Abstract
MYC dimerizes with MAX to bind DNA, with a preference for the E-box consensus CACGTG and several variant motifs. In cells, MYC binds DNA preferentially within transcriptionally active promoter regions. Although several thousand promoters are bound under physiological (low MYC) conditions, these represent only a fraction of all accessible, active promoters. MYC overexpression—as commonly observed in cancer cells—leads to invasion of virtually all active promoters, as well as of distal enhancer elements. We summarize here what is currently known about the mechanisms that may guide this process. We propose that binding site recognition is determined by low-affinity protein–protein interactions between MYC/MAX dimers and components of the basal transcriptional machinery, other chromatin-associated protein complexes, and/or DNA-bound transcription factors. DNA binding occurs subsequently, without an obligate requirement for sequence recognition. Local DNA scanning then leads to preferential stabilization of the MYC/MAX dimer on high-affinity DNA elements. This model is consistent with the invasion of all active promoters that occurs at elevated MYC levels, but posits that important differences in affinity persist between physiological target sites and the newly invaded elements, which may not all be bound in a productive regulatory mode. The implications of this model for transcriptional control by MYC in normal and cancer cells are discussed in the light of the latest literature.
Under normal conditions, MYC binds to only a fraction of the accessible, active promoters. However, in cancer cells, MYC is overexpressed and may bind to virtually all active promoters.
Specific binding of transcription factors (TFs) to their functional sites in the genome is a fundamental step in transcriptional regulation. The ENCODE project (ENCODE Project Consortium 2012) has recently revealed the amplitude and complexity of the regulatory lexicon that tells TFs where to bind in different cellular contexts. Mutations in regulatory regions have emerged as a key process in evolution and disease, as relevant—if not more so—as mutations in coding regions (ENCODE Project Consortium 2012; Schaub et al. 2012). Coordinated efforts at both the computational and experimental levels in the last decade have tried to model and rationalize the way in which low-abundance proteins such as TFs selectively recognize a specific set of binding sites in the genome and how this could be subverted during disease (reviewed in Segal and Widom 2009; Guertin and Lis 2012). We will focus here on what is known about the interactions with the genome of a particular factor, MYC, encoded by the c-MYC (herein MYC) proto-oncogene. However, the basic principles we will describe are shared by other TFs.
Direct oncogenic activation of MYC occurs through structural alterations that cause its deregulated expression, most dramatically through gene translocation in Burkitt’s B-cell lymphomas (Küppers and Dalla-Favera 2001), as well as amplifications in a range of different tumor types (see Roussel and Robinson 2013; Schmitz et al. 2014). Most importantly, MYC is frequently overexpressed in cancer even if not structurally altered, being induced or stabilized by growth-regulatory pathways that are themselves targets of activating mutations (e.g., Ras, Wnt, Notch signaling). In this setting, deregulated MYC expression directly contributes to the growth-promoting and oncogenic potential of the mutant pathway (Efstratiadis et al. 2007; Sansom et al. 2007; Sharma et al. 2007; Conacci-Sorrell et al. 2014). Thus, even when not mutated itself, MYC is believed to be a general driver of tumor progression and maintenance. This has led to the concept that MYC, and/or the genes that it controls, might represent important therapeutic targets. Indeed, in MYC-driven mouse tumor models, MYC inactivation can elicit tumor regression (Felsher and Bishop 1999; Jain et al. 2002; Shachaf et al. 2004; Soucek et al. 2008; Felsher 2010). In addition, targeting endogenous MYC caused regression of tumors driven by a ras oncogene (Wilkins and Sansom 2008; Soucek et al. 2013; Gabay et al. 2014).
MYC is a TF of the basic helix-loop-helix-leucine zipper (bHLH-LZ) family. These proteins form specific homo- or heterodimers via the HLH-LZ domains, as a prerequisite for DNA binding to the general “E-box” consensus CANNTG, mediated by the basic regions (Blackwell and Weintraub 1990). MYC has only one known dimerization partner, MAX (Blackwood and Eisenman 1991), and binds the E-box CACGTG, or variants thereof (Blackwell et al. 1990, 1993; Solomon et al. 1993). Although MAX can also homodimerize or interact with MXD or MNT proteins forming repressor complexes (Ayer et al. 1993; Zervos et al. 1993; Hurlin et al. 1995, 1997), MYC cannot homodimerize or bind other partners, at least under physiological conditions. As a consequence, its interaction with MAX is crucial for MYC-dependent gene regulation, cell-cycle progression, apoptosis, and transformation (Amati et al. 1992, 1993a,b; Kretzner et al. 1992; Mukherjee et al. 1992). Interestingly, heterodimerization with MAX is required not only when MYC binds E boxes, to which MYC/MAX heterodimers bind directly, but also for binding to “nonconsensus” sites (Mao et al. 2003). MYC/MAX dimers are presumably recruited to these alternative sites through protein–protein interaction with other DNA-binding factors such as Miz-1 (Seoane et al. 2001; Staller et al. 2001), Sp1 (Gartel et al. 2001), NF-Y (Izumi et al. 2001), Smad2/3 (Feng et al. 2002), and YY1 (Shrivastava et al. 1993). One potential explanation, at least for Miz-1 binding, is that dimerization with MAX is required for the correct folding of MYC (Adhikary and Eilers 2005; Wiese et al. 2013; Conacci-Sorrell et al. 2014).
THE GENOMIC ERA: INTERACTION OF MYC WITH PROMOTERS AND ENHANCERS
ChIP-seq analyses for MYC in different human cell types (including normal and cancer cells) as part of the ENCODE project (Lee et al. 2012) revealed a considerable variation in the proportion of binding events that occur in promoters, ranging from 45% to 75% depending on the cell type. These observations confirm and extend previous analyses that reported the existence of MYC-bound regions also outside of annotated promoters or CAGE tags (cap analysis of gene expression), marking transcriptional initiation sites (Seitz et al. 2011; Perna et al. 2012). Recent work based on chromatin profiling has revealed that most of these distal MYC-binding sites can be classified as active enhancers, based on the presence of the histone marks H3K4me1 and H3K27ac (Lin et al. 2012; Nie et al. 2012; A Sabò, T Kress, M Pelizzola, et al., submitted).
Based on a B-cell line in which MYC overexpression is driven by a Tet-repressible promoter (P493-6), Lin et al. (2012) found that when expressed at low levels MYC binds almost exclusively promoter regions (of expressed genes), whereas when overexpressed it basically invades all “open” promoters and enhancers. Of notice, we obtained similar results in a transgenic mouse model of MYC-driven lymphomagenesis, showing that during tumor progression MYC gradually invades those promoters and enhancers that are already active in naïve B cells, which never experienced MYC overexpression (A Sabò, T Kress, M Pelizzola, et al., submitted). DNA binding in conditions of “high MYC,” even if widespread, does not become sequence independent, but is less selective, including variant sites that have lower affinity for MYC/MAX in vitro (Lin et al. 2012; A Sabò, T Kress, M Pelizzola, et al., submitted). MAX is cobound to the same regions, indicating that DNA binding still occurs through the MYC/MAX heterodimer.
These findings confirm and expand earlier studies. Based on quantitative ChIP-PCR analysis of selected DNA regions, Fernandez et al. (2003) showed that when expressed at relatively low levels, MYC preferentially interacted with promoters containing E boxes, relative to either promoters without E boxes or E boxes outside a promoter context. When overexpressed, MYC not only associated at increased frequency with low-affinity E-box targets but also, at extreme levels, with other sequences, suggesting that some binding could be less sequence specific (Fernandez et al. 2003; Orian 2003; Guccione et al. 2006; Zeller et al. 2006). Most importantly, however, “high-affinity” and “low-affinity” binding sites could still be distinguished by differential MYC levels, indicating that relative binding affinities were intrinsic properties of those sites, and were not altered on overexpression of MYC.
An important feature enriched among MYC-binding sites is the CpG island (Fernandez et al. 2003; Zeller et al. 2006), which is known to be associated with active chromatin (Deaton and Bird 2011). Indeed, MYC-targeted promoters are invariably associated with an active chromatin profile (H3K4me1, me2, me3, H3K27ac, and others) and in no case does MYC appear to bind compacted, heterochromatic domains even in the presence of a canonical E-box motif (Guccione et al. 2006; Kim et al. 2008; Lin et al. 2012; A Sabò, T Kress, M Pelizzola, et al., submitted). In this regard, important observations were made during induced pluripotent stem cell (iPSC) reprogramming (Soufi et al. 2012). Here MYC cooperated with Oct4, Sox2, and Klf4 (OSK) but required prior OSK activity to access closed chromatin. In other words, MYC was not acting as a pioneer transcription factor, but required prior opening of chromatin domains by OSK (although, once bound, MYC also enhanced the interaction of OSK with chromatin). In summary, sequence recognition by MYC comes second to recognition of (and/or access to) an open chromatin context, and E boxes outside of such context are not significantly bound.
While in prokaryotes DNA sequence elements are the only determinants of transcription factor binding, in eukaryotes these elements can be masked by nucleosomes, and need to be positioned in accessible regions to be recognized by the corresponding TF. Indeed, computational modeling revealed that the presence of binding motifs together with chromatin accessibility (defined as DNAseI hypersensitive sites) is enough to predict the vast majority of transcription factor binding sites that are mapped experimentally through ChIP-seq experiments (Kaplan et al. 2011; Pique-Regi et al. 2011; Arvey et al. 2012). Other mechanisms that may restrict access of TFs to selected chromatin domains include higher-order chromatin organization and nuclear compartmentalization, as well as interactions with proteins that recognize specific histone marks, the so-called “readers” (Kouzarides 2007). These mechanisms were nicely illustrated by work comparing the binding profiles for HSF (heat-shock factor) in the genome of live cells and in naked genomic DNA incubated with recombinant HSF in vitro (Guertin et al. 2012). Although a subset of sites was bound only in vitro (which could be explained by their inaccessibility in vivo), others were bound only in vivo, presumably owing to structural properties of the chromatin in the nuclear environment and/or protein–protein interactions that are missing in the in vitro experiment.
The clustering of active regulatory elements (promoters and enhancers) in domains of high RNA polymerase II (RNA Pol II) concentration inside the nucleus, called transcription factories (reviewed in Edelman and Fraser 2012), could favor the spreading of overexpressed MYC to sites that were not previously occupied. Indeed, TSS-associated MYC-binding sites are premarked not only by open chromatin marks such as H3K4me3, but also by the presence of RNA Pol II. In cells with high MYC expression, MYC basically covers all the RNA Pol II-bound promoters similarly to a general transcription factor (Li 2003; Guccione et al. 2006; Martinato et al. 2008; Lee et al. 2012; Lin et al. 2012; Nie et al. 2012; A Sabò, T Kress, M Pelizzola, et al., submitted). Of notice, MYC has been reported to interact with RNA Pol II itself (Koch et al. 2007) as well as with general transcriptional regulators such as TFII-I (Roy et al. 1993), TBP (Hateboer et al. 1993), and P-TEFb (Eberhardy and Farnham 2001; Eberhardy 2002; Rahl et al. 2010; Rahl and Young 2014).
MYC interacts with many other transcriptional cofactors that lack sequence specificity (see Hann 2014). One of these is TRRAP (McMahon et al. 1998), a subunit of the histone acetyltransferase (HAT) complexes TFTC/STAGA and NuA4, which contain the HAT subunits GCN5/PCAF and Tip60, respectively (Allard et al. 1999; Doyon et al. 2004; Nagy and Tora 2007). Through the STAGA complex, MYC also interacts with Mediator (Liu et al. 2008). Moreover, MYC associates with the HATs CBP and p300 (Vervoorts et al. 2003), the H3K4me3 demethylases JARID1A/KDM5A and JARID1B/KDM5B (Secombe et al. 2007), the ASH2-MLL methyl-transferase complex (Luscher-Firzlaff et al. 2008), the ATPase ATAD2/ANCCA (Ciro et al. 2009), and different components of the SWI/SNF chromatin-remodeling complex (Cheng et al. 1999; Park et al. 2002; Pal et al. 2003). Furthermore, MYC interacts with corepressor complexes containing the histone deacetylases HDAC1 (Satou 2001; Jiang et al. 2007; Matsuoka et al. 2008) or HDAC3 (Kurland and Tansey 2008), or the DNA methyltransferase DNMT3a (Brenner et al. 2005). In addition to the commonly accepted concept that MYC, or other TFs, can recruit these cofactors to chromatin (Frank et al. 2001, 2003; Bouchard et al. 2004), a particularly interesting scenario is that the interactions with cofactors and with the basal transcription machinery may constitute the rate-limiting steps by which MYC recognizes the restricted set of genomic regions (active or poised promoters) in which it will ultimately bind DNA. Both scenarios may also coexist, leading to a mutual stabilization of the various interacting partners (TFs and cofactors) on chromatin. Whether the interaction of MYC with such a plethora of positive and negative coregulators underlies recognition and regulation of selected gene sets largely remains to be elucidated.
Another plausible mechanism of interaction with the genome is the indirect tethering of MYC to DNA elements distinct from E boxes, driven by the interactions with other DNA-binding proteins (in particular, TFs). Analysis of sequence motifs underlying ChIP-seq signals in ENCODE data sets confirmed that MYC/MAX-binding sites are enriched for the consensus E-box motif, but revealed that a surprisingly vast portion of them (especially when outside promoters) lack an E box, compatible with indirect or assisted DNA binding (Neph et al. 2012). Searching for alternative enriched motifs and comparing with ChIP-seq data for other TFs, the most probable candidates for tethering factors appear to be EGR1, CTCF, CTCFL, GABPA, and AP1 (Neph et al. 2012; Wang et al. 2012), although their functional significance in MYC/MAX recruitment has not been investigated as yet.
Protein–protein interactions between MYC and other sequence-specific TFs were reported in earlier work, and were evoked to explain biological findings that could not be ascribed to the direct binding of MYC to E boxes. For example, although MYC-induced apoptosis depends on dimerization with MAX (Amati et al. 1993b), it appears to require an additional function of the MYC dimerization (HLH-LZ) domain. Indeed, although a chimeric MYC protein with the basic domain of Mad1 was functionally equivalent to wild-type MYC (Nikiforov 2003), a chimera with the whole bHLH-LZ region of Mad1 retained the ability to dimerize with MAX and bound DNA with unaltered sequence specificity, but was unable to induce apoptosis (James and Eisenman 2002). Besides dimerization with MAX, the MYC HLH-LZ region mediates binding to Miz-1 (Herold et al. 2002) or YY1 (Shrivastava et al. 1993). In various studies, interfering with the Miz-1/MYC interaction led to an impairment of some of the biological activities of MYC, including apoptosis, down-regulation of the CDK inhibitors p15INK4b and p21Cip1, cell-cycle progression, or stimulation of self-renewal (Seoane et al. 2001; Staller et al. 2001; Herold et al. 2002; Patel 2005; Kerosuo et al. 2008; van Riggelen et al. 2010; Wiese et al. 2013).
Although the interaction with Miz-1 occurs in the context of MYC/MAX dimers (Staller et al. 2001), and possibly of their direct contact with DNA, other TFs can interact with monomeric MYC, as shown for the retinoic acid (RA) receptor RARα during RA-induced differentiation of leukemic cells (Uribesalgo et al. 2011). These and other interactions point to specialized functions of MYC within specific signaling pathways, which may coexist with its more general role in transcriptional control. These aspects are beyond the scope of the present review, and will not be discussed in further detail here.
Finally, other activities of MYC on chromatin may not relate to transcription of RNA Pol II-dependent promoters and may be largely MAX independent. Several genes that are transcribed by RNA polymerase III (and that lack discernible E boxes in their regulatory regions) can be induced by MYC also in the absence of MAX or when MYC is truncated and unable to bind MAX (Steiger et al. 2008) probably owing to interaction with the general transcription factor TFIIIB (Gomez-Roman et al. 2003; Gallant 2013). MYC has also been found in prereplicative complexes (where MAX was present only in small, substoichiometric amounts) suggesting that MYC activity in DNA replication could be at least in part MAX (and E-box) independent (Dominguez-Sola et al. 2007; Gallant and Steiger 2009).
In summary, protein–protein interactions with a variety of TFs and cofactors or even proteins of the general transcription or replication machineries could mediate indirect binding of MYC to DNA. Notwithstanding the indications that MYC may be tethered to DNA in the absence of MAX (see above), its main occurrence in cells is as MYC/MAX dimers. Those dimers form in solution, and are most likely directed as such to the correct genomic locations.
TOWARD AN INTEGRATED VIEW OF MYC-GENOME INTERACTIONS
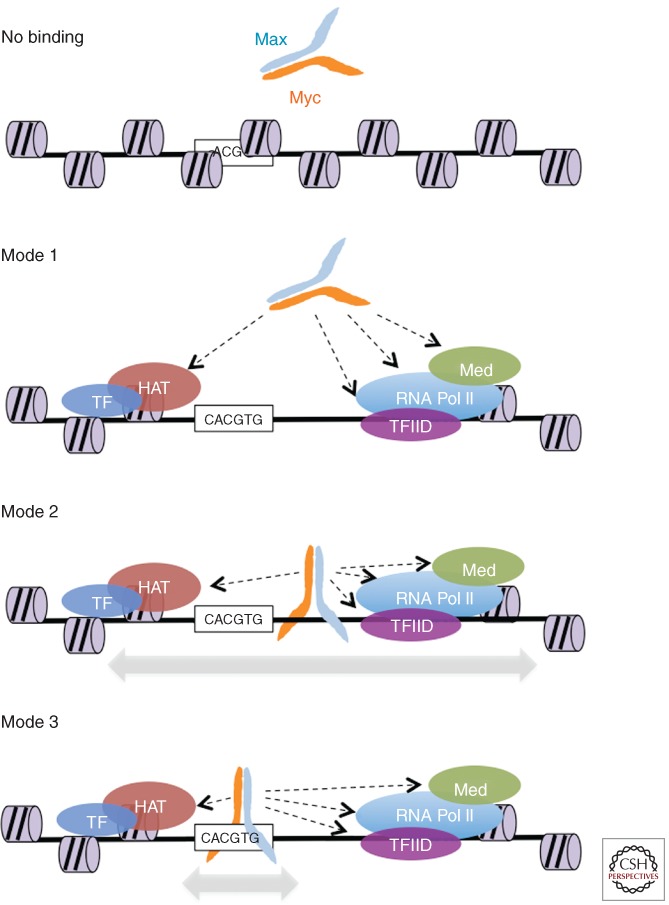
Mechanisms that restrict and regulate the portion of the genome “visible” to TFs appear to be fundamental in higher organisms with large genome, for at least two reasons. If all of the genome were to be accessible, (1) a large excess of genomic DNA would have to be scanned by any given TF before finding the relevant binding sites, and (2) the TF would frequently bind spurious consensus elements at nonfunctional locations in the genome. Based on the evidence discussed so far, we propose that this conundrum is solved by the existence of different modes of interaction of the TF with the genome, and that these modes may occur in succession, restricting sequence-specific DNA binding by the TF to a subset of all potential binding motifs in the genome. This is schematically illustrated here for MYC/MAX dimers (Fig. 1):
No binding, even in the presence of potential high-affinity sequence elements: This applies either to silent/repressed regulatory elements or to genomic locations with no regulatory function. MYC/MAX exclusion may be owing to either packing of DNA into an inaccessible heterochromatic state, the lack of positive determinants for recruitment (histone marks and reader proteins), or selective subnuclear compartmentalization.
Binding mode 1: Binding to open chromatin via protein–protein interactions, but without direct DNA contacts. We hypothesize that this is the initial step in recruiting MYC to its direct DNA-binding sites in the genome, preceding modes 2 and 3, below. In cases in which MYC is tethered by TFs, we surmise that it may also remain in this state, without progressing to modes 2 or 3. This, however, is a hypothetical state, the existence of which remains to be shown.
Binding mode 2: Binding to open chromatin via protein–protein interactions, accompanied by either nonspecific or low-affinity DNA contacts. We hypothesize that this corresponds to a “scanning” mode on DNA, through which MYC/MAX dimers find high-affinity binding sites (mode 3).
Binding mode 3: Same as above, but stabilized by high-affinity DNA contacts (E box or variants). The transition from mode 2 to 3 is supported by structural and biophysical studies of bHLH and bHLH-LZ proteins, and in particular of MAX-containing homo- or heterodimers. Once formed, the dimers expose the basic regions (which are short α-helical stretches) in the proper conformation for insertion into the DNA major groove (see Conacci-Sorrell et al. 2014). This interaction with DNA can occur without any sequence requirement. Available data are consistent with a two-step binding mechanism characterized by the fast and unspecific association of the dimers to DNA (low-affinity binding, or mode 2), followed by a slow conformational rearrangement when specific side chains within the basic region recognize their target DNA sequence (high-affinity binding, or mode 3) (Cohen et al. 1995; Sha et al. 1995; Cave et al. 2000; Sauvé et al. 2007).
Figure 1.
Different modes of MYC/MAX interaction with DNA. Modes 1–3 not only represent different possible modalities of interaction with genomic sites, but are also postulated to constitute the succession of events by which MYC/MAX dimers are driven to their physiological binding sites (mode 3), as discussed in the text. No binding: access of MYC/MAX to potential sites may be restricted by chromatin or nuclear organization, or by the lack of positive cues for selective recruitment (see text). Indeed, it is important to consider that a large majority of E-box motifs in the genome shows no significant MYC/MAX binding. Mode 1: binding to open chromatin via protein–protein interactions, but without direct DNA contacts. Examples of potentially interacting proteins or protein complexes are schematically represented (HAT, histone acetyltransferase complex; Med, Mediator complex). Mode 2: as mode 1 with additional DNA binding, but in a nonsequence-specific, low-affinity manner. Mode 3: same as above, but stabilized by high-affinity DNA contacts.
We thus propose that modes 1, 2, and 3 occur in succession (Fig. 1), with MYC/MAX heterodimers being first recruited to selected chromatin domains (i.e., active promoters or enhancers) via protein–protein interactions (mode 1). This step may be extremely transient or even negligible, being almost simultaneously accompanied by DNA contacts with no requirement for sequence specificity (mode 2). Scanning of the DNA sequence locally would then lead to stabilization of the dimer on medium/high-affinity sites (mode 3). It is noteworthy that higher-order chromatin structures (e.g., transcription factories) may also favor the spreading of MYC to open chromatin domains that occur in close spatial proximity to high-affinity MYC-binding sites.
It is important to note that in ChIP-seq profiles, which provide an average readout for whole cell populations, the aforementioned binding modes would lead to a continuum of DNA-binding intensities, as indeed observed (Nie et al. 2012; Perna et al. 2012). This ranges from barely detectable ChIP signals corresponding to mode 1 (that most probably occur randomly in different domains in each cell and thus contribute to low/dilute signals at the population level) to strong signals corresponding to mode 3. Thus, a homogeneous binding distribution, should not a priori be interpreted as a progressive unimodal interaction of MYC with the genome (Nie et al. 2012), but may instead conceal different binding modes, such as those proposed above. Consistent with this hypothesis, when compared with promoters with no MYC at all, promoters with the lowest MYC signals in serum-stimulated fibroblasts were already enriched for CpG islands (taken here as surrogate marker of active chromatin) but not for E boxes. Higher levels of MYC binding, instead, enriched both for CpG islands and E boxes, indicative of sequence-specific DNA binding (Perna et al. 2012). At the lower end of the scale, modes 1 and 2 may include promoters that are not significantly bound at physiological MYC levels, and should probably not be called as “targets.” That these promoters show enhanced cross-linking in cells with overexpressed MYC is anticipated by our model, but does not per se prove that MYC binds those low-affinity sites in a productive manner leading to transcriptional modulation.
Finally, it is noteworthy here that the X-ray structure of MYC/MAX dimers suggests that these may form heterotetramers (Nair and Burley 2003), which in turn may simultaneously bind to two sites on DNA. This would provide a rationale for the existence of cooperative DNA binding, although biochemical results in this regard remain contradictory (Walhout et al. 1997; Vervoorts and Luscher 1999; Lebel et al. 2007). Another appealing effect of MYC/MAX tetramerization would be to control the formation of long-range chromatin loops, potentially mediating interactions between promoters and enhancers.
IMPLICATIONS FOR TRANSCRIPTIONAL CONTROL
The binding dynamics of a TF with DNA, rather than levels of occupancy, may determine transcriptional output. For yeast Rap1, for example, long residence times at promoters were associated with transcriptional activation, whereas fast-binding turnover coincided with low transcriptional output, even in conditions of similar overall occupancy (Lickwar et al. 2012). Standard ChIP experiments (whether ChIP-PCR, ChIP-chip, or ChIP-seq) do not provide information on binding dynamics, and such information is lacking altogether for MYC/MAX. Yet, it is tempting to speculate that increased residence time on high-affinity DNA elements (i.e., decreased off-rate) may lead to enhanced transcriptional output. Low-affinity binding sites may not necessarily be less important in dictating transcriptional output, in particular when present in clusters. Such clusters may lead to either cooperative binding, which can induce sharp (i.e., digital) transcriptional responses as seen during developmental processes (Segal et al. 2008), or noncooperative binding, which can lead to gradual (i.e., analogic) transcriptional effects, as seen in response to environmental stimuli (Giorgetti et al. 2010). Interestingly, E-box motifs are quite abundant and can occur in proximity of each other. From ENCODE data, for example, we can deduce that ∼10% of MYC-binding sites can be found within 100 bp from other MYC/MAX sites in nonrepetitive regions (deduced from Fig. 2b and Table S4 in Wang et al. 2012). An intriguing hypothesis, which has been proven for Spi-1/Pu.1 (Ridinger-Saison et al. 2012), would be that clustered binding sites mark genes that are transcriptionally induced, versus repressed or nonregulated genes. In this regard, a weak degree of correlation has been observed between the numbers of E boxes on promoters and gene expression (Nie et al. 2012). It is noteworthy that both for Spi-1/Pu.1 and MYC, the distance of binding motifs from the TSS seemed to correlate with target gene expression (Nie et al. 2012; Ridinger-Saison et al. 2012).
An important conundrum to be resolved is whether MYC directly controls the transcriptional output of most, if not of all, active promoters in cells (Lin et al. 2012; Nie et al. 2012) and whether it does so either at physiological or pathological levels (see Levens 2013; Rahl and Young 2014). Although it is clear that a sizable proportion of active promoters is already bound at physiological (low) MYC levels, in our view rigorous peak calling in ChIP-seq data does not allow qualifying all active promoters as “targets” (A Sabò, T Kress, M Pelizzola, et al., submitted), making it difficult to view MYC as a general transcriptional amplifier in those conditions (Nie et al. 2012). MYC overexpression, on the other hand, leads to “invasion” of all active promoters (see above), accompanied—at least in the cell types examined so far—by a general increase in transcriptional activity (Lin et al. 2012; A Sabò, T Kress, M Pelizzola, et al., submitted). However, whether MYC productively interacts with all those promoters to directly amplify their transcriptional output remains to be conclusively shown. An alternative interpretation, not ruled out by presently available data, is that transcriptional amplification may be an indirect consequence of MYC-induced alterations in cellular physiology and/or signaling pathways, themselves regulated by restricted groups of target genes, which may be either up- or down-regulated by MYC. For example, MYC is well known to positively affect cell growth (i.e., cell size) (Iritani and Eisenman 1999; Johnston et al. 1999; Schuhmacher et al. 1999) and mitochondrial energy metabolism (Li et al. 2005; Graves et al. 2012). An important body of parallel literature has correlated these cellular parameters with general increases in transcriptional activity, a phenomenon observed across the eukaryotic evolutionary spectrum, well before the appearance of MYC (reviewed in Marguerat and Bähler 2012).
CONCLUSIONS AND FUTURE PERSPECTIVES
We have presented here the model that in our view best approximates the existing data on MYC-chromatin interactions at the genome-wide level (Fig. 1). This model also highlights some questions that remain to be addressed for a better understanding of these interactions and of their consequences for transcriptional control. We can outline the following:
An important missing link regards the nature of the forces that drive the transcription factor to the correct subset of genomic domains, whether protein–protein interactions (Fig. 1, mode 1), chromatin accessibility, higher-order nuclear organization, or a combination thereof. In particular, it remains unclear whether the rate-limiting steps lie prevalently in the interaction with general components of the transcriptional machinery—and with which of these components exactly—or with specific TFs, perhaps in a context-dependent manner on specific subsets of genes (e.g., Cheng et al. 2006).
Another important question is whether sequence-specific DNA binding (Fig. 1, mode 3) is systematically required for the engagement of MYC into selective and productive gene-regulatory interactions, or whether it simply stabilizes a state that can occur on any generic DNA segment (mode 2). If recognition of chromatin context is indeed preliminary to sequence recognition, MYC mutants that are impaired in the latter should still distribute to active promoters when overexpressed, but should no longer show preferential binding to high-affinity promoters. From earlier work, such mutants are impaired in E-box-dependent transactivation and cellular transformation (Amati et al. 1992, 1993a,b; Kretzner et al. 1992), but may retain some regulatory and biological activity (Cowling and Cole 2007), as also suggested by the existence of MAX-independent functions of MYC in Drosophila (Steiger et al. 2008; Gallant 2013).
Another key question—not discussed in this review—regards the mechanisms through which MYC regulates transcription. Consistent with the fact that it binds active promoters with preloaded RNA Pol II, MYC promotes transcriptional elongation (see Rahl and Young 2014). MYC also regulates histone acetylation (e.g., Bouchard et al. 2001; Frank et al. 2001) and possibly nucleosome remodeling (e.g., Cheng et al. 1999). How these mechanisms come together to determine MYC-dependent gene regulation remains to be determined.
Finally, a major question regards the scale of transcriptional regulation by MYC. In particular, it remains unclear whether MYC acts as a rheostat, directly amplifying the transcriptional output from all active promoters (Lin et al. 2012; Nie et al. 2012), or whether it achieves this effect indirectly, through selective up- and down-regulation of specific target genes (Eilers and Eisenman 2008; Herkert and Eilers 2010) that in turn cause the observed changes in general transcriptional activity. In this regard, it is important to consider that general transcriptional amplification and specific regulatory cues (either positive or negative) are not contradictory, and may well coexist in cells. Although the net outcomes of direct versus indirect amplification would be very similar, the underlying molecular mechanisms are dramatically different. New quantitative data and predictive modeling will be required to discriminate between those alternative scenarios.
ACKNOWLEDGMENTS
We thank Stefano Campaner and Theresia Kress for critical reading of the manuscript. Work in the Amati group is supported by grants from the European Commission (FP7 consortium MODHEP), the European Research Council (ERC), the Italian Association for Cancer Research (AIRC), the Association for International Cancer Research (AICR), and the Italian Health Ministry.
Footnotes
Editors: Chi V. Dang and Robert N. Eisenman
Additional Perspectives on MYC and the Pathway to Cancer available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Adhikary S, Eilers M 2005. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6: 635–645 [DOI] [PubMed] [Google Scholar]
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J 18: 5108–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B, Dalton S, Brooks MW, Littlewood TD, Evan GI, Land H 1992. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature 359: 423–426 [DOI] [PubMed] [Google Scholar]
- Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H 1993a. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell 72: 233–245 [DOI] [PubMed] [Google Scholar]
- Amati B, Littlewood TD, Evan GI, Land H 1993b. The c-Myc protein induces cell cycle progression and apoptosis through dimerization with Max. EMBO J 12: 5083–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvey A, Agius P, Noble WS, Leslie C 2012. Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome Res 22: 1723–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer DE, Kretzner L, Eisenman RN 1993. Mad: A heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell 72: 211–222 [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Weintraub H 1990. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science 250: 1104–1110 [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H 1990. Sequence-specific DNA binding by the c-Myc protein. Science 250: 1149–1151 [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Huang J, Ma A, Kretzner L, Alt FW, Eisenman RN, Weintraub H 1993. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol 13: 5216–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood EM, Eisenman RN 1991. Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 251: 1211–1217 [DOI] [PubMed] [Google Scholar]
- Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Luscher B 2001. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev 15: 2042–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Marquardt J, Brás A, Medema RH, Eilers M 2004. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J 23: 2830–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner C, Deplus R, Didelot C, Loriot A, Viré E, De Smet C, Gutiérrez A, Danovi D, Bernard D, Boon T, et al. 2005. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J 24: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cave JW, Kremer W, Wemmer DE 2000. Backbone dynamics of sequence specific recognition and binding by the yeast Pho4 bHLH domain probed by NMR. Protein Sci 9: 2354–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV 1999. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet 22: 102–105 [DOI] [PubMed] [Google Scholar]
- Cheng ASL, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, Leu Y-W, Chan MWY, Plass C, Nephew KP, et al. 2006. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-α responsive promoters. Mol Cell 21: 393–404 [DOI] [PubMed] [Google Scholar]
- Ciro M, Prosperini E, Quarto M, Grazini U, Walfridsson J, McBlane F, Nucifero P, Pacchiana G, Capra M, Christensen J, et al. 2009. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res 69: 8491–8498 [DOI] [PubMed] [Google Scholar]
- Cohen SL, Ferré-D’Amaré AR, Burley SK, Chait BT 1995. Probing the solution structure of the DNA-binding protein Max by a combination of proteolysis and mass spectrometry. Protein Sci 4: 1088–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Conacci-Sorrell M, McFerrin L, Eisenman RN 2014. An overview of MYC and its interactome. Cold Spring Harb Perspect Med 4: a014327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling VH, Cole MD 2007. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol Cell Biol 27: 2059–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A 2011. CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R 2007. Non-transcriptional control of DNA replication by c-Myc. Nature 448: 445–451 [DOI] [PubMed] [Google Scholar]
- Doyon Y, Selleck W, Lane WS, Tan S, Côté J 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol 24: 1884–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardy SR 2002. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J Biol Chem 277: 40156–40162 [DOI] [PubMed] [Google Scholar]
- Eberhardy SR, Farnham PJ 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J Biol Chem 276: 48562–48571 [DOI] [PubMed] [Google Scholar]
- Edelman LB, Fraser P 2012. Transcription factories: Genetic programming in three dimensions. Curr Opin Genet Dev 22: 110–114 [DOI] [PubMed] [Google Scholar]
- Efstratiadis A, Szabolcs M, Klinakis A 2007. Notch, Myc and breast cancer. Cell Cycle 6: 418–429 [DOI] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN 2008. Myc’s broad reach. Genes Dev 22: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 488: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsher DW 2010. MYC inactivation elicits oncogene addiction through both tumor cell-intrinsic and host-dependent mechanisms. Genes Cancer 1: 597–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM 1999. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell 4: 199–207 [DOI] [PubMed] [Google Scholar]
- Feng X-H, Liang Y-Y, Liang M, Zhai W, Lin X 2002. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-β-mediated induction of the CDK inhibitor p15Ink4B. Mol Cell 9: 133–143 [DOI] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B 2003. Genomic targets of the human c-Myc protein. Genes Dev 17: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev 15: 2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan H-M, Livingston DM, Amati B 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep 4: 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Gabay M, Li Y, Felsher DW 2014. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Gallant P 2013. Myc function in Drosophila. Cold Spring Harb Perspect Med 3: a014324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant P, Steiger D 2009. Myc’s secret life without Max. Cell Cycle 8: 3848–3853 [DOI] [PubMed] [Google Scholar]
- Gartel AL, Ye X, Goufman E, Shianov P, Hay N, Najmabadi F, Tyner AL 2001. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc Natl Acad Sci 98: 4510–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Siggers T, Tiana G, Caprara G, Notarbartolo S, Corona T, Pasparakis M, Milani P, Bulyk ML, Natoli G 2010. Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol Cell 37: 418–428 [DOI] [PubMed] [Google Scholar]
- Gomez-Roman N, Grandori C, Eisenman RN, White RJ 2003. Direct activation of RNA polymerase III transcription by c-Myc. Nature 421: 290–294 [DOI] [PubMed] [Google Scholar]
- Graves JA, Wang Y, Sims-Lucas S, Cherok E, Rothermund K, Branca MF, Elster J, Beer-Stolz D, Van Houten B, Vockley J, et al. 2012. Mitochondrial structure, function and dynamics are temporally controlled by c-Myc. PLoS ONE 7: e37699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall Olio V, Zardo G, Nervi C, Bernard L, Amati B 2006. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol 8: 764–770 [DOI] [PubMed] [Google Scholar]
- Guertin MJ, Lis JT 2012. Mechanisms by which transcription factors gain access to target sequence elements in chromatin. Curr Opin Genet Dev 23: 116–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin MJ, Martins AL, Siepel A, Lis JT 2012. Accurate prediction of inducible transcription factor binding intensities in vivo ed. M. Snyder. PLoS Genet 8: e1002610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hann S 2014. Myc protein interactions. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hateboer G, Timmers HT, Rustgi AK, Billaud M, van ’t Veer LJ, Bernards R 1993. TATA-binding protein and the retinoblastoma gene product bind to overlapping epitopes on c-Myc and adenovirus E1A protein. Proc Natl Acad Sci 90: 8489–8493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkert B, Eilers M 2010. Transcriptional repression: The dark side of myc. Genes Cancer 1: 580–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz H-P, Haenel F, Eilers M 2002. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell 10: 509–521 [DOI] [PubMed] [Google Scholar]
- Hurlin PJ, Queva C, Koskinen PJ, Steingrímsson E, Ayer DE, Copeland NG, Jenkins NA, Eisenman RN 1995. Mad3 and Mad4: Novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J 14: 5646–5659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlin PJ, Queva C, Eisenman RN 1997. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev 11: 44–58 [DOI] [PubMed] [Google Scholar]
- Iritani BM, Eisenman RN 1999. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci 96: 13180–13185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi H, Molander C, Penn LZ, Ishisaki A, Kohno K, Funa K 2001. Mechanism for the transcriptional repression by c-Myc on PDGF β-receptor. J Cell Sci 114: 1533–1544 [DOI] [PubMed] [Google Scholar]
- Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW 2002. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 297: 102–104 [DOI] [PubMed] [Google Scholar]
- James L, Eisenman RN 2002. Myc and Mad bHLHZ domains possess identical DNA-binding specificities but only partially overlapping functions in vivo. Proc Natl Acad Sci 99: 10429–10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Espeseth A, Hazuda DJ, Margolis DM 2007. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus Type 1 promoter. J Virol 81: 10914–10923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P 1999. Drosophila myc regulates cellular growth during development. Cell 98: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan T, Li X-Y, Sabo PJ, Thomas S, Stamatoyannopoulos JA, Biggin MD, Eisen MB 2011. Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early Drosophila development. PLoS Genet 7: e1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerosuo L, Piltti K, Fox H, Angers-Loustau A, Häyry V, Eilers M, Sariola H, Wartiovaara K 2008. Myc increases self-renewal in neural progenitor cells through Miz-1. J Cell Sci 121: 3941–3950 [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132: 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch HB, Zhang R, Verdoodt B, Bailey A, Zhang C-D, Yates JR, Menssen A, Hermeking H 2007. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle 6: 205–217 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kretzner L, Blackwood EM, Eisenman RN 1992. Myc and Max proteins possess distinct transcriptional activities. Nature 359: 426–429 [DOI] [PubMed] [Google Scholar]
- Küppers R, Dalla-Favera R 2001. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene 20: 5580–5594 [DOI] [PubMed] [Google Scholar]
- Kurland JF, Tansey WP 2008. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res 68: 3624–3629 [DOI] [PubMed] [Google Scholar]
- Lebel R, McDuff F-O, Lavigne P, Grandbois M 2007. Direct visualization of the binding of c-Myc/Max heterodimeric b-HLH-LZ to E-Box sequences on the hTERT promoter. Biochemistry 46: 10279–10286 [DOI] [PubMed] [Google Scholar]
- Lee B-K, Bhinge AA, Battenhouse A, McDaniell RM, Liu Z, Song L, Ni Y, Birney E, Lieb JD, Furey TS, et al. 2012. Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. Genome Res 22: 9–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Levens D 2013. Cellular MYCro economics: Balancing MYC function with MYC expression. Cold Spring Harb Perspect Med 3: a014233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z 2003. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci 100: 8164–8169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim J-W, Yustein JT, Lee LA, Dang CV 2005. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol 25: 6225–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD 2012. Genome-wide protein–DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 484: 251–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Vorontchikhina M, Wang Y-L, Faiola F, Martinez E 2008. STAGA recruits Mediator to the MYC oncoprotein to stimulate transcription and cell proliferation. Mol Cell Biol 28: 108–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher-Firzlaff J, Gawlista I, Vervoorts J, Kapelle K, Braunschweig T, Walsemann G, Rodgarkia-Schamberger C, Schuchlautz H, Dreschers S, Kremmer E, et al. 2008. The human trithorax protein hASH2 functions as an oncoprotein. Cancer Res 68: 749–758 [DOI] [PubMed] [Google Scholar]
- Mao DYL, Watson JD, Yan PS, Barsyte-Lovejoy D, Khosravi F, Wong WW-L, Farnham PJ, Huang THM, Penn LZ 2003. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr Biol 13: 882–886 [DOI] [PubMed] [Google Scholar]
- Marguerat S, Bähler J 2012. Coordinating genome expression with cell size. Trends Genet 28: 560–565 [DOI] [PubMed] [Google Scholar]
- Martinato F, Cesaroni M, Amati B, Guccione E, Abraham E 2008. Analysis of Myc-induced histone modifications on target chromatin. PLoS ONE 3: e3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y, Fukamachi K, Uehara N, Tsuda H, Tsubura A 2008. Induction of a novel histone deacetylase 1/c-Myc/Mnt/Max complex formation is implicated in parity-induced refractoriness to mammary carcinogenesis. Cancer Sci 99: 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94: 363–374 [DOI] [PubMed] [Google Scholar]
- Mukherjee B, Morgenbesser SD, DePinho RA 1992. Myc family oncoproteins function through a common pathway to transform normal cells in culture: Cross-interference by Max and trans-acting dominant mutants. Genes Dev 6: 1480–1492 [DOI] [PubMed] [Google Scholar]
- Nagy Z, Tora L 2007. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26: 5341–5357 [DOI] [PubMed] [Google Scholar]
- Nair SK, Burley SK 2003. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112: 193–205 [DOI] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. 2012. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 488: 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem Cells. Cell 151: 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov MA 2003. The Mad and Myc basic domains are functionally equivalent. J Biol Chem 278: 11094–11099 [DOI] [PubMed] [Google Scholar]
- Orian A 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev 17: 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, Tempst P, Sif S 2003. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol 23: 7475–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Wood MA, Cole MD 2002. BAF53 forms distinct nuclear complexes and functions as a critical c-Myc-interacting nuclear cofactor for oncogenic transformation. Mol Cell Biol 22: 1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JH 2005. Targeting of Miz-1 is essential for Myc-mediated apoptosis. J Biol Chem 281: 3283–3289 [DOI] [PubMed] [Google Scholar]
- Perna D, Fagà G, Verrecchia A, Gorski MM, Barozzi I, Narang V, Khng J, Lim KC, Sung W-K, Sanges R, et al. 2012. Genome-wide mapping of Myc binding and gene regulation in serum-stimulated fibroblasts. Oncogene 31: 1695–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK 2011. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res 21: 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Rahl PB, Young RA 2014. Myc and transcription elongation. Cold Spring Harb Perspect Med 4: a014340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, Mccuine S, Burge CB, Sharp PA, Young RA 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridinger-Saison M, Boeva V, Rimmele P, Kulakovskiy I, Gallais I, Levavasseur B, Paccard C, Legoix-Ne P, Morle F, Nicolas A, et al. 2012. Spi-1/PU.1 activates transcription through clustered DNA occupancy in erythroleukemia. Nucleic Acids Res 40: 8927–8941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Roussel MF, Robinson GW 2013. Role of MYC in medulloblastoma. Cold Spring Harb Perspect Med 3: a014308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AL, Carruthers C, Gutjahr T, Roeder RG 1993. Direct role for Myc in transcription initiation mediated by interactions with TFII-I. Nature 365: 359–361 [DOI] [PubMed] [Google Scholar]
- Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR 2007. Myc deletion rescues Apc deficiency in the small intestine. Nature 446: 676–679 [DOI] [PubMed] [Google Scholar]
- Satou A 2001. A novel transrepression pathway of c-Myc. Recruitment of a transcriptional corepressor complex to c-Myc by MM-1, a c-Myc-binding protein. J Biol Chem 276: 46562–46567 [DOI] [PubMed] [Google Scholar]
- Sauvé S, Naud J-F, Lavigne P 2007. The mechanism of discrimination between cognate and non-specific DNA by dimeric b/HLH/LZ transcription factors. J Mol Biol 365: 1163–1175 [DOI] [PubMed] [Google Scholar]
- Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M 2012. Linking disease associations with regulatory information in the human genome. Genome Res 22: 1748–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW, Eick D, Kohlhuber F 1999. Control of cell growth by c-Myc in the absence of cell division. Curr Biol 9: 1255–1258 [DOI] [PubMed] [Google Scholar]
- *.Schmitz R, Ceribelli M, Pittaluga S, Wrigth G, Staudt LM 2014. Oncogenic mechanisms in Burkitt lymphoma. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombe J, Li L, Carlos L, Eisenman RN 2007. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev 21: 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J 2009. From DNA sequence to transcriptional behaviour: A quantitative approach. Nat Rev Genet 10: 443–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U 2008. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature 451: 535–540 [DOI] [PubMed] [Google Scholar]
- Seitz V, Butzhammer P, Hirsch B, Hecht J, Gütgemann I, Ehlers A, Lenze D, Oker E, Sommerfeld A, Wall Von Der E, et al. 2011. Deep sequencing of MYC DNA-binding sites in Burkitt lymphoma. PLoS ONE 6: e26837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massagué J 2001. TGFβ influences Myc, Miz-1, and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol 3: 400–408 [DOI] [PubMed] [Google Scholar]
- Sha M, Ferré-D’Amaré AR, Burley SK, Goss DJ 1995. Anti-cooperative biphasic equilibrium binding of transcription factor upstream stimulatory factor to its cognate DNA monitored by protein fluorescence changes. J Biol Chem 270: 19325–19329 [DOI] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, et al. 2004. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 431: 1112–1117 [DOI] [PubMed] [Google Scholar]
- Sharma VM, Draheim KM, Kelliher MA 2007. The Notch1/c-Myc pathway in T cell leukemia. Cell Cycle 6: 927–930 [DOI] [PubMed] [Google Scholar]
- Shrivastava A, Saleque S, Kalpana GV, Artandi S, Goff SP, Calame K 1993. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science 262: 1889–1892 [DOI] [PubMed] [Google Scholar]
- Solomon DL, Amati B, Land H 1993. Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers. Nucleic Acids Res 21: 5372–5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI 2008. Modelling Myc inhibition as a cancer therapy. Nature 455: 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield JR, Sodir NM, Massó-Vallés D, Serrano E, Karnezis AN, Swigart LB, Evan GI 2013. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev 27: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151: 994–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Möröy T, Bartek J, Massagué J, Hänel F, et al. 2001. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol 3: 392–399 [DOI] [PubMed] [Google Scholar]
- Steiger D, Furrer M, Schwinkendorf D, Gallant P 2008. Max-independent functions of Myc in Drosophila melanogaster. Nat Genet 40: 1084–1091 [DOI] [PubMed] [Google Scholar]
- Uribesalgo I, Buschbeck M, Gutiérrez A, Teichmann S, Demajo S, Kuebler B, Nomdedéu JF, Martín-Caballero J, Roma G, Benitah SA, et al. 2011. E-box-independent regulation of transcription and differentiation by MYC. Nat Cell Biol. [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Muller J, Otto T, Beuger V, Yetil A, Choi PS, Kosan C, Möröy T, Felsher DW, Eilers M 2010. The interaction between Myc and Miz1 is required to antagonize TGF-dependent autocrine signaling during lymphoma formation and maintenance. Genes Dev 24: 1281–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervoorts J, Luscher B 1999. DNA binding of Myc/Max/Mad network complexes to oligonucleotides containing two E box elements: c-Myc/Max heterodimers do not bind DNA cooperatively. Biol Chem 380: 1121–1126 [DOI] [PubMed] [Google Scholar]
- Vervoorts J, Lüscher-Firzlaff JM, Rottmann S, Lilischkis R, Walsemann G, Dohmann K, Austen M, Lüscher B 2003. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep 4: 484–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJ, Gubbels JM, Bernards R, van der Vliet PC, Timmers HT 1997. c-Myc/Max heterodimers bind cooperatively to the E-box sequences located in the first intron of the rat ornithine decarboxylase (ODC) gene. Nucleic Acids Res 25: 1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. 2012. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res 22: 1798–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wiese K, Walz S, von Eyss B, Wolf E, Athineos D, Sansom O, Eilers M 2013. The role of Miz1 in the Myc-dependent tumorigenesis. Cold Spring Harb Perspect Med 3: a014290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JA, Sansom OJ 2008. C-Myc is a critical mediator of the phenotypes of Apc loss in the intestine. Cancer Res 68: 4963–4966 [DOI] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CWH, Chiu K-P, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. 2006. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci 103: 17834–17839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervos AS, Gyuris J, Brent R 1993. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell 72: 223–232 [DOI] [PubMed] [Google Scholar]