183F3 = DAOM 241034).
Additional strains examined. Australia, New South Wales, Katandra Nature Reserve, insect larva, 2003, collected by A.L. Markovina, received via J.I. Pitt, CBS 134557 (= DTO 57A8). – South Africa, Western Cape Province, Stellenbosch, Protea repens infructescence, collected by C.M. Visagie, cultures CBS 134555 (= CV 112 = DTO 181A5 = DAOM 241032), CBS 134556 (= CV 227 = DTO 181C9 = DAOM 241033), CBS 134212 (= CV 401 = DTO 181F7 = DAOM 241031); Struisbaai, P. repens infructescence, collected by C.M. Visagie, CBS 134210 (= CV 1722 = DTO 183D1); CBS 134211 (= CV 204 = DTO 181C6).
Notes — Penicillium amaliae is characterised by monoverticillate conidiophores that have a vesiculate stipe and produce spheroidal rough walled conidia. It grows relatively well on all media. Conidial colour en masse is pale green, a character also observed in P. lilacinoechinulatum. Phylogenetically, it resolves as a distinct clade closely related to P. lilacinoechinulatum, P. adametzii and P. brocae (Fig. 2). The numerous phialides of P. amaliae distinguish it from P. adametzii and P. brocae. Also, the growth of P. brocae is more restricted than the new species, whereas P. adametzii grows faster at 30 and 37 °C. Penicillium lilacinoechinulatum is its closest relative, distinguished by its faster growth at 30 °C, slower growth at 37 °C, and stronger acid produced on CREA.
Penicillium alexiae Visagie, Houbraken, Samson, sp. nov. — MycoBank MB803785; Fig. 3, 4, 10
Fig. 10.
Penicillium alexiae (CBS 134558T). a. Colonies on CYA, MEA and YES from left to right (top = obverse, bottom = reverse); b. texture on CYA; c, d. texture on MEA; e. sclerotia; f–k. conidiophores; l. conidia. — Scale bar in e = 100 μm; k = 10 μm, applies to f–l.
ITS barcode. KC790400.
Alternative markers. KC773778 (β-tubulin), KC773803 (calmodulin).
Etymology. Latin, alexiae: Named after Alexia Juliana Marcela Laurentien ‘Prinses der Nederlanden, Prinses van Oranje-Nassau’ (translated from Dutch as: ‘Princess of the Netherlands, Princess of Orange-Nassau’) and second daughter of Princess Máxima of the Netherlands and Prince Willem-Alexander of the Netherlands.
Diagnosis — Colonies on CYA 18–21 mm, CYA 30 °C 8–15 and CYA 37 °C no growth; brown soluble pigment commonly produced on most media. Acid not produced on CREA. White sclerotia produced on CYA and MEA. Conidiophores monoverticillate; stipes smooth walled; phialides 8.5–10.5 × 2.5–3.5 μm; conidia smooth walled, subspheroidal, 2.5–3 × 2–3 μm.
Colony morphology — Colony diam, 7 d, in mm: CYA 18–21; CYA 30 °C 8–15; CYA 37 °C no growth; MEA 18–21; YES 21–26; DG18 15–17; CYAS 15–18; CREA 10–14.
CYA, 25 °C, 7 d: Colonies raised high at centre, radially sulcate, white sclerotia present; margins low, narrow, irregular; mycelia white; texture velutinous; sporulation moderately dense, conidial colour en masse greyish green (25B4); exudate absent, soluble pigment reddish brown, reverse pigmentation brown (7E8) at centre, paler brown (6E8) elsewhere. CYA, 30 °C, 7 d: Colonies crateriform, having a greyish colour, with no sporulation. MEA, 25 °C, 7 d: Colonies low, radially sulcate, white sclerotia present; margins low, wide, entire; mycelia white; texture velutinous; sporulation moderately dense, conidial colour en masse greyish green (25B4); exudate absent, soluble pigment absent, reverse pigmentation brownish orange (6C7). YES, 25 °C, 7 d: Colonies low, raised at centre, radially sulcate; margins low, narrow, entire; mycelia white; texture velutinous; sporulation moderately dense, conidial colour en masse greyish green (25B4); exudate absent, soluble pigment pale brown, reverse pigmentation brownish orange (6C7) at centre, yellowish grey elsewhere (2B2); CREA, 25 °C, 7 d: Acid not produced.
Conidiophores strictly monoverticillate; stipes smooth walled, 50–185 × 2.5–3 μm, vesicle 3.5–4.5 μm (4.0 ± 0.3); phialides ampulliform, 5–12 per stipe, 8.5–10.5 × 2.5–3.5 μm (9.6 ± 0.6 × 3.0 ± 0.3); conidia smooth walled, subspheroidal, 2.5–3 × 2–3 μm (2.6 ± 0.1 × 2.4 ± 0.2), average width/length = 0.91 ± 0.04, n = 45; sclerotia white, 70–165 × 70–135 μm.
Specimen examined. Tunisia, Tabarka, Quercus suber forest soil, 2 Feb. 2009, collected by C. Silva Pereira, CBS H-21142, holotype, culture ex-type CBS 134558 (= DTO 118H8 = FERS1SF_05).
Notes — Penicillium alexiae typically produces slow growing colonies with a brown soluble pigment on most media. This is a character also observed for P. bilaiae and P. adametzioides. The faster growth of P. bilaiae at 30 and 37 °C on CYA and strong acid production makes it distinct. Acid is not produced by P. alexiae, in comparison with P. adametzioides, which exhibits weak acid production. One of the most striking features of P. alexiae is the production of the white sclerotia. Considering these species micromorphology, P. alexiae is easily distinguished from both P. bilaiae and P. adametzioides, because its non-swollen vesicles result in a compact conidiophore.
Penicillium arianeae Visagie, Houbraken, Samson, sp. nov. — MycoBank MB803786; Fig. 3, 4, 11
Fig. 11.
Penicillium arianeae (CBS 134559T). a. Colonies on CYA, MEA and YES from left to right (top = obverse, bottom = reverse); b. texture on CYA; c, d. texture on MEA; e–m. conidiophores; n. conidia. — Scale bar in n = 10 μm, applies to e–n.
ITS barcode. KC773833.
Alternative markers. KC773784 (β-tubulin), KC773811 (calmodulin).
Etymology. Latin, arianeae: named after Ariane Wilhelmina Máxima Ines ‘Prinses der Nederlanden, Prinses van Oranje-Nassau’ (translated from Dutch as: ‘Princess of the Netherlands, Princess of Orange-Nassau’) and third daughter of Princess Máxima of the Netherlands and Prince Willem-Alexander of the Netherlands.
Diagnosis — Colonies restricted on most media, on CYA 9–12 mm, with no growth above 30 °C, MEA 10–14 mm. Conidiophores monoverticillate; stipes smooth walled, with slightly vesiculate ending; phialides 7.5–10 × 3–4 μm; conidia spheroidal and have thick rough walls, 3–3.5 × 3–3.5 μm. Sclerotia are not produced.
Colony morphology — Colony diam, 7 d, in mm: CYA 9–12; CYA 30 °C no growth; CYA 37 °C no growth; MEA 10–14; YES 14–16; DG18 11–15; CYAS 9–10; CREA 3–4.
CYA, 25 °C, 7 d: Colonies crateriform, radially sulcate at margins; margins low, narrow, irregular; mycelia white at the margin; texture floccose; sporulation absent to sparse, conidial colour en masse greenish grey to pale green (25A2–25A3); exudate clear, soluble pigment inconspicuously brownish, reverse pigmentation pale orange (5A3) and greyish yellow (4B3). CYA, 30 °C, 7 d: No growth observed. MEA, 25 °C, 7 d: Colonies crateriform, radially sulcate; margins low, narrow, somewhat irregular; mycelia white; texture floccose; sporulation sparse to sometimes moderately dense, conidial colour en masse greyish green (25D4); exudate absent, soluble pigment absent, reverse pigmentation brownish orange (5C5). YES, 25 °C, 7 d: Colonies crateriform, radially and concentrically sulcate; margins low, narrow, irregular; mycelia white; texture floccose; sporulation absent to sparse, conidial colour en masse greenish white; exudate absent, soluble pigment reddish brown, reverse pigmentation pale yellow (4A4–4A5); CREA, 25 °C, 7 d: No acid produced.
Conidiophores strictly monoverticillate; stipes smooth walled, 30–180 × 2.5–3 μm, vesicle 4–7 μm (5.5 ± 1.0); phialides ampulliform, 6–12 per stipe, 7.5–10 × 3–4 μm (8.5 ± 0.7 × 3.5 ± 0.2); conidia thick, rough walled, spheroidal, 3–3.5 × 3–3.5 μm (3.2 ± 0.2 × 3.2 ± 0.2), average width/length = 0.98 ± 0.03, n = 30; sclerotia not produced.
Specimen examined. The Netherlands, Bussum, Spanderswoud, soil, Apr. 2006, collected by J. Houbraken, L. Janson & R. Samson, CBS H-21143, holotype, culture ex-type CBS 134559 (= DTO 20B8).
Notes — Penicillium arianeae has restricted growth on all media. This character easily distinguishes it from its closest relatives, P. amaliae, P. lilacinoechinulatum and P. adametzii. Its thick, rough walled conidia are also unique in this section.
ACCEPTED SPECIES IN SECTION SCLEROTIORA HOUBRAKEN & SAMSON
In: Penicillium subgenus Aspergilloides.
Type species. Penicillium sclerotiorum.
We accept the following names in the section Sclerotiora. As discussed at more length below, P. nodositatum, previously classified in this section (Houbraken & Samson 2011) is removed and does not belong in Penicillium s.str. In this list, GenBank accession numbers to ITS barcodes and alternative identification markers are provided (BT = β-tubulin; CMD = calmodulin).
Penicillium adametzii K.M. Zalessky, Bull. Int. Acad. Polon. Sci., Cl. Sci. Math., Ser. B, Sci. Nat. 1927: 507. 1927. (MB119777). — Herb.: IMI 39751. Ex-type: CBS 209.28 = ATCC 10407 = IMI 039751 = MUCL 29106 = NRRL 737. ITS barcode: JN714929. (Alternative markers: BT = JN625957; RPB2 = JN121455; CMD = KC773796).
Penicillium adametzioides S. Abe ex G. Sm., Trans. Brit. Mycol. Soc. 46: 335. 1963 ≡ Penicillium adametzioides S. Abe, J. Gen. Appl. Microbiol., Tokyo 2: 68. 1956 (nom. inval., Art. 36). (MB302372). — Herb.: CBS 313.59. Ex-type: CBS 313.59 = ATCC 18306 = FAT1302 = IFO 6055 = IMI 068227 = NRRL 3405 = QM 7312. ITS barcode: JN686433. (Alternative markers: BT = JN799642; RPB2 = JN406578; CMD = JN686388).
Penicillium alexiae Visagie, Houbraken & Samson (this study). (MB 803785). — Herb.: CBS-H 21142. Ex-type: CBS 134558 = DTO 118H8 = FERS1SF_05. ITS barcode: KC790400. (Alternative markers: BT = KC773778; CMD = KC773803).
Penicillium amaliae Visagie, Houbraken & Jacobs (this study). (MB803784). — Herb.: CBS-H 21141. Ex-type: CBS 134209 = CV 1875 = DTO 183F3 = DAOM 241034. ITS barcode: JX091443. (Alternative markers: BT = JX091563; CMD = JX141557).
Penicillium angulare S.W. Peterson, E.M. Bayer & Wicklow, Mycologia 96: 1289. 2004. (MB487891). — Herb.: BPI 842268. Ex-type: CBS 130293 = IBT 27051 = NRRL 28157. ITS barcode: (representative strain NRRL 28140) AY313613. (Alternative markers: BT = KC773779; RPB2 = JN406554; CMD = KC773804).
Penicillium arianeae Visagie, Houbraken & Samson (this study). (MB 803786). — Herb.: CBS-H 21143. Ex-type: CBS 134559 = DTO 20B8. ITS barcode: KC773833. (Alternative markers: BT = KC773784; CMD = KC773811).
Penicillium bilaiae Chalab., Bot. Mater. Otd. Sporov. Rast. 6: 165. 1950. (MB302379). — Herb.: IMI 113677. Ex-type: CBS 221.66 = ATCC 22348 = ATCC 48731 = CCRC 31675 = FRR 3391 = IJFM 5025 = IMI 113677 = MUCL 31187 = VKMF-854. ITS barcode: JN714937. (Alternative markers: BT = JN625966; RPB2 = JN406610; CMD = JN626009).
Penicillium brocae S.W. Peterson, Jeann. Pérez, F.E. Vega & Infante, Mycologia 95: 143. 2003. (MB373658). — Herb.: BPI 841763. Ex-type: CBS 116113 = IBT 26293 = NRRL 31472. ITS barcode: AF484398. (Alternative markers: BT = KC773787; RPB2 = JN406639; CMD = KC773814).
Penicillium cainii K.G. Rivera & Seifert, Stud. Mycol. 70: 147. 2011. (MB563159). — Herb.: DAOM 239914. Ex-type: CCFC 239914. ITS barcode: JN686435. (Alternative markers: BT = JN686366; CMD = JN686389).
Penicillium guanacastense K.G. Rivera, Urb & Seifert, Mycotaxon 119: 324. 2012. (MB563044). — Herb.: DAOM 239912. Ex-type: CCFC 239912. ITS barcode: JN626098. (Alternative markers: BT = JN625967; CMD = JN626010).
Penicillium herquei Bainier & Sartory, Bull. Soc. Mycol. France 28: 121. 1912. (MB536431). — Herb.: IMI 28809. Ex-type: CBS 336.48 = NRRL 1040 = ATCC 10118 = BIOURGE 452 = FRR 1040 = IFO 31747 = IMI 28809 = MUCL 29213 = NCTC 1721 = QM 1926 = Thom 4640.447. ITS barcode: JN626101. (Alternative markers: BT = JN625970; RPB2 = JN121494; CMD = JN626013).
Penicillium hirayamae Udagawa, J. Agric. Sci. (Tokyo) 5: 6. 1959. (Teleomorphic synonym: Eupenicillium hirayamae D.B. Scott & Stolk). (MB302402). — Herb.: IMI 78255. Ex-type: CBS 229.60 = ATCC 18312 = IFO 6435 = IMI 078255 = IMI 078255ii = NHL 6046 = NRRL 143 = QM 7885. ITS barcode: JN626095. (Alternative markers: BT = JN625955; RPB2 = JN121459; CMD = JN626003).
Penicillium jacksonii K.G. Rivera & Seifert, Stud. Mycol. 70: 151. 2011. (MB563160). — Herb.: DAOM 239937. Ex-type: CCFC 239937. ITS barcode: JN686437. (Alternative markers: BT = JN686368; CMD = JN686391).
Penicillium johnkrugii K.G. Rivera & Seifert, Stud. Mycol. 70: 151. 2011. (MB563161). — Herb.: DAOM 239943. Ex-type: CCFC 239943. ITS barcode: JN686447. (Alternative markers: BT = JN686378; CMD = JN686401).
Penicillium jugoslavicum C. Ramírez & Munt.-Cvetk., Mycopathologia 88: 65. 1984. (MB124173). — Herb.: CBS 192.87. Ex-type: CBS 192.87 = IJFM 7785 = IMI 314508. ITS barcode: KC411688. (Alternative markers: BT = KC773789; RPB2 = JN406618; CMD = KC773815).
Penicillium lilacinoechinulatum S. Abe ex G. Sm., Trans. Brit. Mycol. Soc. 46: 335. 1963 ≡ Penicillium lilacinoechinulatum S. Abe, J. Gen. Appl. Microbiol., Tokyo 2: 54, 1956 (nom. inval., Art. 36). (MB120793). — Herb.: CBS 454.93. Ex-type: CBS 454.93 = ATCC 18309 = FAT 84 = FRR 3451 = IFO 6231 = IMI 068211 = QM 7289. ITS barcode: AY157489. (Alternative markers: BT = KC773790; CMD = KC773816).
Penicillium malachiteum (Yaguchi & Udagawa) Houbraken & Samson, Stud. Mycol. 70: 47. 2011 ≡ Chromocleista malachitea Yaguchi & Udagawa, Trans. Mycol. Soc. Japan 34: 102. 1993 (Houbraken & Samson 2011). (MB561971). — Herb.: CBS 647.95. Ex-type: CBS 647.95 = IBT 17515. ITS barcode: KC773838. (Alternative markers: BT = KC773794; CMD = KC773820).
Penicillium mallochii K.G. Rivera, Urb & Seifert, Mycotaxon 119: 322. 2012. (MB563043). — Herb.: DAOM 239917. Ex-type: CCFC 239917. ITS barcode: JN626104. (Alternative markers: BT = JN625973; CMD = JN626016).
Penicillium maximae Visagie, Houbraken & Samson (this study). (MB803783). — Herb.: CBS-H21144. Ex-type: CBS 134565 = NRRL2060 = DTO 244C7. ITS barcode: EU427298. (Alternative markers: BT = KC773795; CMD = KC773821).
Penicillium sclerotiorum J.F.H. Beyma, Zentralbl. Bakteriol., 2. Abt., 96: 418. 1937. (MB277708). — Herb.: IMI 40569. Ex-type: CBS 287.36 = ATCC 10494 = IFO 6105 = IMI 040569 = NRRL 2074 = QM 1938 = VKMF-353. ITS barcode: JN626132. (Alternative markers: BT = JN626001; RPB2 = JN406585; CMD = JN626044).
Penicillium vanoranjei Visagie, Houbraken & Samson (this study). (MB803782). — Herb.: CBS-H 21145. Ex-type: CBS 134406 = DTO 99H6 = AHS3SF_13. ITS barcode: KC695696. (Alternative markers: BT = KC695686; CMD = KC695691).
Penicillium viticola Nonaka & Masuma, Mycoscience 52: 339. 2011. (MB516048). — Herb.: TNS-F38702. Ex-type: JCM 17636 = FKI-4410. ITS barcode: AB606414. (Alternative markers: BT = AB540174; CMD = JN686393 (representative strain DAOM 239933).
DISCUSSION
Houbraken & Samson (2011) introduced Penicillium section Sclerotiora for their ‘clade 2’, based on a four-gene phylogeny (RPB1, RPB2, Tsr1, Cct8) and accepted 17 species. The section corresponds to clade 3 of Peterson (2000). Rivera & Seifert (2011) and Rivera et al. (2012) recently revised the type species for the section, P. sclerotiorum, introduced five new species, redefined the concept of P. sclerotiorum, and provided a dichotomous key to the species of this clade. As such, we did not further consider known species from this clade here, although P. vanoranjei and P. maximae belong there.
Morphological characters that distinguish species of Penicillium section Sclerotiora are summarised in Table 2, mostly modified from data provided by Rivera & Seifert (2011), and illustrated in Fig. 3, 4, 5. This data is meant to facilitate species identification in conjunction with sequence data. GenBank accession numbers for ITS barcodes and supplementary identification markers to all known species of the section are given in the taxonomy section above. Although phenotypic characters that define the three clades of the section could not be identified, species identification using only morphology is possible. Colony morphology of strains grown under standardised conditions was especially informative. Conidiophore morphology among the different species is very similar. There are exceptions, however, with P. herquei and P. malachiteum both producing biverticillate conidiophores. Houbraken & Samson (2011) distinguished these two species by production of cleistothecia in P. malachiteum. We found growth at 30 °C a more useful character, with P. malachiteum showing restricted growth compared to P. herquei. It should be noted, however, that preliminary data show that this clade represents a species complex, with several species undescribed. As such, for this study we considered only type strains and a couple of representative strains with identical sequences. For the monoverticillate species in the P. adametzii complex (clade 2), growth rates at 30 and 37 °C are taxonomically informative. For example, P. arianeae and P. jugoslavicum do not grow at 30 °C, whereas only P. adametzii, P. amaliae, P. bilaiae, P. hirayamae and P. lilacinoechinulatum were able to grow at 37 °C. Also, acid production on CREA is a reliable taxonomic character. In general, most species in section Sclerotiora produce conidiophores with smooth walled stipes and spheroidal to subspheroidal conidia.
SEM observations of P. vanoranjei revealed a sheet-like extracellular matrix that might act as a kind of glue or protection against adverse conditions including drying. Penicillium vanoranjei produces large amounts of exudates in some colonies. We have often observed the drying of exudate droplets in older cultures, leaving a thin, sheet-like coloured matrix behind. This mechanism could be responsible for the covering of the sclerotia shown in the SEM micrographs (Fig. 7b–e). Conidial wall texture is informative in section Sclerotiora but difficult to interpret with light microscopy. SEM photographs show the development of P. vanoranjei conidia in basipetal chains, and the development of conidial roughening as conidia expand (Fig. 7f–i).
Grigorieva-Manoilova & Poradielova (1915) originally isolated P. multicolor from soil in Russia and described it as having short stipes ending in vesicles. They reported that colony colours range from yellow to orange to red. Raper & Thom (1949) used several strains isolated from America as representative for P. multicolor, but noted that their identification of these strains (i.e. NRRL 2060) might be doubtful. Pitt (1979) considered P. multicolor a confused name and did not accept it in his treatment of the genus. Peterson & Horn (2009) treated NRRL 2060 as the ex-type of P. multicolor. However, this strain has features inconsistent with the original description by Grigorieva-Manoilova & Poradielova (1915). Based on sequence data, it was shown that the ex-type for P. multicolor (CBS 501.73 = ATCC 24723 = IMI 174716) is closely related to P. fellutanum, in section Charlesia (Houbraken & Samson 2011). Rivera & Seifert (2011) considered it as a tentative synonym of P. fellutanum. To resolve this confusion, we introduce and describe P. maximae above for NRRL 2060, depositing the herbarium material as CBS-H21144 (= CBS 134565 ex-type). Penicillium multicolor in the sense of CBS 501.73 will be included in a future study of section Charlesia. However, we recommend KC790402 as the species ITS DNA barcode, with the alternative β-tubulin marker JN799645, for identification of the species.
Valla et al. (1989) originally described P. nodositatum and deposited strains in CBS. Houbraken & Samson (2011) included the ex-type strain (CBS 330.90) in their phylogenies, reporting it had identical sequences to the ex-type strain (CBS 221.66) of P. bilaiae. However, they noted problems with the synonymy of P. nodositatum with P. bilaiae because it produces biverticillate conidiophores (Valla et al. 1989), in contrast to the monoverticillate conidiophores of P. bilaiae (Chalabuda 1950, Pitt 1979, Savard et al. 1994). We traced the problem to incorrect accession numbers in the CBS database. Although the herbarium specimen was submitted with accession number CBS 330.90, the accepted species list endorsed by the International Commission of Penicillium and Aspergillus (ICPA, http://www.aspergilluspenicillium.org/) included the living ex-type strain of the herbarium specimen as CBS 333.90. The source of this error in the database cannot be traced. However, the living culture CBS 330.90 represents P. bilaiae with monoverticillate conidiophores, whereas P. nodositatum (CBS 333.90T) is closely related to P. kabunicum and does not belong in Penicillium s.str. (Houbraken & Samson 2011). Herbarium material for P. nodisitatum has subsequently been lost. In future, the strain will be lectotypified and epitypified in a study focused on the classification of species rejected from Penicillium s.str. by Houbraken & Samson (2011).
The isolation of P. amaliae from both South Africa and Australia is intriguing. The South African isolates were obtained from Protea repens infructescences as part of an ecological study focused on mite dispersal of Penicillium in this niche. The occurrence of this species in Australia from insect larvae thus suggests a pattern. Similarly, a closely related species, P. brocae, was described from coffee-berry borers and their faeces and guts in Mexico (Peterson et al. 2003). The idea of Penicillium species being transported or vectored by mites and insects is not new (Peterson et al. 2003, Hubert et al. 2004, Seifert et al. 2004, Visagie et al. 2009). However, it has never been demonstrated as a mutualistic relationship. In a similar ecological study from Fynbos, a mutualistic association was shown between Ophiostoma species and mites (Roets et al. 2007). Our preliminary data does suggest that at least some Penicillium species have a similar mutualistic association (Visagie 2012). Visagie (2012) showed that Penicillium populations in the diverse Fynbos biome in South Africa are distinct at the different sites sampled. Postulating a mutualistic relationship and positive correlation for the dispersal of Penicillium populations in this niche may help explain the distribution patterns of these species and make us re-evaluate the idea of water and wind as the only primary dispersal mechanisms in Penicillium.
Acknowledgments
We acknowledge the South African Biosystematics Initiative (SABI) of the National Research Foundation (NRF) and The Alfred P. Sloan Foundation and the grant NATO (ESP.MD.SFPP 981674) for financial support during this project. We are grateful to J.I. Pitt and the NRRL who provided strains used for this study.
REFERENCES
- Chalabuda TV. 1950. Species novae e genere Penicillium Link. Notulae Systematicae e sectione Cryptogamic Instituti Botanici Academiae Scientiarum URSS 6: 161–169 [Google Scholar]
- Frisvad JC, Samson R. 2004. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Studies in Mycology 49: 1–174 [Google Scholar]
- Gascuel O. 1997. BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Molecular Biology and Evolution 14: 685–695 [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied and Environmental Microbiology 61: 1323–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieva-Manoilova OC, Poradielova NN. 1915. Concerning a new pigment producing mold belonging to the genus Penicillium (trans. text). Archives des Sciences Biologiques Leningrad 19: 117–131 [Google Scholar]
- Hong SB, Cho HS, Shin HD, Frisvad JC, Samson RA. 2006. Novel Neosartorya species isolated from soil in Korea. International Journal of Systematics and Evolutionary Microbiology 56: 477–486 [DOI] [PubMed] [Google Scholar]
- Houbraken J, Samson RA. 2011. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology 70: 1–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert J, Stejskal V, Munzbergova Z, Kubatova A, Vanova M, Zdarkova E. 2004. Mites and fungi in heavily infested stores in the Czech Republic. Journal of Economic Entomology 97: 2144–2153 [DOI] [PubMed] [Google Scholar]
- Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods in Molecular Biology 537: 39–64 [DOI] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. 1967. Methuen handbook of color. Denmark, Sankt Jørgen Tryk [Google Scholar]
- McNeill J, Barrie FR, Buck WR, Demoulin V, Greuter W, et al. 2012. International code of nomenclature for algae, fungi, and plants (Melbourne code). Regnum vegetabile 154: 1–240 Gartner Verlag kg. http://www.iapt-taxon.org/nomen/main.php [Google Scholar]
- Nonaka K, Masuma R, Iwatsuki M, Shiomi K, Otoguro K, Ōmura S. 2011. Penicillium viticola, a new species isolated from a grape in Japan. Mycoscience 52: 338–343 [Google Scholar]
- Okuda T, Klich MA, Seifert KA, Ando K. 2000. Media and incubation effects on morphological characteristics of Penicillium and Aspergillus. In: Samson RA, Pitt JI. (eds), Integration of modern taxonomic for Penicillium and Aspergillus classification: 83–99 Harwood Academic Publishers, Amsterdam, the Netherlands [Google Scholar]
- Peterson SW. 2000. Phylogenetic analysis of Penicillium species based on ITS and LSU-rDNA nucleotide sequences. In: Samson RA, Pitt JI. (eds), Integration of modern taxonomic for Penicillium and Aspergillus classification: 163–178 Harwood Academic Publishers, Amsterdam, the Netherlands [Google Scholar]
- Peterson SW, Bayer E, Wicklow D. 2004. Penicillium thiersii, Penicillium angulare and Penicillium decaturense, new species isolated from wood-decay fungi in North America and their phylogenetic placement from multilocus DNA sequence analysis. Mycologia 96: 1280–1293 [PubMed] [Google Scholar]
- Peterson SW, Horn BW. 2009. Penicillium parvulum and Penicillium georgiense, sp. nov., isolated from the conidial heads of Aspergillus species. Mycologia 101: 71–83 [DOI] [PubMed] [Google Scholar]
- Peterson SW, Pérez J, Vega F, Infante F. 2003. Penicillium brocae, a new species associated with the coffee berry borer in Chiapas, Mexico. Mycologia 95: 141–147 [PubMed] [Google Scholar]
- Pitt JI. 1979. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. Academic Press Inc., England [Google Scholar]
- Pitt JI, Samson RA, Frisvad JC. 2000. List of accepted species and their synonyms in the family Trichocomaceae. In: Samson RA, Pitt JI. (eds), Integration of modern taxonomic for Penicillium and Aspergillus classification: 9–79 Harwood Academic Publishers, Amsterdam, the Netherlands [Google Scholar]
- Raper KB, Thom C. 1949. A manual of the Penicillia. Baltimore, The Williams & Wilkins Company [Google Scholar]
- Rivera KG, Chavarría-Díaz F, Garcia M, Urb M, Thorn RG, et al. 2012. Penicillium mallochii and P. guanacastense, two new species isolated from Costa Rican caterpillars. Mycotaxon 119: 315–328 [Google Scholar]
- Rivera KG, Seifert KA. 2011. A taxonomic and phylogenetic revision of the Penicillium sclerotiorum complex. Studies in Mycology 70: 139–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roets F, Crous PW, Wingfield MJ, Dreyer LL. 2007. Discovery of fungus-mite-mutualism within a unique niche of the Cape Floral Kingdom. Environmental Entomology 36: 1226–1237 [DOI] [PubMed] [Google Scholar]
- Samson RA, Yilmaz N, Houbraken J, Spierenburg H, Seifert KA, et al. 2011. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Studies in Mycology 70: 159–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savard ME, Miller JD, Blais LA, Seifert KA, Samson RA. 1994. Secondary metabolites of Penicillium bilaii strain PB-50. Mycopathologia 127: 19–27 [DOI] [PubMed] [Google Scholar]
- Seifert KA, Hoekstra ES, Frisvad JC, Louis-Seize G. 2004. Penicillium cecidicola, a new species on cynipid insect galls on Quercus pacifica in the western United States. Studies in Mycology 50: 517–523 [Google Scholar]
- Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Sunderland, MA [Google Scholar]
- Valla G, Capellano A, Hugueney R, Moiroud A. 1989. Penicillium nodositatum Valla, a new species inducing myconodules on Alnus roots. Plant and Soil 114: 142–146 [Google Scholar]
- Visagie CM. 2012. The polyphasic taxonomy of Penicillium and Talaromyces spp. isolated from the diverse Fynbos biome. PhD dissertation, University of Stellenbosch, South Africa [Google Scholar]
- Visagie CM, Roets F, Jacobs K. 2009. A new species of Penicillium, P. ramulosum sp. nov., from the natural environment. Mycologia 101: 888–895 [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct identification of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR Protocols: a guide to methods and applications: 315–322 Academic Press, USA [Google Scholar]