Abstract
We have performed a phenotypic and phylogenetic study of a set of fungi, mostly of veterinary origin, morphologically similar to the Chrysosporium asexual morph of Nannizziopsis vriesii (Onygenales, Eurotiomycetidae, Eurotiomycetes, Ascomycota). The analysis of sequences of the D1-D2 domains of the 28S rDNA, including representatives of the different families of the Onygenales, revealed that N. vriesii and relatives form a distinct lineage within that order, which is proposed as the new family Nannizziopsiaceae. The members of this family show the particular characteristic of causing skin infections in reptiles and producing hyaline, thin- and smooth-walled, small, mostly sessile 1-celled conidia and colonies with a pungent skunk-like odour. The phenotypic and multigene study results, based on ribosomal ITS region, actin and β-tubulin sequences, demonstrated that some of the fungi included in this study were different from the known species of Nannizziopsis and Chrysosporium and are described here as new. They are N. chlamydospora, N. draconii, N. arthrosporioides, N. pluriseptata and Chrysosporium longisporum. Nannizziopsis chlamydospora is distinguished by producing chlamydospores and by its ability to grow at 5 °C. Nannizziopsis draconii is able to grow on bromocresol purple-milk solids-glucose (BCP-MS-G) agar alkalinizing the medium, is resistant to 0.2 % cycloheximide but does not grow on Sabouraud dextrose agar (SDA) with 3 % NaCl. Nannizziopsis arthrosporioides is characterised by the production of very long arthroconidia. Nannizziopsis pluriseptata produces 1- to 5-celled sessile conidia, alkalinizes the BCP-MS-G agar and grows on SDA supplemented with 5 % NaCl. Chrysosporium longisporum shows long sessile conidia (up to 13 μm) and does not produce lipase.
Keywords: animal infections, ascomycetes, Chrysosporium, mycoses, Nannizziopsiaceae, Nannizziopsis, Onygenales, reptiles
INTRODUCTION
The genus Chrysosporium comprises a large number of ubiquitous anamorphic species, which are predominantly found in soil, marine and freshwater sediments, decaying wood, feathers, skin and hair of mammals, reptiles and birds (Rees 1967a, b, c, de Hoog et al. 2000, Hubalek 2000, Mandeel et al. 2009). Chrysosporium is usually characterised by whitish to pale colonies and conidia sessile or arising on short stalks from the fertile hyphae. The conidia are broader than the diameter of the hyphae, and they are usually subglobose, pyriform or claviform and are released rhexolytically (Sigler 1997, de Hoog et al. 2000). Due to the large number of species of Chrysosporium (approximately seventy), the poor morphological differentiation of its species and, in some cases, the absence of an associated sexual morph, they are not easy to identify and the distinction from similar genera such as Geomyces, Malbranchea, or Sporotrichum, among others, is difficult (Vidal et al. 2000).
Based on the analysis of the sequences of the internal transcribed spacer region (ITS), Vidal et al. (2000) demonstrated that Chrysosporium is polyphyletic and the phylogenetic relationships of Chrysosporium merdarium, the type species of the genus, revealed that it belongs to the Gymnoascaceae (Onygenales). Those same authors also indicated that some morphological characters traditionally used in taxonomy such as the colour of the colony, the growth rate at different temperatures, conidiogenesis, and conidial morphology, are subject to homoplasy and, in some cases, are not useful to resolve the species boundaries.
Some Chrysosporium species develop teleomorphs belonging to very diverse genera in the families Arthrodermataceae (Currah 1985), Ascosphaeriaceae (van Oorschot 1980), Chaetomiaceae (Vidal et al. 2000), Gymnoascaceae (van Oorschot 1980, Currah 1985), Lasiosphaeriaceae (Mouchacca & Gams 1993, Ueda 1994), Onygenaceae (van Oorschot 1980, Currah 1985), Monascaceae (Pettersson et al. 2011) and Myxotrichaceae (Vidal et al. 2000).
Most of the Chrysosporium isolates found in the clinical laboratory are contaminants but some species occasionally infect humans. Most produce skin and nail lesions although some deep infections, mainly in immunocompromised patients, have also been reported (Sigler 1997, Sigler et al. 1998, Roilides et al. 1999, de Hoog et al. 2000, Stebbins et al. 2004, Abdel-Razik & Zaki 2008). In most of those reports; however the etiologic agent has been identified only at the genus level. One of the most relevant pathogenic species is Nannizziopsis vriesii, which has a Chrysosporium anamorph, causing severe and often fatal dermatomycosis in different species of reptiles (Paré et al. 1997, Nichols et al. 1999, Thomas et al. 2002, Bertelsen et al. 2005, Mitchell et al. 2006, Paré et al. 2006, Bowman et al. 2007, Paré & Jacobson 2007, Han et al. 2010, Hedley et al. 2010, Hellebuyck et al. 2010, Allender et al. 2011, Johnson et al. 2011). However, N. vriesii also produces infections in humans (Stebbins et al. 2004, Brandt et al. 2005, Steininger et al. 2005). It has been suggested that other Chrysosporium species, morphologically similar to N. vriesii, could also be involved in human and animal infections (Brandt et al. 2005, Abarca et al. 2008, 2009, 2010). The recent description of C. guarroi, which infects reptiles (Abarca et al. 2010) and is phylogenetically related to N. vriesii, suggests the existence of a complex of morphologically similar species.
Using phenotypic and molecular methods, we have studied a set of clinical Chrysosporium isolates from different reptiles and humans that are morphologically similar to N. vriesii, in order to better characterise these fungi and to determine their phylogenetic boundaries.
MATERIALS AND METHODS
Fungal isolates
The clinical isolates and reference strains included in the study are detailed in Table 1. Only two strains of N. vriesii were included in this study. This is the type species of the genus, i.e., the type strain and a clinical strain from Germany. Of the other species of Nannizziopsis (N. albicans, N. hispanica, N. mirabilis, N. patagonica and N. tropicalis) only live cultures of N. albicans are available, but this species is phylogenetically related to Amauroascus (Onygenaceae) (Solé et al. 2002) and was not included in the study.
Table 1.
Fungi included in this study.
| Species1 |
Origin | GenBank accession no. |
||||
|---|---|---|---|---|---|---|
| D1-D2 | ACT | ITS | TUB | |||
| Nannizziopsis vriesii | RKI 04-0104 | Human, brain abscess, Nigerian man, Germany | HF547853 | HF547877 | HF547869 | HF547878 |
| Nannizziopsis chlamydospora (Chrysosporium sp. 1) | UTHSC 04-2056 | Pogona vitticeps, USA | HF547854 | HF547879 | HF547870 | HF547880 |
| Nannizziopsis chlamydospora (Chrysosporium sp. 1) | UTHSC 06-1419 | Pogona vitticeps, USA | HF547855 | HF547881 | HF547871 | HF547882 |
| Nannizziopsis draconii (Chrysosporium sp. 2) | CCFVB CH12 | Pogona vitticeps, Spain | HF547856 | HF547883 | EU883993* | HF547884 |
| Nannizziopsis arthrosporioides (Chrysosporium sp. 3) | UTHSC R-4263 | Physignathus sp. (water dragon), USA | HF547857 | HF547885 | HF547872 | HF547886 |
| Chrysosporium longisporum (Chrysosporium sp. 4) | UTHSC R-4380 | Snake, multifocal dermatitis, USA | HF547858 | HF547887 | HF547873 | HF547888 |
| Nannizziopsis pluriseptata (Chrysosporium sp. 5) | UTHSC 10-1045 | Skink lizard, USA | HF547859 | HF547889 | HF547874 | HF547890 |
| Chrysosporium ophiodiicola | CBS 122913T | Snake, subcutaneous granuloma, USA | EU15820* | HF547891 | EU15819* | HF547892 |
| Nannizziopsis vriesii | IMI 149994T | Ameiva sp., skin and lungs, USA | AY176715* | HF547893 | AJ131687* | HF547894 |
| Nannizziopsis guarroi | CBS 124553T | Iguana iguana, Spain | FJ839684* | HF547895 | EU018451* | HF547896 |
| CCFVB CH11 | Iguana iguana, Spain | HF547860 | ||||
| CCFVB CH14 | Iguana iguana, Spain | HF547861 | ||||
| CCFVB CH15 | Iguana iguana, Spain | HF547862 | ||||
| CCFVB CH16 | Iguana iguana, Spain | HF547863 | ||||
| UTHSC R-4309 | Snake, USA | HF547864 | ||||
| UTHSC 05-1370 | Pogona vitticeps , USA | HF547865 | ||||
| UTHSC 06-3993 | Agama agama , USA | HF547866 | HF547897 | HF547875 | HF547898 | |
| UTHSC 07-3227 | Pogona vitticeps, USA | HF547867 | ||||
| UTHSC R-4317 | Human, disseminated disease, Nigerian man, USA | HF547868 | HF547899 | HF547876 | HF547900 | |
| Uncinocarpus reesii | ATCC 34533 | Feathers, Australia | AY176724* | |||
| Amauroascus niger | ATCC 22339T | Soil, USA | AY176706* | |||
| Chrysosporium tropicum | MUCL 10068T | Woolen overcoat, Solomon Islands | AY176731* | |||
| Aphanoascus mephitalis | ATCC 22144 T | Wolf dung, Canada | AY176725* | |||
| Chrysosporium keratinophilum | CBS 392.67 | Soil, New Zealand | AY176730* | |||
| Arthroderma cajetani | OMH H1-10 | Human, Canada | AY176736* | |||
| Arthroderma otae | UAMH 2338 | Human, skin scrapings and hair, Canada | AY176735* | |||
| Arthroderma ciferrii | ATCC 24447T | Soil, USA | EF413625* | |||
| Ctenomyces serratus | CBS 187.61T | Soil, Australia | AY176733* | |||
| Chrysosporium vallenarense | UAMH 6914 | Dung of Alopex lagopus, Chile | AY176732* | |||
| Gymnoascus littoralis | CBS 454.73 | Conch shell, Canada | FJ358272* | |||
| Gymnascus aurantiacus | ATCC 22394 T | Soil, Russia | AY176747* | |||
| Gymnascus ruber | CBS 352.90 | Soil, England | AY176746* | |||
| Ascosphaera subglobosa | Voucher A.A. Wynns 5004(C) | Megachile rotundata, USA | HQ540517* | |||
| Ascosphaera apis | CBS 252.32 | Apis mellifica, Denmark | AY004344* | |||
| Paracoccidioides brasiliensis | IMTSP 556 | Human, Brazil | U81263* | |||
| Ajellomyces dermatitidis | ATCC 18187 T | Human, USA | AY176704* | |||
| Ajellomyces capsulatus | UAMH 7141 | Soil, USA | AF038353* | |||
| Byssochlamys nivea | CBS 100.11T | Geastrum coronatum, Sweden | AY176750* | |||
| Eurotium herbariorum | ATCC 16469 | Unpainted board, USA | AY176751* | |||
| Petromyces alliaceus | ATCC 16891 | Soil, Australia | AY176752* | |||
| Arachnomyces nodosetosus | CCF 3957 | Human, nail infection, Czech Republic | HM205103* | |||
| Arachnomyces glareosus | CBS 116129 | Human, thumb nail, Canada | FJ358273* | |||
| Arachnomyces minimus | CBS 324.70 | Decayed wood, Canada | FJ358274* | |||
| Eremascus fertilis | KVL 10-09 | Pollen, Denmark | HQ540515* | |||
| Lecythophora hoffmannii | CBS 140.41 | Sewage water, England | AB261976* | |||
| Bettsia alvei | KVL 10-08 | Pollen, Denmark | HQ540516* | |||
T Ex-type strain.
* sequences retrieved from the GenBank database.
1 ATCC: American Type Culture Collection, USA; CBS: Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; CCF: Culture Collection of Fungi, Department of Botany, Charles University in Prague, Czech Republic; CCFVB: Culture Collection of the Veterinary Mycology Group, Bellaterra, Barcelona, Spain; IMI: International Mycological Institute Culture Collection, Surrey, United Kingdon; IMTSP: Instituto de Medicina Tropical de São Paulo Culture Collection, São Paulo, Brazil; KVL: Entomopathogenic Fungal Culture Collection at Department of Agriculture and Ecology, Faculty of Life Sciences, University of Copenhagen, Denmark; MUCL: Mycotheque de l’Universite Catholique de Louvain, Louvain la Neuve, Belgium; OMH: Ontario Ministry of Health, Toronto, Ontario, Canada; RKI: Robert Koch Institute, Berlin, Germany; UAMH: Microfungus Collection and Herbarium, University of Alberta, Canada; UTHSC: Fungus Testing Laboratory, University of Texas Health Science Center at San Antonio, USA.
Molecular study
DNA was extracted according to Perdomo et al. (2011). Detailed protocols for the amplification of D1 and D2 domains of the 28S rDNA (D1-D2), the internal transcribed spacer region (ITS), and a fragment of actin (ACT) and β-tubulin (TUB) genes were described in Cano et al. (2004) (ITS), Voigt & Wöstemeyer (2000) (ACT), and Gilgado et al. (2005) (D1-D2 and TUB). PCR products were purified and sequenced at Macrogen Corp. Europe (Amsterdam Zuid-Oost, The Netherlands) with a 3730XL DNA analyzer (Applied Biosystems). The program SeqMan (Lasergene, Madison, Wisconsin) was used to obtain consensus sequences of each isolate. DNA sequences were aligned with the program ClustalX v. 1.8 (Thompson et al. 1997) with default parameters, followed by manual adjustments with a text editor. A D1-D2 and an ITS BLAST were carried out with the ex-type strains of N. vriesii and C. guarroi in order to select the closest species and to include it in the phylogenetic study.
Sequences retrieved from GenBank and included in the phylogenetic analysis are in Table 1. Phylogenetic analysis of the D1-D2 encompassed representatives of the clinical isolates and reference strains of the families within the order Onygenales (Ajellomycetaceae, Arachnomycetaceae, Arthrodermataceae, Ascosphaeraceae, Gymnoascaceae and Onygenaceae) as well as some representatives of Eurotiales. Eremascus fertilis (HQ540515) and Lecythophora hoffmannii (AB261976) were used as outgroups. The combined dataset (ITS, ACT and TUB), included representatives of the clinical isolates and the type strains of C. guarroi, C. ophiodiicola and N. vriesii. The phylogenetic analyses were conducted using MEGA v. 5.05 (Tamura et al. 2011) with maximum likelihood (ML) algorithm, using Tamura 3-parameter substitution model with gamma distribution (D1-D2) and Tamura-Nei with gamma distribution (combined dataset). The robustness of branches was assessed by bootstrap analysis of 1 000 replicates. Bayesian analyses (BA) were carried out using MrBayes v. 3.1 (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003). Bayesian analyses were performed by running 1 000 000 generations in four chains, saving the current tree every 100 generations. The last 18 000 trees were used to construct a 50 % majority-rule consensus tree and to determine the posterior probabilities of the branches. The sequences generated in this study and the alignments used in the phylogenetic analyses were deposited in GenBank (Table 1) and TreeBASE (accession URL: TB2:S13558), respectively.
Morphological studies
Colonial features were examined after 14 days of incubation on malt extract agar (MEA; Difco Laboratories, Detroit, MI, USA), oatmeal agar (OA; 30 g filtered oat flakes, 20 g agar, 1 L distilled water), potato dextrose agar (PDA; Pronadisa S.A., Spain), phytone yeast extract agar (PYE; Becton Dickinson Microbiology Systems, Cockeysville, MD, USA) and Sabouraud dextrose agar (SDA; Pronadisa S.A., Spain). Colonial growth rates were determined at different temperatures (from 5 °C to 40 °C, in 5 °C intervals). To induce the formation of ascomata, the isolates were grown on OA and incubated at 25 °C and 30 °C for up to three months. Colour notations (in parenthesis) are from Kornerup & Wanscher (1978). Microscopic features were studied on PDA and PYE slide cultures incubated for 7–14 d at 30 °C, and mounted in lactic acid.
Physiological studies
Production of urease was determined in Christensen’s urea broth after incubation at 30 °C for 7 d. Lipase activity was tested by growing on Tween 80 opacity test medium (TOTM) according to Slifkin (2000), incubating the Petri dishes at 30 °C for 14 d. Growth on dermatophyte test medium (DTM) and colour changes from yellow (acidic) to red (basic) were recorded after incubating the Petri dishes at room temperature (20 °C to 30 °C) for 14 d. Hydrolysis of milk solids was detected on bromocresol purple-milk solids-glucose agar (BCP-MS-G) Petri dishes, according Kane et al. (1997), after incubation at 30 °C for 14 d. Cycloheximide tolerance was evaluated by growing isolates on SDA supplemented with 0.2 % of cycloheximide (Sigma, USA) at 30 °C for 14 d. Tolerance to NaCl was evaluated by growth of isolates on SDA, amended with 3 % and 5 % w/v NaCl, after incubation for 14 d at 30 °C. Hemolysis was evaluated by culturing isolates on blood agar (BioMérieux, France) for 14 d at 30 °C.
RESULTS
Molecular analysis
With the primers used, we were able to amplify and sequence 485–502 bp (D1-D2), 428–507 bp (ITS), 628–645 bp (ACT) and 402–419 bp (TUB). None of the isolates showed an ITS sequence identity higher than 95 % with the species of Chrysosporium represented in GenBank, with the exception of eight reptilian isolates that showed 100 % identity with the type strain of C. guarroi and one isolate from a human systemic infection in the USA (UTHSC R-4317) that showed a 97.8 % identity also with this species. Maximum likelihood (ML) and Bayesian analyses of D1-D2 dataset produced phylogenetic trees with similar topologies. Fig. 1 shows the D1-D2 ML tree including the bootstrap support (bs) and the posterior probabilities (pp). Three main clades could be distinguished within the ingroup corresponding to the orders Eurotiales (97 % bs/1 pp), Arachnomycetales (97 % bs/1 pp) and Onygenales (72 % bs/1 pp), respectively. The Onygenales encompassed five well-supported clades, corresponding to the families Ascosphaeraceae (99/1), Gymnoascaceae (99/1), Arthrodermataceae (96/1) and Onygenaceae (90/1), and the fifth one (99/1) that embraced the type strains of C. guarroi and N. vriesii, and most of our clinical isolates, and a poorly-supported group (66/-) that included some members of the family Ajellomycetacaeae. This analysis did not resolve the taxonomic position of the type strain of C. ophiodiicola and of the clinical isolate UTHSC R-4380 (provisionally named Chrysosporium sp. 4), which consisted of two different branches between the Onygenaceae and the Nannizziopsis group.
Fig. 1.
Maximum-likelihood (ML) tree based on Tamura three-parameter corrected nucleotide distances among the D1 and D2 domains of the 28S rRNA gene sequences of taxa included in Table 1. Numbers on the branches are bootstrap ML values above 55 %, followed by Bayesian posterior probabilities (Bpp) above 0.6. Branch lengths are proportional to distance. Sequences not generated in this study and obtained from the GenBank database are indicated in parentheses. Ex-type strains of the different species are indicated with T. New species proposed in this study are indicated in bold. N. = Nannizziopsis. C. = Chrysosporium.
In the combined ITS-ACT-TUB ML tree (Fig. 2) several terminal well-supported branches representing undescribed species were shown. These were Chrysosporium sp. 1, that included the isolates UTHSC 04-2056 and UTHSC 06-1419, Chrysosporium sp. 2 (isolate CCFVB CH12), Chrysosporium sp. 3 (isolate UTHSC R-4263) and Chrysosporium sp. 5 (isolate UTHSC 10-1045). Additionally, three strains of C. guarroi also formed a terminal clade (84 % bs) that included the type strain of this species, one reptilian isolate and a human clinical strain (UTHSC R-4317), which was slightly separated from the other two. The ex-type strain of N. vriesii, and a human clinical strain, morphologically identified as N. vriesii (RKI 04-0104), were separated from the rest of fungi included in the tree but also separated between them. The ex-type strain of C. ophiodiicola (CBS 122913) and the isolate UTHSC R-4380 (Chrysosporium sp. 4) were placed away from the others and in fact acted as outgroups.
Fig. 2.
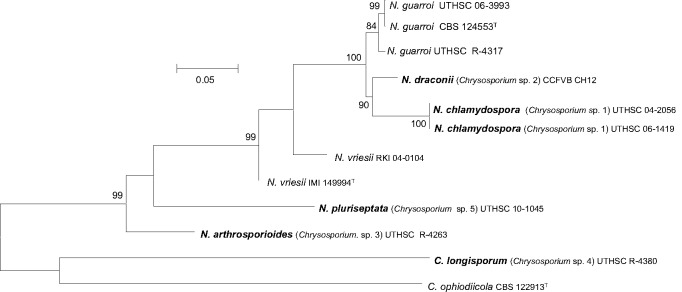
Maximum-likelihood (ML) tree obtained from the combined DNA sequence data from three loci (ITS, actin and β-tubulin). Bootstrap support values above 70 % are indicated at the nodes.
Morphological study
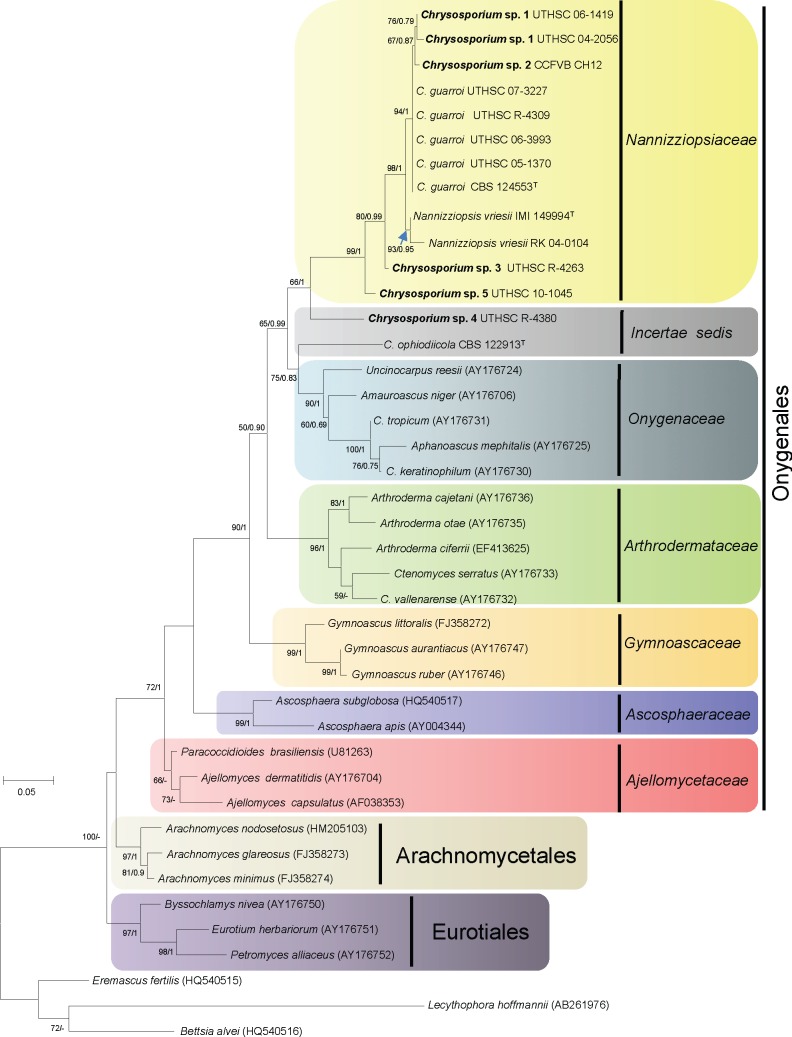
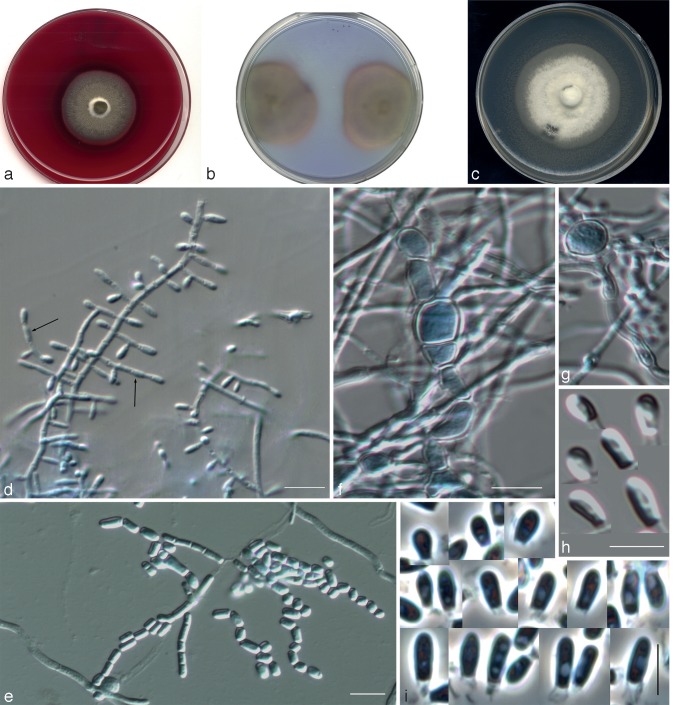
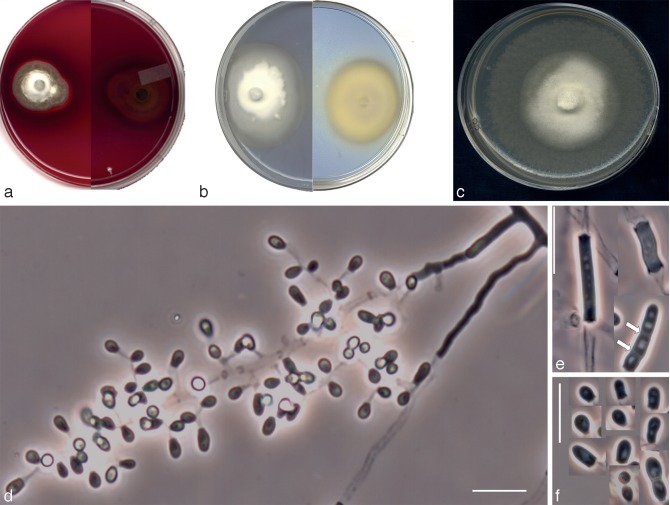
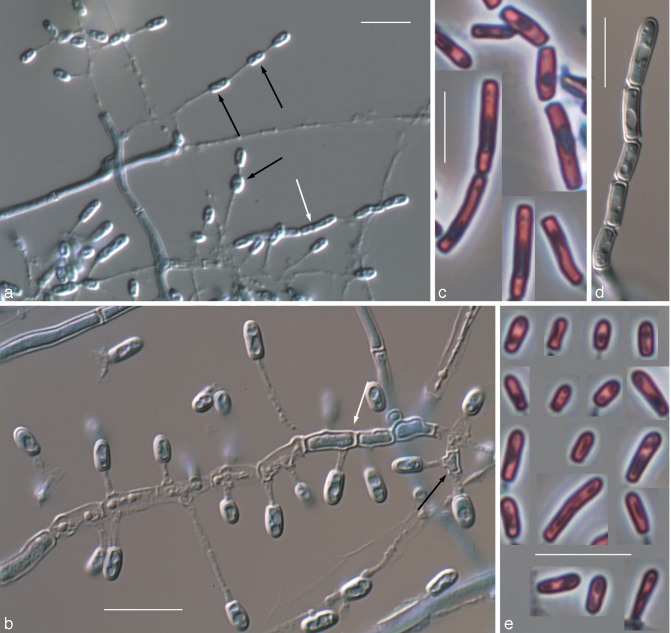
The isolates included in this study were characterised by the production of thin- and smooth-walled, small, mostly sessile and 1-celled conidia. The only species able to produce chlamydospores was Chrysosporium sp. 1 (Fig. 3f, g). Single intercalary conidia were formed by all the isolates, with those of Chrysosporium sp. 3 being the longest (Fig. 5e). All the species, with the exception of Chrysosporium sp. 4, produced arthroconidia in chains, usually terminal (Fig. 3e, 6f, g). In all the isolates the sessile and terminal conidia were similar in size, although some conidia of Chrysosporium sp. 4 (Fig. 7e) and Chrysosporium sp. 5 (Fig. 6h) were considerably longer (above 10 μm). Chrysosporium sp. 5 is the only species that produced up to 5-celled conidia (Fig. 6h). On PYE at 25 °C Chrysosporium guarroi showed the slowest growth rate (17–22 mm in 14 d), whereas Chrysosporium sp. 4 was the fastest (40–46 mm in 14 d).
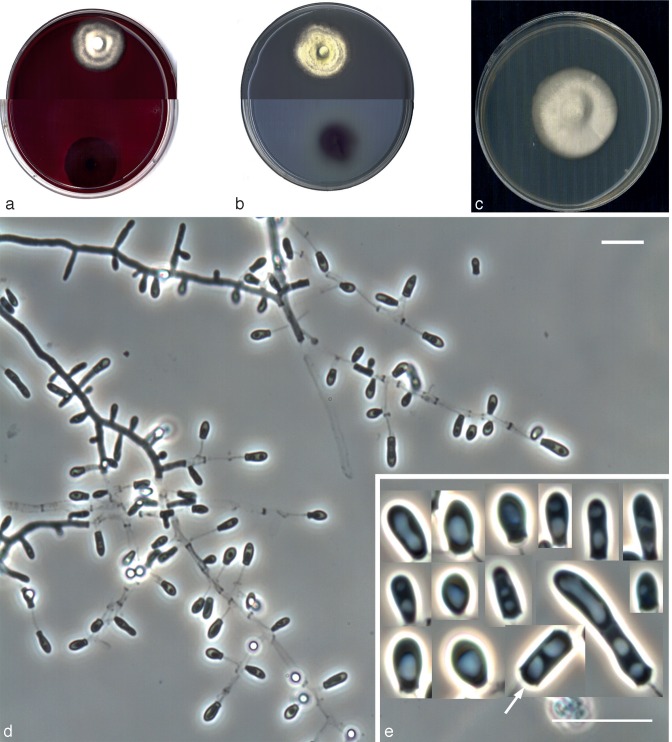
Fig. 3.
Nannizziopsis chlamydospora UTHSC 04-2056 (= Chrysosporium sp. 1). a. Colony on blood agar; b. colonies on BCP-MS-G (reverse); c. colony on TOTM; d. conidiophores bearing sessile and intercalary conidia (black arrow), and conidia on side branches; e. long chains of lateral and terminal arthroconidia; f. chlamydospores in chains; g. a solitary chlamydospore and thick-walled hyphae; h, i. conidia. — Scale bars: d–g = 10 μm; h, i = 5 μm (d–h, differential interference contrast; i, phase contrast).
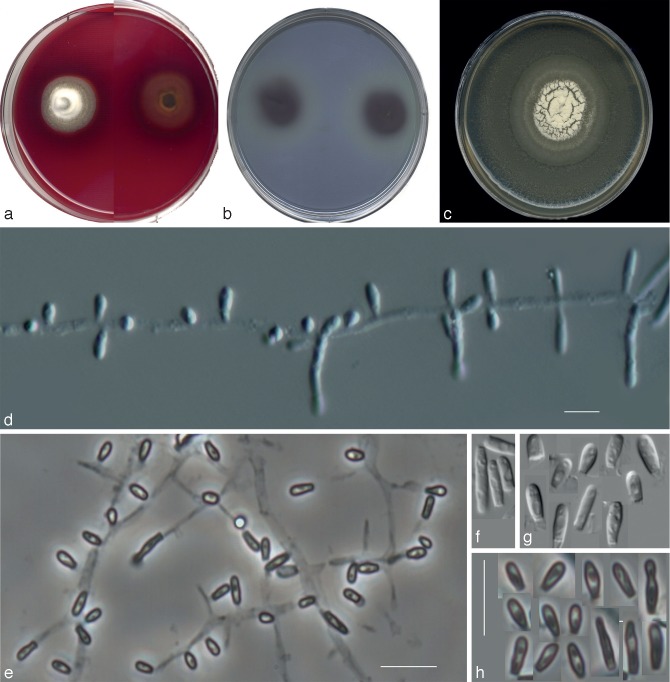
Fig. 5.
Nannizziopsis arthrosporioides UTHSC R-4263 (= Chrysosporium sp. 3). a. Colony on blood agar (surface and reverse); b. colony on BCP-MS-G (surface and reverse); c. colony on TOTM; d. fertile hyphae bearing sessile conidia; e. two singly intercalary conidia and a terminal chain of arthroconidia (white arrows show the septa); f. sessile (some 2-celled) conidia. — Scale bars = 10 μm (d–f, differential interference contrast).
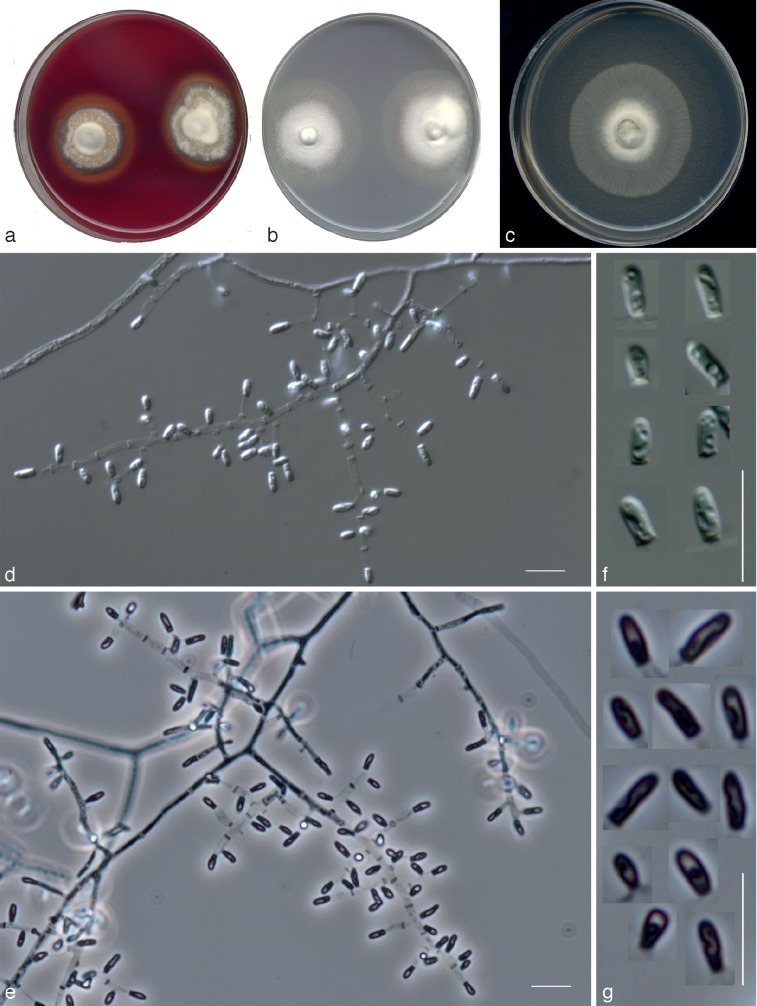
Fig. 6.
Nannizziopsis pluriseptata UTHSC 10-1045 (= Chrysosporium sp. 5). a. Colonies on blood agar (the arrow shows the b-hemolysis halus); b. colony on BCP-MS-G (surface and reverse); c. colony on TOTM; d, e. fertile hyphae bearing mostly sessile conidia (arrow showing a intercalary conidium); f, g. arthroconidia; h. sessile conidia (observe the presence of up to 5-celled propagules). — Scale bars = 10 μm (d, f, differential interference contrast; e, g, h, phase contrast).
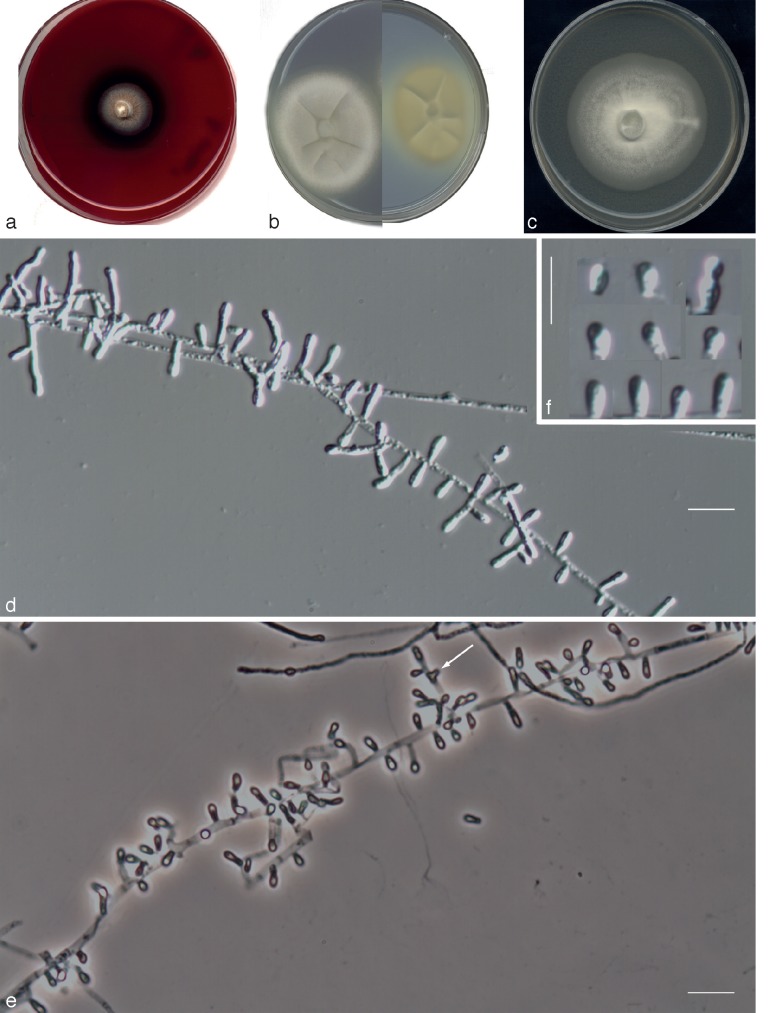
Fig. 7.
Chrysosporium longisporum UTHSC R-4380 (= Chrysosporium sp. 4). a. Colony on blood agar (surface and reverse); b. colony on BCP-MS-G (surface and reverse); c. colony on TOTM; d. fertile hyphae bearing mostly sessile conidia; e. sessile and intercalary (arrow) conidia. — Scale bars = 10 μm (d, e, phase contrast).
Physiological characterisation
The urease test was strain dependent, however the majority of the isolates were positive. All the fungi tested grew on DTM changing the colour of the medium from yellow to red (data not shown). The results of the other physiological tests are summarised in Table 2. With the exception of C. ophiodiicola, all the strains included in the study produced hemolysis on blood agar (Fig. 3a, 4a, 5a, 6a, 7a, 8a, 9a). All of them, with the exception of Chrysosporium sp. 4, showed lipolytic activity (Fig. 3c, 4c, 5c, 6c, 8c, 9c). The isolates were grown on BCP-MS-G agar to test the acidification or alkalisation of this medium, and the milk solids hydrolysis. Nannizziopsis vriesii, C. ophiodiicola, Chrysosporium sp. 3 and Chrysosporium sp. 4 acidified the medium, whereas C. guarroi, Chrysosporium sp. 1, Chrysosporium sp. 2 and Chrysosporium sp. 5 produced alkalisation, and only Chrysosporium sp. 2 and Chrysosporium sp. 5 showed a strong hydrolysis of milk solids. On SDA medium with 3 % NaCl, Chrysosporium sp. 2 did not grow and nor did four isolates of C. guarroi. On the same medium with 5 % NaCl, growth was scarce for all these isolates, except for Chrysosporium sp. 5 that showed good growth.
Table 2.
Key physiological features of fungi included in this study.
| Fungi | BCP-MS-G agar |
Hemolysis (on blood agar) | Lipase | SDA plus 3 % NaCl | SDA plus 5 % NaCl | Cycloheximide tolerance (SDA plus 0.2 %) | Growth on PYE at 15 °C | Growth on PYE at 40 °C | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Milk solids hydrolysis | pH reaction |
|||||||||
| Acidification | Alkalisation | |||||||||
| Nannizziopsis vriesii IMI 149994T | − | + | − | + | + | + | (+) | + | − | − |
| Nannizziopsis vriesii RKI 04-0104 | (+) | + | − | + | + | + | (+) | − | + | + |
| Nannizziopsis guarroi CBS 124553T | − | − | + | + | + | − | − | + | + | − |
| Nannizziopsis guarroi UTHSC R-4309 | − | − | + | + | + | − | − | + | + | − |
| Nannizziopsis guarroi UTHSC 05-1370 | − | − | + | + | + | − | − | + | + | − |
| Nannizziopsis guarroi UTHSC R-4262 | − | − | + | + | + | − | − | + | + | − |
| Nannizziopsis guarroi UTHSC R-4317 | (+) | − | + | + | + | + | − | + | + | + |
| Chrysosporium ophiodiicola CBS 122913T | (+) | + | − | − | + | + | (+) | + | + | − |
| Nannizziopsis chlamydospora UTHSC 04-2056 (Chrysosporium sp. 1) | (+) | − | + | + | + | + | − | + | + | + |
| Nannizziopsis chlamydospora UTHSC 06-1419 (Chrysosporium sp. 1) | (+) | − | + | + | + | + | − | + | + | + |
| Nannizziopsis draconii CCFVB CH12 (Chrysosporium sp. 2) | + | − | + | + | + | − | − | + | + | − |
| Nannizziopsis arthrosporioides UTHSC R-4263 (Chrysosporium sp. 3) | (+) | + | − | + | + | + | (+) | + | + | + |
| Chrysosporium longisporum UTHSC R-4380 (Chrysosporium sp. 4) | − | + | − | + | − | + | (+) | + | + | − |
| Nannizziopsis pluriseptata UTHSC 10-1045 (Chrysosporium sp. 5) | + | − | + | + | + | + | + | + | + | + |
* Reactions on BCP-MS-G agar: − = absence; + = positive; (+) = scarce positive. Growth on SDA plus 5% NaCl: − = absence; + = positive; (+) = scarce positive growth.
Fig. 4.
Nannizziopsis draconii CCFVB CH12 (= Chrysosporium sp. 2). a. Colony on blood agar (surface and reverse); b. colonies on BCP-MS-G (reverse); c. colony on TOTM; d, e. conidiophores bearing sessile and terminal conidia; f. arthroconidia; g, h. sessile and terminal conidia. — Scale bars = 10 μm (d, f, g, differential interference contrast; e, h, phase contrast).
Fig. 8.
Nannizziopsis guarroi CBS 124553. a. Colonies on blood agar; b. colonies on BCP-MS-G; c. colony on TOTM; d, e. fertile hyphae bearing sessile conidia; f, g. sessile conidia. — Scale bars = 10 μm (d, f, differential interference contrast; e, g, phase contrast).
Fig. 9.
Chrysosporium anamorph of Nannizziopsis vriesii IMI 149994. a. Colony on blood agar; b. colony on BCP-MS-G (surface and reverse); c. colony on TOTM; d, e. fertile hyphae bearing mostly sessile conidia (arrow shows an intercalary conidium); f. sessile conidia. — Scale bars = 10 μm (d, f, differential interference contrast; e, phase contrast).
The combination of both phenotypic and molecular results demonstrated that five Chrysosporium species (Chrysosporium sp. 1, Chrysosporium sp. 2, Chrysosporium sp. 3, Chrysosporium sp. 4 and Chrysosporium sp.5) represent new species. The type species of the genus Chrysosporium, C. merdarium, is phylogenetically unrelated to N. vriesii (Vidal et al. 2000), and phenotypically very different; it is not keratinolytic, produces very variable colonies (yellow, pink or green), and sparsely echinulate, subglobose conidia. Therefore, we propose to accomodate the species Chrysosporium sp. 1, Chrysosporium sp. 2, Chrysosporium sp. 3, and Chrysosporium sp. 5, which in the combined tree were placed in the Nannizziopsis clade, in the genus Nannizziopsis, while Chrysosporium sp. 4 is described here as a new species of Chrysosporium.
Nannizziopsis chlamydospora Stchigel, D.A. Sutton, Cano & Guarro, sp. nov. (Chrysosporium sp.1) — MycoBank MB801986; Fig. 3a–i
Etymology. From Greek chlamydos-, cloak, and from Latin -spora, spore.
Colonies on PYE at 30 °C attaining a diameter of 41–48 mm after 14 d, yellowish white (M. 4A2), elevated at the centre and radially folded, compact, with an irregular margin; reverse yellowish white (M. 4A2). Hyphae hyaline, septate, smooth-walled, straight or twisted, 1–3(–4) μm wide. Conidia unicellular, sessile, on short protrusions or on side branches, less frequently terminal, hyaline, thin- and smooth-walled, pyriform, claviform, or cylindrical, 3–9 × 1.5–2 μm; intercalary conidia, cylindrical to doliiform, 6–10 × 1.5–2 μm; arthroconidia catenate, cylindrical to doliiform, 4–10 × 2–4 μm. Chlamydospores globose, broadly ellipsoidal or irregular, smooth- and thick-walled, 5–15(–20) μm diam. Sexual morph not observed. Fetid (skunk-like) odour produced on all the culture media tested.
Minimum and maximum temperature of growth — 5 °C and 40 °C, respectively. Colonies reaching a diameter of 33–39 mm on PDA, 37–41 mm on SDA, 35–37 mm on MEA and 25–32 mm on OA after 14 d at 25 °C.
Specimens examined. USA, ex Pogona vitticeps dermal lesion, holotype CBS H-21115, cultures ex-type CBS 133985, UTHSC 04-2056, FMR 10835; ex Pogona vitticeps dermal lesion, UTHSC 06-1419.
Nannizziopsis draconii J. Cabañes, Abarca, Stchigel, Cano & Guarro, sp. nov. (Chrysosporium sp. 2) — MycoBank MB801987; Fig. 4a–h
Etymology. From Latin draco, dragon, referring to the source (a lizard) from where the fungus was isolated.
Colonies on PYE at 30 °C attaining a diameter of 32–38 mm after 14 d, yellowish white (M. 1A2), felted, slightly elevated at centre, with regular margin; reverse yellowish white (M. 2A2) to pale yellow at centre (M. 4A3). Hyphae hyaline, septate, smooth-walled, 1–3(–5) μm wide. Conidia unicellular, mostly sessile, also produced on short protrusions or on side branches, or terminal, hyaline, thin- and smooth-walled, claviform or cylindrical, 4–7 × 1.5–2(–2.5) μm; intercalary conidia scarce, cylindrical, 4–9 × 1.5–2 μm; arthroconidia catenate, mostly cylindrical or doliiform, scarcely produced, 5–9 × 1.5–2.5 μm. Chlamydospores absent. Sexual morph not observed. Fetid (skunk-like) odour produced on all the culture media tested.
Minimum and maximum temperature of growth — 15 °C and 35 °C, respectively. Colonies reaching a diameter of 32–35 mm on PDA, 34–37 mm on SDA, 35–43 mm on MEA and 32–40 mm on OA after 14 d at 25 °C.
Specimen examined. Spain, ex Pogona vitticeps, holotype CBS H-21116, cultures ex-type CBS 133987, CCFVB CH12, FMR 10859.
Nannizziopsis arthrosporioides Stchigel, D.A. Sutton, Cano & Guarro, sp. nov. (Chrysosporium sp. 3) — MycoBank MB801988; Fig. 5a–f
Etymology. From the Greek arthron-, articulation, and from Latin -spora, spore.
Colonies on PYE at 30 °C attaining a diameter of 34–37 mm after 14 d, yellowish white (M. 1A2), zonate, felted, slightly cottony at centre, with lobate margins; reverse yellowish white (M. 4A2). Hyphae hyaline, septate, smooth-walled, 1–4 μm wide, straight or twisted. Conidia 1(–2)-celled, mostly sessile, also produced on short protrusions or terminal, hyaline, thin- and smooth-walled, subglobose, pyriform, obovate, or claviform to cylindrical, 2.5–7 × 1.5–3 μm; intercalary conidia present, similar to the arthroconidia in shape and size; arthroconidia arranged in short terminal and intercalary chains, doliiform to cylindrical or irregularly-shaped, 5–15 × 1.5–4 μm. Chlamydospores absent. Sexual morph not observed. Fetid (skunk-like) odour present on all the culture media tested.
Minimum and maximum temperature of growth — 15 °C and 30 °C, respectively. Colonies reaching a diam of 34–38 mm on PDA, 28–32 mm on SDA, 42–45 mm on MEA, and 30–33 mm on OA, after 14 d at 25 °C.
Specimen examined. USA, ex Physignathus sp., holotype CBS H-21117, cultures ex-type CBS 133988, UTHSC R-4263, FMR 10842.
Nannizziopsis pluriseptata Stchigel, D.A. Sutton, Cano & Guarro, sp. nov. (Chrysosporium sp. 5) — MycoBank MB801989; Fig. 6a–h
Etymology. From the Latin pluri-, many, and -septum, septum.
Colonies on PYE at 30 °C attaining a diameter of 38–40 mm after 14 d, white to orange white (M. 5A2), zonate, felted, slightly cottony at the centre, with regular margins; reverse orange white (M. 5A2). Hyphae hyaline, septate, smooth-walled, 1–5 μm wide, straight. Conidia 1(–5)-celled, mostly sessile, also produced on short protrusions or on side branches, or terminal, hyaline, thin- and smooth-walled, pyriform, obovate, claviform to cylindrical, 2.5–8(–15) × 1.5–2.5 μm; intercalary conidia occasionally present, cylindrical to doliiform or irregularly shaped, 2.5–5 × 2–2.5 μm; arthroconidia, disposed in lateral or terminal short chains, cylindrical to doliiform, 4–7 × 2.5–3.5 μm, usually bearing sessile conidia. Chlamydospores and sexual morph absent. Fetid (skunk-like) odour present on all culture media tested.
Minimum and maximum temperature of growth — 20 °C and 40 °C, respectively. Colonies reaching a diam of 32–36 mm on PDA, 33–35 mm on SDA, 35–37 mm on MEA and 23–25 mm on OA after 14 d at 25 °C.
Specimen examined. USA, ex skin of a skink (Eumeces inexpectatus Taylor), holotype CBS H-21118, cultures ex-type CBS 133989, UTHSC 10-1045, FMR 12084.
Chrysosporium longisporum Stchigel, D.A. Sutton, Cano & Guarro, sp. nov. (Chrysosporium sp. 4) — MycoBank MB801990; Fig. 7a–e
Etymology. From the Latin longo-, long, and -spora, spore.
Colonies on PYE at 25 °C attaining a diameter of 40–46 mm after 14 d, white to pale orange (M. 6A3), zonate, felted, slightly cottony at centre, with regular margins; reverse pale orange (M. 5A2). Hyphae hyaline, septate, smooth-walled, 1–5 μm wide, straight. Conidia 1(–2)-celled, mostly sessile, or produced on short protrusions or on side branches or terminal, hyaline thin- and smooth-walled, pyriform, obovate, claviform to cylindrical, 3–13 × 2–3.5 μm; intercalary conidia present, cylindrical to doliiform, 3–6 × 2–3 μm, usually bearing sessile conidia; arthroconidia in chains absent. Chlamydospores and sexual morph absent. Fetid (skunk-like) odour present on all the culture media tested.
Minimum and maximum temperature of growth — 5 °C and 25 °C, respectively. Colonies reaching a diam of 40–44 mm on PDA, 40–46 mm on SDA, 45–50 mm on MEA and 25–50 mm on OA after 14 d at 25 °C.
Specimen examined. USA, ex dermic lesion of a tentacled snake (Erpeton tentaculatum Lacépède), holotype CBS H-21139, cultures ex-type CBS 133990, UTHSC R-4380, FMR 10617.
Nannizziopsis guarroi (J.Cabañes & Abarca) J.Cabañes, Abarca, Guarro, Stchigel & Cano, comb. nov. — MycoBank MB801991; Fig. 8a–g
Basionym. Chrysosporium guarroi J.Cabañes & Abarca, Med. Mycol. 48: 370. 2010.
Notes — All the clinical isolates, with the exception of C. longisporum, in the D1-D2 tree (Fig. 1) formed a well-supported clade (100 % bs/1 pp) within the Onygenales, and were phylogenetically separated from the other families of the order. All these species are phenotypically similar and share the ability to cause dermal lesions in reptiles. These characteristics support the proposal of a new family.
Nannizziopsiaceae Guarro, Stchigel, D.A. Sutton & Cano, fam. nov. — MycoBank MB802007
Type genus. Nannizziopsis (Apinis) Currah, Mycotaxon 24: 164. 1985.
Ascomycota, Pezizomycotina, Eurotiomycetes, Eurotiomycetidae, Onygenales. Ascomata (when present) discrete, spherical, whitish, with a peridium composed of a network of loosely interwoven, verrucose, hyaline hyphae which are constricted at the septa. Asci spherical, 8-spored, soon evanescent. Ascospores spherical, hyaline, thick- and smooth-walled under light microscope, spiny to reticulate under scanning electron microscope. Chrysosporium anamorph consisting of sessile conidia, rarely intercalary, solitary, hyaline, smooth- and thin-walled, pyriform, obovate, obovoid, clavate or cylindrical, 1-celled, rarely 2–5-celled, usually with broad basal scars; arthroconidia 1-celled, intercalary or terminally disposed, in chains. Fetid (skunk-like) odour is present in all the members of this family.
DISCUSSION
The order Onygenales comprises six families: Ajellomycetaceae, Arachnomycetaceae, Arthodermataceae, Ascosphaeraceae, Gymnascaceae and Onygenaceae (Lumbsch & Huhndorf 2010). Until now, these families, with the exception of the Onygenaceae, which is clearly polyphyletic (Sugiyama et al. 1999, Herr et al. 2001, Sugiyama & Mikawa 2001, Gibas et al. 2002, Untereiner et al. 2002, 2004), were well delimited. In our D1-D2 phylogenetic tree (Fig. 1), the members of the Arachnomycetaceae were located outside the Onygenales. The data supports the revalidation of the order Arachnomycetales proposed by Gibas et al. (2002). On the other hand, two species of Ascosphaera (Ascosphaerales), i.e., A. apis and A. subglobosa, were included within the Onygenales, but the third member of this family used in our phylogenetic study, Bettsia alvei, was located out the Onygenales, Arachnomycetales and Eurotiales, and in fact acted as outgroup in the tree. This agrees with Wynns et al. (2012), in which that species together with Eremascus fertilis (Eremascaceae, Coryneliales) formed a group separated from the clade made up by the species of Ascosphaera. Our analysis demonstrated that several fungi phylogenetically related and morphologically similar to the Chrysosporium anamorph of Nannizziopsis vriesii (Fig. 9) constitute a new lineage within the Onygenales, clearly differentiated and phylogenetically distant from the members of the other families of the order. This lineage is considered a new family. This family includes the genus Nannizziopsis, with the type species N. vriesii, C. guarroi, which is here included in the genus Nannizziopsis, and four of the five new species described here. Chrysosporium guarroi was recently described by Abarca et al. (2010) based on several isolates that caused different cases of dermatomycosis in pet green iguanas in Spain (Fig. 8). During the course of our study we also identified four isolates of that species from snakes, iguanas and bearded dragons, and one from a human specimen. The human isolate differed from those infecting reptiles in some molecular and physiological features; i.e., 20 bp/1514 bp in the combined dataset analysis and it showed positive growth on SDA plus 3 % NaCl and milk solid hydrolysis. The conidia of C. guarroi are similar to those of Chrysosporium anamorph of N. vriesii, but in the latter they are usually sessile, while those in C. guarroi are mostly borne at the ends of narrow stalks. Other recently described species morphologically similar to Nannizziopsis spp. is C. ophiodiicola (Fig. 10), which was isolated from a mycotic granuloma of a black rat snake. This species is distinguished by its narrow and cylindrical conidia, mostly on long stalks, and because it was neither able to split urea nor produce hemolysis. In our phylogenetic tree, the taxonomic position of this species was unresolved.
Fig. 10.
Chrysosporium ophiodiicola CBS 122913. a, b. Fertile hyphae bearing sessile conidia, intercalary conidia (black arrows) and intercalary chains of arthroconidia (white arrows); c, d. arthroconidia; e. sessile conidia. — Scale bars = 10 μm (a, b, d, differential interference contrast; c, e, phase contrast).
The genus Nannizziopsis was reviewed, although only on the basis of morphological criteria, by Guarro et al. (1991), and they accepted the species N. albicans, N. hispanica and N. vriesii. Nannizziopsis mirabilis (Uchiyama et al. 1995), N. tropicalis (Cano et al. 1997) and N. patagonica (Udagawa & Uchiyama 1999) were later described on the same criteria. With the exception of N. hispanica, all species of Nannizziopsis produce a chrysosporium-like anamorph. Unfortunately, with the exception of N. albicans, living cultures of these species are not available. More recently, in different phylogenetic studies, N. albicans was placed very far from the type strain of N. vriesii, being later accommodated in the genus Amauroascus (Vidal et al. 2000, Solé et al. 2002). These data are congruent with the ornamentation of the ascospores, which is considered a useful criterion in the taxonomy of the Onygenales. Although, the sexual morph of N. vriesii is rarely produced in culture, its ascospores have been described as echinulate (Apinis 1970, Guarro et al. 1991), while those of Amauroascus spp. are clearly reticulate, as are those of N. albicans. As we mentioned above, we could not include in our molecular analysis, apart from N. vriesii, the type strains of the other previously described species of Nannizziopsis; however, on the basis of the characteristics reported in their descriptions, we can infer that probably they would be better accommodated in Amauroascus.
The new species described here were phenotypically very similar. Nannizziopsis chlamydospora can be distinguished from the other species of the genus because it produces chlamydospores and grows at 5 °C. Nannizziopsis draconii can be differentiated from the other species by the combination of several features, i.e. the ability to grow on BCP-MS-G agar alkalinizing the medium, tolerance to 0.2 % cycloheximide, and the inability to grow on SDA with 3 % NaCl. Nannizziopsis arthrosporioides produces abundant long arthroconidia. Nannizziopsis pluriseptata produces from 1- to 5-celled sessile conidia, alkalinizes the BCP-MS-G agar and grows on SDA supplemented with 5 % NaCl. These pluriseptate conidia have some resemblance to those of the dermatophytes Trichophyton erinacei, Trichophyton thuringiense and Trichophyton terrestre (family Arthrodermataceae). However, the macroconidia of T. erinacei (20–50 × 5–7 μm), T. thuringiense (8–30 × 3–5 μm) and of T. terrestre (9–50 × 4–5 μm) are larger than those of N. pluriseptata (5–15 × 1.5–2.5 μm). Furthermore, N. pluriseptata presents terminal and lateral chains of arthroconidia, which are absent in Trichophyton spp. Chrysosporium longisporum is morphologically similar to the species of Nannizziopsis but it is characterized by producing long sessile conidia (up to 13 μm), and because is the only species unable to produce lipases.
The clinical isolate RKI 04-0104 and the type strain of N. vriesii only produced the Chrysosporium anamorph in culture, which is easily recognized by the production of very narrow sessile conidia (2–3 μm). Its teleomorph was only obtained in the original description of the species (Currah 1985, Guarro et al. 1991). In our study, all the attempts to induce the formation of ascomata on numerous media containing different sterile vegetable materials and horse hairs failed. The production of asperulate hyphae in culture, similar to those that constitute the ascomatal peridium, and considered typical of this species (Thomas et al. 2002), was also negative in our study. Although in the combined dataset tree, the clinical isolate of N. vriesii was separated from the type strain (IMI 149994) of this species, it was considered as belonging to that species. Both strains were morphologically very similar and the ACT and TUB sequences of the two strains were practically identical. Their separation in the phylogenetic tree was due to the presence of some differences (mainly insertions) in the ITS region.
Nannizziopsis is considered a primary pathogen causing dermal infections in different classes of reptiles, such as chameleons (Paré et al. 1997, 2006), crocodiles (Thomas et al. 2002), lizards (Martel et al. 2006, Mitchell et al. 2006, Bowman et al. 2007, Abarca et al. 2008, 2009, 2010, Han et al. 2010, Hedley et al. 2010, Hellebuyck et al. 2010, van Waeyenberghe et al. 2010, Johnson et al. 2011), and snakes (Nichols et al. 1999, Bertelsen et al. 2005, Rajeev et al. 2009, Eatwell 2010, Allender et al. 2011). The infections have consisted of single cases in pets or captive individuals but also in free-living animals, although different outbreaks in different species of reptiles have also been identified. The infection generally starts on the skin and progress rapidly involving subcutaneous soft tissues causing cutaneous ulcers and granulomas with infection of deeper tissues. Finally, the fungus can disseminate producing a fatal outcome. Cases involving these fungi have been reported in Australia, Belgium, Canada, Spain, UK and USA. Occasionally, Nannizziopsis spp. can infect humans causing severe lesions, as the case of lung infiltration and brain abscess described in a Nigerian man by the strain RKI 04-0104 included in this study (Steininger et al. 2005).
Various treatment options for these fungi include the use of itraconazole, ketoconazole or terbinafine combined with surgical debridement or amputation. The most promising treatment, however, appears to be voriconazole, which has demonstrated efficacy both in humans (Steininger et al. 2005) and in reptiles (Hellebuyck et al. 2010, van Waeyenberghe et al. 2010).
Most of the infections caused by these fungi have been described in the last 10 yr, and is unclear if this could be attributed to recent climatic changes that could have affected the environment where these animals live or that previous infections had been overlooked or misidentified. It has been suggested that the different species of Nannizziopsis are associated with specific hosts (Bertelsen et al. 2005, Bowman et al. 2007). However, our study seems to not confirm this hypothesis, because, for example, N. guarroi infected lizards as well as snakes. Further studies utilising more clinical isolates are required to more fully assess the host boundaries for these species.
Acknowledgments
This project was supported by the Spanish Ministerio de Ciencia e Innovación, grant CGL 2009-08698/BOS.
REFERENCES
- Abarca ML, Castellá G, Martorell J, Cabañes FJ. 2010. Chrysosporium guarroi sp. nov. a new emerging pathogen of pet green iguanas (Iguana iguana). Medical Mycology 48: 365–372 [DOI] [PubMed] [Google Scholar]
- Abarca ML, Martorell J, Castellá G, Ramis A, Cabañes FJ. 2008. Cutaneous hyalohyphomycosis caused by a Chrysosporium species related to Nannizziopsis vriesii in two green iguanas (Iguana iguana). Medical Mycology 46: 349–354 [DOI] [PubMed] [Google Scholar]
- Abarca ML, Martorell J, Castellá G, Ramis A, Cabañes FJ. 2009. Dermatomycosis in a pet inland bearded dragon (Pogona vitticeps) caused by a Chrysosporium species related to Nannizziopsis vriesii. Veterinary Dermatology 20: 295–299 [DOI] [PubMed] [Google Scholar]
- Abdel-Razik M, Zaki SM. 2008. Experimental pathogenicity and molecular characterization of an environmental isolate of Chrysosporium zonatum Al-Musallam and Tan (Family: Onygenaceae, Order: Onygenales). International Journal of Agriculture and Biology 10: 273–277 [Google Scholar]
- Allender MC, Dreslik M, Wylie S, Phillips C, Wylie DB, et al. 2011. Chrysosporium sp. infection in eastern massasauga rattlesnakes. Emerging Infectious Diseases 17: 2383–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apinis AE. 1970. Concerning Rollandina. Transactions of the British Mycological Society 50: 500–502 [Google Scholar]
- Bertelsen MF, Crawshaw GJ, Sigler L, Smith DA. 2005. Fatal cutaneous mycosis in tentacled snakes (Erpeton tentaculatum) caused by the Chrysosporium anamorph of Nannizziopsis vriesii. Journal of Zoo and Wildlife Medicine 36: 82–87 [DOI] [PubMed] [Google Scholar]
- Bowman MR, Paré JA, Sigler L, Naeser JP, Sladky KK, et al. 2007. Deep fungal dermatitis in three inland bearded dragons (Pogona vitticeps) caused by the Chrysosporium anamorph of Nannizziopsis vriesii. Medical Mycology 45: 371–376 [DOI] [PubMed] [Google Scholar]
- Brandt ME, Gaunt D, Iqbal N, McClinton S, Hambleton S, Sigler L. 2005. False-positive Histoplasma capsulatum Gen-Probe chemiluminescent test result caused by a Chrysosporium species. Journal of Clinical Microbiology 43: 1456–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano J, Guarro J, Gené J. 2004. Molecular and morphological identification of Colletotrichum species of clinical interest. Journal of Clinical Microbiology 42: 2450–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano J, Ulfig K, Guillamon JM, Vidal P, Guarro J. 1997. Studies on keratinophilic fungi. IX. Neoarachnotheca gen. nov. and a new species of Nannizziopsis. Antonie van Leeuwenhoek 72: 149–158 [DOI] [PubMed] [Google Scholar]
- Currah RS. 1985. Taxonomy of the Onygenales: Arthrodermataceae, Gymnascaceae, Myxotrichaceae and Onygenaceae. Mycotaxon 24: 1–216 [Google Scholar]
- Eatwell K. 2010. Suspected fatal Chrysosporium anamorph of Nannizziopsis vriesii (CANV) dermatitis in an albino Boa constrictor (Constrictor constrictor). Journal of Small Animal Practice 51: 290. [DOI] [PubMed] [Google Scholar]
- Gibas CFC, Sigler L, Summerbell RC, Currah RS. 2002. Phylogeny of the genus Arachnomyces and the establishment of Arachnomycetales, a new eurotiomycete order in the Ascomycota. Studies in Mycology 47: 131–139 [Google Scholar]
- Gilgado F, Cano J, Gené J, Guarro J. 2005. Molecular phylogeny of the Pseudallescheria boydii species complex: Proposal of two new species. Journal of Clinical Microbiology 43: 4930–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarro J, Cano J, Vroey C de. 1991. Nannizziopsis (Ascomycotina) and related genera. Mycotaxon 42: 193–200 [Google Scholar]
- Han JI, Lee SJ, Na KJ. 2010. Necrotizing dermatomycosis caused by Chrysosporium spp. in three captive green iguanas (Iguana iguana) in South Korea. Journal of Exotic Pet Medicine 19: 240–244 [Google Scholar]
- Hedley J, Eatwell K, Hume L. 2010. Necrotising fungal dermatitis in a group of bearded dragons (Pogona vitticeps). Veterinary Record 166: 464–465 [DOI] [PubMed] [Google Scholar]
- Hellebuyck T, Baert K, Pasmans F, Waeyenberghe L van, Beernaert L, et al. 2010. Cutaneous hyalohyphomycosis in a girdled lizard (Cordylus giganteus) caused by the Chrysosporium anamorph of Nannizziopsis vriesii and successful treatment with voriconazole. Veterinary Dermatology 21: 429–433 [DOI] [PubMed] [Google Scholar]
- Herr RA, Taracha EJ, Taborda PR, Taylor JW, Ajello L, Mendoza L. 2001. Phylogenetic analysis of Lacazia loboi places this previously uncharacterized pathogen within the dimorphic Onygenales. Journal of Clinical Microbiology 39: 309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoog GS de, Guarro J, Figueras MJ, Gené J. 2000. Atlas of clinical fungi, 2nd ed Baarn / Reus, Centraalbureau voor Schimmelcultures / Universitat Rovira I Virgili, The Netherlands, Spain [Google Scholar]
- Hubalek Z. 2000. Keratinophilic fungi associated with free-living mammals and birds. In: Kushwaha RKS, Guarro J. (eds), Biology of dermatophytes and other keratinophilic fungi: 93–103 Revista Iberoamericana de Micología, Bilbao, Spain [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Johnson RSP, Sangster CR, Sigler L, Hambleton S, Paré JA. 2011. Deep fungal dermatitis caused by the Chrysosporium anamorph of Nannizziopsis vriesii in captive coastal bearded dragons (Pogona barbata). Australian Veterinary Journal 89: 515–519 [DOI] [PubMed] [Google Scholar]
- Kane J, Summerbell R, Sigler L, Krajden S, Land G. (eds). 1997. Laboratory handbook of dermatophytes: a clinical guide and laboratory manual of dermatophytes and other filamentous fungi from skin, hair, and nails. Star Publishing Press, USA [Google Scholar]
- Kornerup A, Wanscher JH. 1978. Methuen handbook of colour. Methuen & Co Ltd, UK [Google Scholar]
- Lumbsch HT, Huhndorf SB. 2010. Myconet volume 14. Part one. Outline of Ascomycota – 2009. Fieldiana Life and Earth Sciences 1: 1–247 [Google Scholar]
- Mandeel Q, Nardoni S, Mancianti F. 2009. Keratinophilic fungi on feathers of common clinically healthy birds in Bahrain. Mycoses 54: 71–77 [DOI] [PubMed] [Google Scholar]
- Martel A, Fonteyne PA, Chiers K, Decostere A, Pasmans F. 2006. Nasal Nannizziopsis vriesii granuloma in an ameiva lizard (Ameiva chaitzami). Flemish Veterinary Journal 75: 306–307 [Google Scholar]
- Mitchell MA, Diaz-Figueroa O, Bernstein J, Walden M, Wickes B, et al. 2006. Chrysosporium anamorph of Nannizziopsis vriesii dermatitis in a bearded dragon (Pogona vitticeps). Proceedings of the Association of Reptile and Amphibian Veterinarian, Baltimore, Maryland [Google Scholar]
- Mouchacca J, Gams W. 1993. The hyphomycete genus Cladorrhinum and its teleomorph connections. Mycotaxon 48: 415–440 [Google Scholar]
- Nichols DK, Weyant RS, Lamirande EW, Sigler L, Mason RT. 1999. Fatal mycotic dermatitis in captive brown tree snakes (Boiga irregularis). Journal of Zoo and Wildlife Medicine 30: 111–118 [PubMed] [Google Scholar]
- Oorschot CAN van. 1980. A revision of Chrysosporium and allied genera. Studies in Mycology 20: 1–89 [Google Scholar]
- Paré JA, Coyle KA, Sigler L, Maas AK, III, Mitchell RL. 2006. Pathogenicity of the Chrysosporium anamorph of Nannizziopsis vriesii for veiled chameleons (Chamaeleo calyptratus). Medical Mycology 44: 25–31 [DOI] [PubMed] [Google Scholar]
- Paré JA, Jacobson ER. 2007. Mycotic diseases of reptiles. In: Jacobson ER. (ed), Infectious Diseases and Pathology of Reptiles: 527–570 CRC Press, USA [Google Scholar]
- Paré JA, Sigler L, Hunter DB, Summerbell RC, Smith DA, Machin KL. 1997. Cutaneous mycoses in chameleons caused by the Chrysosporium anamorph of Nannizziopsis vriesii (Apinis) Currah. Journal of Zoo and Wildlife Medicine 28: 443–453 [PubMed] [Google Scholar]
- Perdomo H, Sutton DA, García D, Fothergill AW, Cano J, et al. 2011. Spectrum of clinically relevant Acremonium species in the United States. Journal of Clinical Microbiology 49: 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson OV, Leong SL, Lantz H, Rice T, Dijksterhuis J, et al. 2011. Phylogeny and intraspecific variation of the extreme xerophile, Xeromyces bisporus. Fungal Biology 115: 1100–1111 [DOI] [PubMed] [Google Scholar]
- Rajeev S, Sutton DA, Wickes BL, Miller DL, Giri D, et al. 2009. Isolation and characterization of a new fungal species, Chrysosporium ophiodiicola, from a mycotic granuloma of a black rat snake (Elaphe obsoleta obsoleta). Journal of Clinical Microbiology 47: 1264–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees RG. 1967a. Keratinophilic fungi from Queensland 1. Isolations from animal hair and scales. Sabouraudia 5: 165–172 [DOI] [PubMed] [Google Scholar]
- Rees RG. 1967b. Keratinophilic fungi from Queensland 2. Isolations from feathers of wild birds. Sabouraudia 6: 14–18 [DOI] [PubMed] [Google Scholar]
- Rees RG. 1967c. Keratinophilic fungi from Queensland 3. Isolations from feathers of domestic fowls. Sabouraudia 6: 19–28 [DOI] [PubMed] [Google Scholar]
- Roilides E, Sigler L, Bibashi E, Katsifa H, Flaris N, Panteliadis C. 1999. Disseminated infection due to Chrysosporium zonatum in a patient with chronic ranulomatous disease and review of non-aspergillus fungal infections in patients with this disease. Journal of Clinical Microbiology 37: 18–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Sigler L. 1997. Chrysosporium and molds resembling dermatophytes. In: Kane J, Summerbell R, Sigler L, Krajden S, Land G. (eds), Laboratory handbook of dermatophytes: a clinical guide and laboratory manual of dermatophytes and other filamentous fungi from skin, hair, and nails: 261–311 Star Publishing Press, USA [Google Scholar]
- Sigler L, Flis AL, Carmichael JW. 1998. The genus Uncinocarpus (Onygenaceae) and its synonym Brunneospora: new concepts, combinations and connections to anamorphs in Chrysosporium, and further evidence of its relationship with Coccidioides immitis. Canadian Journal of Botany 76: 1624–1636 [Google Scholar]
- Slifkin M. 2000. Tween 80 opacity test responses of various Candida species. Journal of Clinical Microbiology 38: 4626–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solé M, Cano J, Guarro J. 2002. Molecular phylogeny of Amauroascus, Auxarthron, and morphologically similar onygenalean fungi. Mycological Research 106: 388–396 [Google Scholar]
- Stebbins WG, Krishtul A, Bottone EJ, Phelps R, Cohen S. 2004. Cutaneous adiaspiromycosis: a distinct dermatologic entity associated with Chrysosporium species. Journal of the American Academy of Dermatology 51: S185–S189 [DOI] [PubMed] [Google Scholar]
- Steininger C, Lunzen J van, Tintelnot K, Sobottka I, Rohde H, Horstkotte MA, Stellbrink HJ. 2005. Mycotic brain abscess caused by opportunistic reptile pathogen. Emerging Infectious Diseases 11: 349–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M, Mikawa T. 2001. Phylogenetic analysis of the non-pathogenic genus Spiromastix (Onygenaceae) and related onygenalean taxa based on large subunit ribosomal DNA sequences. Mycoscience 42: 413–421 [Google Scholar]
- Sugiyama M, Ohara A, Mikawa T. 1999. Molecular phylogeny of onygenalean fungi based on small subunit ribosomal DNA (SSU rDNA) sequences. Mycoscience 40: 251–258 [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AD, Sigler L, Peucker S, Norton JH, Nielan A. 2002. Chrysosporium anamorph of Nannizziopsis vriesii associated with fatal cutaneous mycoses in the salt-water crocodile (Crocodylus porosus). Medical Mycology 40: 143–151 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL X windows interface. Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama S, Kamiya S, Udagawa SI. 1995. A new species of Nannizziopsis from New Jersey soil. Mycoscience 36: 205–209 [Google Scholar]
- Udagawa S, Uchiyama S. 1999. Taxonomic studies on new or critical fungi of non-pathogenic Onygenales 2. Mycoscience 40: 291–305 [Google Scholar]
- Ueda S. 1994. A new Cercophora with a Chrysosporium-like anamorph. Mycoscience 35: 287–290 [Google Scholar]
- Untereiner WA, Scott JA, Naveau FA, Currah RS, Bachewich J. 2002. Phylogeny of Ajellomyces, Polytolypa and Spiromastix (Onygenaceae) inferred from rDNA sequence and non-molecular data. Studies in Mycology 47: 25–35 [Google Scholar]
- Untereiner WA, Scott JA, Naveau FA, Sigler L, Bachewich J, Angus A. 2004. The Ajellomycetaceae, a new family of vertebrate-associated Onygenales. Mycologia 96: 812–821 [DOI] [PubMed] [Google Scholar]
- Vidal P, Vinuesa MA, Sanchez-Puelles JA, Guarro J. 2000. Phylogeny of the anamorphic genus Chrysosporium and related taxa based on rDNA internal transcribed spacer sequences. In: Kushwaha RKS, Guarro J. (eds), Biology of dermatophytes and other keratinophilic fungi: 22–28 Revista Iberoamericana de Micología, Bilbao, Spain [Google Scholar]
- Voigt K, Wöstemeyer J. 2000. Reliable amplification of actin genes facilitates deep-level phylogeny. Microbiological Research 155: 179–195 [DOI] [PubMed] [Google Scholar]
- Waeyenberghe L van, Baert K, Pasmans F, Rooij P van, Hellebuyck T, et al. 2010. Voriconazole, a safe alternative for treating infections caused by the Chrysosporium anamorph of Nannizziopsis vriesii in bearded dragons (Pogona vitticeps). Medical Mycology 48: 880–885 [DOI] [PubMed] [Google Scholar]
- Wynns AA, Jensen AB, Eilenberg J, Rosalind J. 2012. Ascosphaera subglobosa, a new spore cyst fungus from North America associated with the solitary bee Megachile rotunda. Mycologia 104: 108–114 [DOI] [PubMed] [Google Scholar]