Abstract
Burkitt lymphoma is a germinal center B-cell-derived cancer that was instrumental in the identification of MYC as an important human oncogene more than three decades ago. Recently, new genomics technologies have uncovered several additional oncogenic mechanisms that cooperate with MYC to create this highly aggressive cancer. The transcription factor TCF-3 is central to Burkitt lymphoma pathogenesis. TCF-3 is rendered constitutively active in Burkitt lymphoma by two related mechanisms: (1) somatic mutations that inactivate its negative regulator ID3, and (2) somatic mutations in TCF-3 that block the ability of ID3 to bind and interfere with its activity as a transcription factor. TCF-3 is also a master regulator of normal germinal center B-cell differentiation. Within the germinal center, TCF-3 up-regulates genes that are characteristically expressed in the rapidly dividing centroblasts, the putative cell of origin for Burkitt lymphoma, while repressing genes expressed in the less proliferative centrocytes. TCF-3 promotes antigen-independent (tonic) B-cell-receptor signaling in Burkitt lymphoma by transactivating immunoglobulin heavy- and light-chain genes while repressing PTPN6, which encodes the phosphatase SHP-1, a negative regulator of B-cell-receptor signaling. Tonic B-cell-receptor signaling sustains Burkitt lymphoma survival by engaging the PI3 kinase pathway. In addition, TCF-3 promotes cell-cycle progression by transactivating CCND3, encoding a D-type cyclin that regulates the G1–S phase transition. Additionally, CCND3 accumulates oncogenic mutations that stabilize cyclin D3 protein expression and drive proliferation. These new insights into Burkitt lymphoma pathogenesis suggest new therapeutic strategies, which are sorely needed in developing regions of the world where this cancer is endemic.
MYC translocations are a hallmark of Burkitt lymphomas. But additional oncogenic mechanisms and genetic lesions (e.g., constitutive activation of transcription factor TCF-3) cooperate with MYC to create this highly aggressive cancer.
In 1958, Dennis Burkitt described 38 cases of “sarcomas” involving the jaws of African children seen at Mulago Hospital in Kampala Uganda, providing the first clinical description of Burkitt lymphoma (BL) (Burkitt 1958). These tumors that are particularly prevalent in young children in tropical Africa, accounting for 30%–50% of all childhood cancers in equatorial Africa, most frequently affect extranodal sites including the jaws, the abdomen, and endocrine organs. Shortly after the first description of BL, several pathologists described lymphomas in Europe and the United States that were indistinguishable at a histological level from African BL. Thus, it became clear that BL was not uniquely an African disease, although incidence varied markedly in Africa and Europe or the United States. The particular geographical distribution of BL led to the African variety (also common in New Guinea) being referred to as endemic BL (eBL), whereas tumors occurring elsewhere were referred to as sporadic BL (sBL). Aside from the different geographical distribution and prevalence, eBL and sBL differ in their association with the Epstein-Barr virus (EBV), which was originally discovered in an eBL specimen. EBV is present in the tumor cells of virtually all cases of eBL, whereas only 10%–20% of sBL cases harbor this viral DNA. With the advent of the human immunodeficiency virus (HIV), HIV patients were observed to be predisposed to non-Hodgkin lymphomas, including BL. This led to the inclusion of a third BL variant in the WHO classification of hematological malignancies: immunodeficiency-related BL.
In 1975, a characteristic chromosomal translocation, involving the distal region of the long arm of chromosome 14 and the long arm of chromosome 8, was discovered in BL cells (Zech et al. 1976). A few years later, the human MYC gene was identified at the breakpoint of the t(8;14) translocation, juxtaposing the immunoglobulin (IG) heavy-chain locus with MYC (Dalla-Favera et al. 1982; Taub et al. 1982). As a consequence, MYC is brought under the transcriptional control of the IG enhancer elements leading to its constitutive transcriptional deregulation. Although MYC translocations occur less commonly in other lymphomas, such as diffuse large B-cell lymphoma (DLBCL), they are the hallmark of BL, being present in all cases of all three BL subtypes. These translocations typically occur on the background of a simple karyotype lacking additional rearrangements (Hummel et al. 2006).
The oncogenic potential of deregulated MYC in cells of hematopoietic origin was impressively shown only 3 years later. Transgenic mice that expressed the MYC gene under the control of the IG heavy-chain intronic enhancer (Eμ), emulating the chromosomal translocation found in BL, developed B-cell lymphomas with a latency of 4–6 mo (Adams et al. 1985). However, recent understanding of human BL pathogenesis has revealed that these Eμ/MYC transgenic mice are not faithful models for human BL. Human BLs share a gene expression program with normal germinal center (GC) B cells (Dave et al. 2006). Normal GC B cells and BL tumors express the cytidine deaminase (AID), which mediates both IG somatic hypermutation and IG class switch recombination (CSR). As a consequence, human BLs have somatically mutated IG variable regions, and IG/MYC translocations typically involve IG switch regions, suggesting that they arise by aberrant CSR. In contrast, MYC-driven lymphomas in Eμ/MYC transgenic mice typically arise from pre-B or naïve B cells, not GC B cells. A more recent mouse model in which MYC and PI3 kinase (PI3K) are conditionally activated in GC B cells appears to be a faithful mimic of human BL (Sander et al. 2012). These oncogenic alleles are apparently not sufficient to fully transform B cells because the B-cell tumors that arise in this mouse model are clonal, indicating that additional oncogenic events are required.
In this review, we focus on the new picture of BL pathogenesis that has emerged from genomic analysis of human BL tumors.
EPIDEMIOLOGY OF EBL: THE ROLE OF INFECTIOUS AGENTS IN THE PATHOGENESIS OF EBL
When Denis Burkitt mapped the prevalence of BL across sub-Saharan Africa, a geographical coincidence emerged between lymphoma and endemic malarial infections (Burkitt 1961, 1962). Subsequent studies indicated that the geographical distribution of BL corresponds not only to the distribution of malaria, but also to the intensity of malarial infection (Dalldorf et al. 1964; Morrow et al. 1976). Despite this epidemiological evidence, the exact role of malaria in the pathogenesis of BL remains unclear. One potential mechanism could derive from the ability of malaria to induce chronic B-cell activation (Illingworth et al. 2013), which might predispose B cells to the acquisition of IG/MYC translocations. A second mechanism could involve activation of EBV replication by malarial infection. EBV persists in a latent state in resting memory B cells that circulate in the peripheral blood (Thorley-Lawson 2001). The exposure of B cells to malarial antigens during multiple infectious episodes could reactivate the virus from memory B cells, thereby increasing viral load and the number of EBV-infected cells (Donati et al. 2006; Chene et al. 2007).
The contribution of EBV to the pathogenesis of BL is similarly enigmatic. Following the isolation of EBV from an eBL specimen by Epstein and colleagues, the virus was shown to be present in the malignant B cells in all eBL cases and in a minority of sBL cases. EBV induces the proliferation of resting human B cells, leading to the outgrowth of transformed but not malignant lymphobastoid cell lines (LCLs) (Rowe et al. 2009). LCLs express a variety of EBV-encoded latency proteins, many of which modulate key regulatory pathways such as PI3K and NF-κB that have been linked to cancer (for review, see Thorley-Lawson 2001). Given its ability to induce proliferation of LCLs, EBV might be assumed to play a role in BL pathogenesis. However, BL cells express a much more restricted pattern of viral gene products, primarily EBNA1, which is involved in the replication of the EBV genome. A clear oncogenic function of EBNA1 has not been established because initial reports of the induction of B-cell lymphomas by EBNA1 in transgenic mouse models could not be reproduced (Wilson et al. 1996; Kang et al. 2008). Recent data indicate that the expression of EBNA1 is selectively retained in EBV-positive BL cell lines, providing indirect evidence of its role in BL pathogenesis. A dominant-negative EBNA1 that inhibits episomal replication revealed a variable dependence of BL cell lines on the presence of the viral genome (Vereide and Sugden 2011). The lack of expression of most EBV proteins in EBV-infected BL cells may be the result of vigorous selective pressure by T cells that are specific for latent antigens. In the absence of functional T cells, EBV-induced LCLs grow unimpeded, as in the case of posttransplant lymphoproliferative disorder. Thus, it is likely that a “hit-and-run” model explains the role of EBV in BL, in which EBV inhibits the apoptosis of premalignant tumor cells, allowing transforming events to occur, at which point the majority of its latency proteins are no longer necessary.
THE ROLE OF THE GERMINAL CENTER REACTION IN THE DEVELOPMENT OF BL
Many mature B-cell malignancies, including BL, coopt the gene expression program of their normal GC B-cell counterparts and shape it to execute their oncogenic purposes. One important aspect of this GC program is AID, which mediates somatic hypermutation and IG CSR in normal GC cells but in experimental settings can also induce IG/MYC translocations similar to those found in BL (Pasqualucci et al. 2004; Ramiro et al. 2004). GC-restricted transcription factors can have a powerful oncogenic effect when dysregulated because they can simultaneously affect multiple downstream pathways. This results in high dependency—addiction—of lymphoma cells on lineage-restricted transcription factors (Rui et al. 2011). Most famous in this respect is BCL6, which is required for germinal center formation and is recurrently translocated in human lymphomas (for review, see Basso and Dalla-Favera 2012).
GCs are histological structures that form in secondary lymphoid organs in response to antigen encounter by peripheral naïve B cells. These structures are sites of rapid proliferation and selection of B cells expressing high-affinity IGs. Traditionally, GC B cells are subdivided into two cell populations, centroblasts and centrocytes (MacLennan 1994). Centroblasts are GC B cells that undergo rapid proliferation and somatic hypermutation, whereas centrocytes are GC B cells that are selected for high-affinity IGs by their ability to bind antigen displayed on the surface of follicular dendritic cells. Centroblasts and centrocytes are distributed to opposite poles of the GC structure: a centrocyte-rich light zone (LZ) that includes follicular dendritic cells and T follicular helper cells, and a centroblast-rich dark zone (DZ). Gene expression profiling of sorted centroblasts and centrocytes showed that these cells only differ in the expression of a limited number of genes establishing the characteristic activation program of LZ cells and the strong proliferation program of DZ cells (Victora et al. 2012). The similarities between the centroblasts’ and centrocytes’ gene expression fits with a “cyclic reentry” model in which LZ and DZ cells regularly move between the LZ and DZ compartments, shifting their biological phenotypes in subtle but important ways in the process (for review, see Victora and Nussenzweig 2012).
Initial gene expression profiling of BL showed that this lymphoma subtype expresses a subset of the genes that characterize the GC stage of differentiation (Dave et al. 2006). Recent profiling of DZ and LZ cells showed that BL most closely resembles DZ cells, whereas the GC-derived GCB subtype of DLBCL is more akin to LZ cells (Victora et al. 2012). MYC mRNA is more highly expressed in LZ cells than in DZ cells. In fact, MYC protein expression is confined to a discrete population of LZ cells that appear to be activated by antigen and T follicular helper cells and likely undergo positive selection (Calado et al. 2012; Dominguez-Sola et al. 2012). These MYC-positive LZ cells are presumably primed to reenter the DZ, where they adopt a centroblast phenotype and lose expression of MYC, partially owing to direct repression by BCL6 (Dominguez-Sola et al. 2012). These observations suggest that the translocation-dependent expression of MYC in BL represents the unnatural grafting of MYC function onto a centroblast-like gene expression program. Indeed, the t(8;14) translocation of MYC often removes the BCL6-binding sites in the MYC 5′ region, alleviating repression. Thus, the translocation of MYC in BL uncouples MYC expression in the GC from the natural process of positive selection in the LZ. Recent evidence suggests that one function of MYC is to amplify the expression of all genes that are active in a cell (Lin et al. 2012; Nie et al. 2012; Levens 2013; Rahl and Young 2014), and thus the role of MYC in BL may be to augment the centroblast phenotype. In this context, it is noteworthy that TCF-3, which encodes the transcription factor E2A, is characteristically and selectively expressed in centroblasts and is central to BL biology (see below). A nonmutually exclusive alternative is that MYC may transactivate particular target genes that endow BL with new functions that are lacking in centroblasts (for review, see Dang 2012). In particular, MYC alters cellular metabolism to favor the biosynthesis of various macromolecules that are needed for the malignant phenotype.
GENETICS OF BL
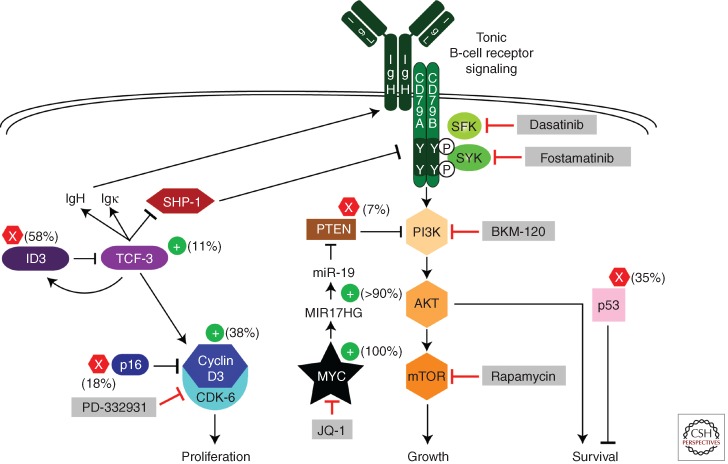
The observed delay in lymphoma onset in transgenic MYC mouse models and the clonality of these tumors suggests that deregulated expression of MYC alone is not sufficient for malignant transformation. Indeed, additional genetic lesions cooperate with MYC to generate human BL, as revealed by high-throughput sequencing approaches (Love et al. 2012; Richter et al. 2012; Schmitz et al. 2012). The unique genetic etiology of BL is underscored by the observation that many genes are recurrently mutated in BL but rarely if ever mutated in DLBCL, whereas several mutated genes that are hallmarks of DLBCL are not altered in BL. The mutational landscape of BL led to a model of its regulatory circuitry that accounts for the abnormal proliferation, growth, and survival of this lymphoma subtype (Fig. 1).
Figure 1.
Schematic of recurrent oncogenic pathways in Burkitt lymphoma. Shown are pathways that regulate proliferation, growth, and survival of Burkitt lymphomas. Gain-of-function mutations are indicated by + signs and loss-of-function mutations by × signs. (Gray boxes) Potential drugs to block these deregulated pathways. See text for details. (From Schmitz et al. 2012; modified, with permission, from the author.)
MYC is the most recurrently mutated gene in BL, sustaining mutations in 70% of cases, which often affect its transactivation domain (Schmitz et al. 2012). Many of these mutations are likely to have been introduced by AID, because of the proximity of the IG and MYC loci in BL. Some transactivation-domain mutants have enhanced transforming ability and are selectively impaired in transactivation of the proapoptotic Bcl2 family member Bim, thus uncoupling the pro-proliferative action of MYC from its ability to induce apoptosis (Hemann et al. 2005). Inactivating mutations in TP53 are also common in BL, occurring in 35% of cases. The selection for these mutations is likely to stem from the fact that overexpression of MYC in primary cells induces TP53-dependent apoptotic pathways (Evan et al. 1992; Meyer et al. 2006).
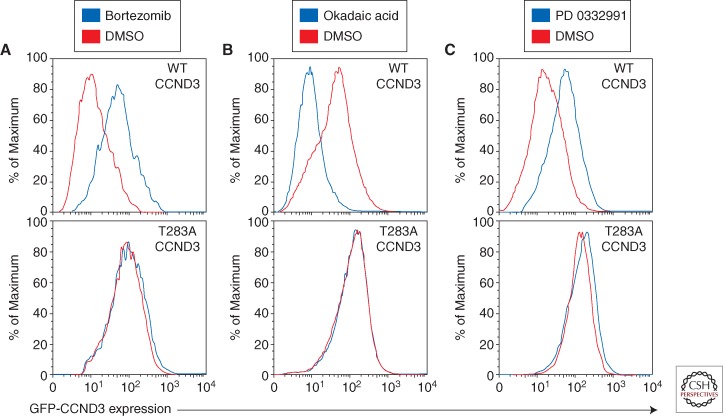
The highly proliferative nature of BL tumors is promoted by mutations in the CCND3 gene encoding cyclin D3, a D-type cyclin that regulates the G1–S cell-cycle transition. In this regard, it is notable that disruption of the Ccnd3 locus in mice impairs the generation and expansion of germinal centers (Peled et al. 2010). CCND3 mutations in BL often alter the threonine residue at position 283 (T283), known to be involved in cyclin D3 phosphorylation, as well as nearby proline (P284) and isoleucine (I290) residues. T283, P284, and I290 are conserved among cyclin D3 homologs in different species and are part of a phosphorylation motif that regulates D-type cyclin phosphorylation and stability (Diehl et al. 1997). In addition, BL tumors acquire various nonsense and frameshift mutations that remove up to 41 amino acids from the cyclin D3 carboxyl terminus, including this phosphorylation motif (Richter et al. 2012; Schmitz et al. 2012). The CCND3 mutations occur in 38% of sBL tumors but are significantly less prevalent among eBL samples (2.6%), indicating a distinct genetic pathogenesis for this BL subtype. The dearth of CCND3 mutations in eBL tumors raises the possibility that another D-type cyclin could be induced by EBV, thus removing the selective pressure for CCND3 mutations. The BL-derived cyclin D3 mutant isoforms accumulate to more than 10-fold higher levels than the wild-type isoform, suggesting an increase in protein stability (Schmitz et al. 2012). Consistently, proteasome inhibition increased the abundance of wild-type cyclin D3 but did not further stabilize the mutant isoforms (Fig. 2A). These findings are supported by previous work linking T283 to cyclin D3 protein stability (Casanovas et al. 2004) and further suggesting that P284 and I290 are also involved in cyclin D3 protein turnover. The degradation of cyclin D3 involves phosphorylation of T283, although the nature of the kinase is unclear. In keeping with this model, treatment of BL cells with the pan-phosphatase inhibitor okadaic acid, which generally increases protein phosphorylation, causes a marked destabilization of wild-type cyclin D3 but not mutant cyclin D3 (Fig. 2B). D-type cyclins accumulate in G1 phase and are destabilized during S phase of the cell cycle. When BL cells are trapped in G1 phase by treatment with the CDK-4/CDK-6 inhibitor PD 0332991, wild-type cyclin D3 levels increase, whereas mutant cyclin D3 isoforms are not further stabilized (Fig. 2C). Hence, the CCND3 mutations serve to uncouple cyclin D3 from the regulatory processes that govern its abundance in normal cells. The G1–S phase transition is additionally regulated by the cyclin-dependent kinase inhibitor p16, encoded by the CDKN2A gene. BLs harbor deletions or inactivating mutations in CDKN2A in 17% of samples, with 8% having both CCND3 and CDKN2A mutations, suggesting that these genetic lesions might cooperatively deregulate the cell cycle in BL (Schmitz et al. 2012).
Figure 2.
CCND3 mutations cause increased protein stability. FACS analysis of lymphoma cell lines transduced with mutant (T283A) or wild-type GFP-CCND3 fusion proteins (Schmitz et al. 2012). (A) Mutant CCND3 proteins are not degraded by the proteasome. GFP-CCND3 transduced HBL-1 (DLBCL) cells were cultured overnight in the presence of 20 nm the proteasomal inhibitor bortezomib (PS-341) and analyzed by FACS. (B) Mutant CCND3 proteins are not destabilized in response to phosphatase inhibition. Gumbus (BL) cells expressing GFP-CCND3 proteins were treated for 30 min with 750 nm pan-phosphatase inhibitor okadaic acid and analyzed by FACS. (C) The stability of mutant CCND3 proteins is not regulated in the cell cycle. Gumbus (BL) cells expressing GFP-CCND3 isoforms were treated overnight by addition of 1.5 mm the CDK-4/6 inhibitor PD 0332991. FACS analysis indicated stabilization of wild-type but not mutant CCND3 in the G1 phase of the cell cycle.
Highly recurrent mutations affecting the transcription factor TCF-3 (E2A) and its negative regulator ID3 reveal an important aspect of BL biology that is shared by its normal counterpart, the centroblast. All three subtypes of BL harbor highly recurrent mutations in the TCF-3 gene (10%–25%) and/or in its negative regulator ID3 (35%–58%). Altogether, 70% of sBL cases have mutations that disrupt this regulatory duo. These mutations are virtually absent in DLBCL, suggesting that the TCF-3/ID3 module plays a defining role in BL pathogenesis (Love et al. 2012; Richter et al. 2012; Schmitz et al. 2012). In certain cases, the distinction between BL and DLBCL can be difficult or impossible using current diagnostic methods (Dave et al. 2006). It is likely that diagnostic accuracy could be improved by incorporating a test for TCF-3 and ID3 mutations.
TCF-3 is a basic helix-loop-helix (B-HLH) transcription factor that homodimerizes via its HLH domain and uses its basic region to contact DNA within the major groove of DNA (Murre et al. 1989). TCF-3 DNA binding is inhibited by heterodimerization with ID3, an HLH protein that lacks the basic region (for review, see Kee 2009). A variety of nonsense and frameshift mutations in ID3 as well as missense mutations in the region encoding the HLH domain reduce or eliminate ID3 protein function in BL. The missense mutations destabilize ID3 protein and partially or totally block its interaction with TCF-3 (Schmitz et al. 2012). The TCF-3 mutations in BL affect the B-HLH domain of the alternatively spliced E47 isoform of TCF-3 while sparing the other alternatively spliced isoform, E12, revealing a nonredundant role for these two isoforms in BL. E47 and E12 differ only in their DNA-binding B-HLH domains and have distinct functions during normal B-cell development (Murre et al. 1989; Beck et al. 2009). The affinity of most of these TCF-3 E47 mutants for wild-type ID3 is reduced or absent, suggesting a unifying model in which the TCF-3 and ID3 mutations in BL relieve TCF-3 from the negative influence of ID3 and promote its constitutive activity as a transcription factor.
One recurrent TCF-3 mutant that does not conform to this paradigm is N551K, which affects a residue in the basic region of TCF-3 that contacts DNA. Genome-wide chromatin precipitation analysis revealed that N551K shares many of the same target genes with wild-type TCF-3 (Schmitz et al. 2012). Nonetheless, a small subset of genes was differentially bound by N551K and wild-type TCF-3. Indeed, the differentially bound sites in the genome harbored distinct variants of the E-box to which TCF-3 binds, with N551K preferring the perfectly palindromic 5′-CAGCTG-3′ motif and wild-type TCF-3 preferring the near palindromic 5′-CACCTG-3′ motif. One notable target gene that is bound and repressed by wild-type but not N551K TCF-3 is AKT1, encoding a kinase in the PI3K pathway (Schmitz et al. 2012). Hence, it is possible that the N551K mutant is selected in BL to aid and abet PI3K signaling, which is central to BL pathogenesis (see below).
All BL cell lines depend on TCF-3 for survival and proliferation, including those without mutations in TCF-3/ID3, confirming the central role for this transcription factor in BL pathogenesis (Richter et al. 2012; Schmitz et al. 2012). The transcriptional program regulated by TCF-3 in BL includes many genes that account for the distinct gene expression profiles of BL and DLBCL (Dave et al. 2006). Moreover, the expression of TCF-3-dependent genes readily distinguishes normal germinal center B cells from blood B cells, supporting the notion that TCF-3 contributes to the BL phenotype by enforcing a GC-derived transcriptional program. In this regard, it is important to note that conditional ablation of TCF-3 in GC B cells impairs GC formation (Kwon et al. 2008).
As mentioned above, the gene expression signature of BL resembles that of a normal centroblasts, and TCF-3 is one of the few transcriptional regulators showing higher gene expression in centroblasts than centrocytes in both human and mouse GCs (Victora et al. 2012). To investigate whether TCF-3 might serve as a master regulator of the centroblast transcriptional program, we performed gene set enrichment analysis (Subramanian et al. 2005) to assess whether genes that are activated or repressed by TCF-3 in BL are differentially expressed in centroblasts and centrocytes. Centroblast signature genes were enriched among TCF-3-activated genes (Wilcoxon p value < 1 × 10−40), and centrocyte signature genes were enriched among TCF-3-repressed genes (Wilcoxon p value = 4 × 10−13) (Fig. 3A,B). Hence, TCF-3 controls a centroblast-restricted gene expression signature that is “inherited” by BL cells and intensified in cases with TCF-3/ID3 aberrations (Schmitz et al. 2012).
Figure 3.
Gene set enrichment analysis of TCF-3 target genes. TCF-3-dependent genes were defined by gene expression profiling in three BL cell lines (BL41, Daudi, and Defauw) following TCF-3 knockdown (day 1 and day 2) or wild-type (wt) ID3 overexpression (day 1 and day 2) (Schmitz et al. 2012). To rank genes based on their TCF-3 dependency, a one-sample RVM t-test model (Wright and Simon 2003) was used to calculate p values against the null hypothesis that the average log-ratio was zero. Kolmogorov-Smirnov curves were generated for centroblast (A) and centrocyte (B) gene signatures (Victora et al. 2012), based on the ordering of BL gene expression data by their one-sided p values. Enrichment results were calculated by comparing the proportion of genes with one-sided p values < 0.01 inside and outside the centroblast and centrocytes signatures.
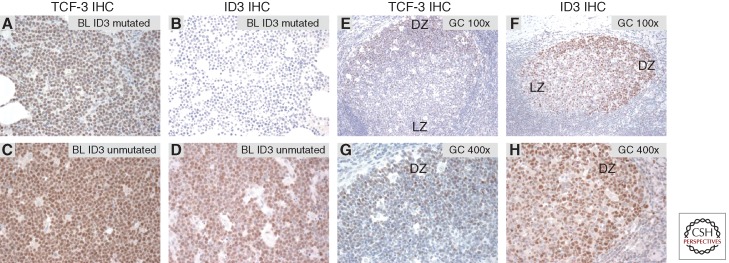
This analysis suggests that TCF-3 is a defining transcriptional regulator of BL and of normal centroblasts. TCF-3 target genes dictate much of BL biology. TCF-3 directly transactivates CCND3, thereby promoting cell-cycle progression. It also induces IG expression while repressing PTPN6, thereby promoting B-cell receptor (BCR) signaling (see below). Interestingly, TCF-3 also transactivates ID3 as well as the related family members ID1 and ID2, thus inducing expression of its own negative regulators. Given the selective acquisition of mutations in ID3 and not ID1 or ID2 in BL, it appears that the TCF-3-ID3 regulatory duo is of greatest functional importance. Immunohistochemical analysis of TCF-3 and ID3 expression revealed that BL tumors with wild-type ID3 uniformly expressed both proteins, whereas tumors with ID3 mutations expressed TCF-3 but showed no detectable ID3 protein (Fig. 4A–D). As expected from the analysis of GC gene expression signatures, immunohistochemical analysis of normal human tonsils revealed that both TCF-3 and ID3 protein were more highly expressed in centroblasts in the DZ than in centrocytes in the LZ (Fig. 4E–H). Future work should focus on the dynamics of TCF-3 regulation in the GC reaction. It is conceivable that the TCF-3–ID3 negative regulatory loop is part of an intrinsic “clock” mechanism that regulates the transition from the centroblast to the centrocyte phenotype.
Figure 4.
(A–D) TCF-3 and ID3 immunohistochemistry could be used as an additional diagnostic tool for BL. (E–H) TCF-3 and its target gene ID3 are specifically expressed in the dark zone (DZ) of the germinal center. (A–D) Immunohistochemical stainings of BL for TCF-3 (Santa Cruz, SC-349) and ID3 (CalBioReagents, clone 17-3). (A,C) TCF-3 is highly expressed in BL. Burkitt lymphomas harboring ID3 mutations (R28*; L70P) do not express ID3 protein (B), whereas ID3 is detectable in BL without ID3 mutations (D). (E–H) Immunohistochemistry for TCF-3 and ID3 in reactive secondary B follicles. Both TCF-3 and ID3 are predominantly expressed in germinal center centroblasts. Original magnification (E,F) 100×, (G,H) 400×. Dark and light zone distinction was performed based on histology using hematoxylin and eosin staining and immunohistochemistry for KI67. DZ, Dark zone; LZ, light zone.
ONCOGENIC SIGNALING PATHWAYS IN BL
The translocation of MYC in BL occurs exclusively to the nonproductively rearranged IG heavy-chain locus, sparing the allele that produces the productive IG heavy chain that is used to construct the BCR (Küppers 2005). This observation suggests that BL may rely on BCR signaling. Moreover, transgenic mouse models suggested a role of BCR signaling in MYC-driven lymphomagenesis (Refaeli et al. 2008). Crossing antigen-specific BCR transgenic mice with MYC transgenic mice affects the rate and outcome of lymphomagenesis initiated by MYC. Conceptually, BCR signaling can be subdivided into “tonic” BCR signaling, which engages the PI3K pathway, and antigen-dependent “active” BCR signaling, which engages multiple pathways, including NF-κB (for review, see Young and Staudt 2013). Compared with other aggressive lymphomas, BL tumors lack expression of NF-κB target genes, suggesting that active BCR signaling does not contribute to this malignancy (Dave et al. 2006). In fact, MYC transgenic mouse models showed that constitutive NF-κB activity is incompatible with the development of MYC-induced lymphomas (Klapproth et al. 2009). The involvement of PI3K signaling in BL pathogenesis was recently supported by analysis of mice engineered to express MYC and a constitutively active form of PI3K specifically in B cells undergoing the GC reaction (Sander et al. 2012). The resulting clonal lymphomas strongly resemble BL by their histology and gene expression profile. Of particular note is the fact that these mouse lymphomas accumulate mutations affecting the phosphorylation motif of cyclin D3 that are also present in human BL (see above), indicating that this mouse model is a faithful mimic of human BL.
A primary source of PI3K signaling in human BL is tonic BCR signaling. In roughly two-thirds of BL cell lines, knockdown of BCR subunits or the BCR-associated tyrosine kinase SYK decreased PI3K activity, as judged by phospho-AKT, and induced apoptosis (Schmitz et al. 2012). The BCR signaling in these BL lines appears to be “tonic” in that inhibition of the NF-κB pathway had no effect on cell viability. PI3K activity in BL is also dependent on TCF-3, suggesting a nexus between oncogenic activation of this transcription factor in BL and tonic BCR signaling (Schmitz et al. 2012). The constitutive activity of TCF-3 in BL fosters dependence on tonic BCR signaling by two mechanisms. First, TCF-3 directly up-regulates BCR expression. TCF-3 was originally identified as a transcription factor (E2A) that binds to regulatory “E-box” motifs in IG heavy- and light-chain enhancers (Murre et al. 1989). In BL, TCF-3 is present at the IG heavy-chain 3′ enhancer, up-regulates expression of the IG heavy and light chains, and increases expression of the BCR on the cell surface (Schmitz et al. 2012). A second mechanism by which TCF-3 augments BCR signaling is by negatively regulating PTPN6, encoding the SHP-1 phosphatase. SHP-1 attenuates BCR signaling by dephosphorylating the ITAM motifs of the CD79A and CD79B signaling subunits of the BCR.
Several additional mechanisms enforce PI3K signaling in BL. MYC contributes in the activation of this prosurvival pathway by inducing expression of the mir-17-92 cluster. Mir-19a and mir-19b reduce expression of PTEN, which counteracts PI3K activity (Xiao et al. 2008; Olive et al. 2009). In a minority of BL cases (7%), PI3K signaling is increased by classical inactivating mutations of PTEN, providing direct genetic evidence that this pathway is key to BL pathophysiology. The reason that PTEN mutations are not more frequent in BL is presumably because tonic BCR signaling and MYC-dependent mir-19 expression mitigate the selective pressure for these genetic alterations. Gene expression signatures of PI3K activity are more highly expressed in human BL biopsies than in other lymphoma subtypes (Sander et al. 2012; Schmitz et al. 2012). This evidence, together with the BL-like phenotype of the conditional MYC–PI3K transgenic mice, strongly suggests that PI3K activity plays a central role in the development and ongoing maintenance of BL.
THERAPEUTIC IMPLICATIONS
Using intensive chemotherapy regimens, BL is often curable (Yustein and Dang 2007). However, the rigors of these BL regimens are often not tolerated by older individuals. Moreover, these regimens suppress the bone marrow and consequently the immune response, placing a premium on supportive care, which is required to detect and treat infections promptly and effectively. This subsequent problem precludes the delivery of these curative regimens in resource-poor environments such as in areas of Africa where BL is endemic (Orem et al. 2008). Consequently, children with BL in Africa often receive lower doses of chemotherapy, which are not as curative as the intensive regimens used in developed countries, leading to an overall cure rate of only 30%–50%. Hence, the development of new treatment regimens that are less immunosuppressive and better tolerated is urgently needed for BL.
Clearly, PI3K signaling inhibitors deserve clinical evaluation in BL. This pathway can be effectively blocked in BL cell lines by the PI3K inhibitor BKM-120, which targets all of the catalytic isoforms of PI3K (Schmitz et al. 2012). Alternatively, rapamycin or its analogs could be used to inhibit the mTORC1 kinase complex, activated by PI3K signaling. Because tonic BCR signaling activates PI3K in a substantial fraction of BL lines, agents that target BCR-proximal kinases might be effective, including inhibitors of SYK (e.g., fostamatinib) or SRC-family kinases (e.g., dasatinib). All of these PI3K-targeting agents are either approved or have been shown to be safe in clinical trials, suggesting that one or more should be evaluated in BL.
The oncogenic activation of cyclin D3 presents another promising target for therapy in BL. Cyclin D3 pairs with CDK-6 to form an active kinase in BL, and knockdown of either subunit is toxic for these cells (Schmitz et al. 2012). Treatment with the CDK-4/6 inhibitor PD 0332991 initially blocks BL cell-cycle progression at the G1–S phase transition, as expected. However, unexpectedly, continuous exposure of BL cells to this agent triggers profound apoptosis. The mechanistic basis for this effect is unclear, but it seems likely that the combination of enforced MYC expression by the t(8;14) translocation with a complete G1 block may trigger a checkpoint response that results in apoptosis. The effect of PD 0332991 against established BL xenografts is impressive, with virtual disappearance of tumor cells by day 10 of treatment (Schmitz et al. 2012). PD 0332991 has been safely administered in early-phase clinical trials and could be evaluated in BL.
Agents that directly target the two most important oncogenic transcription factors in BL—MYC and TCF-3—would be highly rational therapies for this lymphoma subtype. Until recently, transcription factors were deemed to be intractable drug targets. Although small-molecule inhibitors of TCF-3 have yet to be described, chemical inhibitors of MYC expression have been identified. MYC expression requires the action of the BET family of chromatin readers, which can be prevented from binding to chromatin by small-molecule inhibitors that prevent their interaction with acetylated lysines in histone tails (Filippakopoulos et al. 2010; Delmore et al. 2011; Zuber et al. 2011). Indeed, BET inhibitors kill BL cell lines and down-regulate their MYC expression (Mertz et al. 2011). Although MYC undoubtedly plays roles in normal proliferating cells, it is possible that the addiction of BL tumors to MYC may provide an acceptable therapeutic window for the development of BET inhibitors in this disease.
ACKNOWLEDGMENTS
We thank Moez Dawood for helpful discussion. R.S. is supported by the Dr. Mildred Scheel Stiftung für Krebsforschung (Deutsche Krebshilfe).
Footnotes
Editors: Chi V. Dang and Robert N. Eisenman
Additional Perspectives on MYC and the Pathway to Cancer available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL 1985. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318: 533–538 [DOI] [PubMed] [Google Scholar]
- Basso K, Dalla-Favera R 2012. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev 247: 172–183 [DOI] [PubMed] [Google Scholar]
- Beck K, Peak MM, Ota T, Nemazee D, Murre C 2009. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J Exp Med 206: 2271–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkitt D 1958. A sarcoma involving the jaws in African children. Br J Surg 46: 218–223 [DOI] [PubMed] [Google Scholar]
- Burkitt DP 1961. Observations on the geography of malignant lymphoma. East Afr Med J 38: 511–514 [PubMed] [Google Scholar]
- Burkitt D 1962. A children’s cancer dependent on climatic factors. Nature 194: 232–234 [DOI] [PubMed] [Google Scholar]
- Calado DP, Sasaki Y, Godinho SA, Pellerin A, Kochert K, Sleckman BP, de Alboran IM, Janz M, Rodig S, Rajewsky K 2012. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol 13: 1092–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanovas O, Jaumot M, Paules AB, Agell N, Bachs O 2004. P38SAPK2 phosphorylates cyclin D3 at Thr-283 and targets it for proteasomal degradation. Oncogene 23: 7537–7544 [DOI] [PubMed] [Google Scholar]
- Chene A, Donati D, Guerreiro-Cacais AO, Levitsky V, Chen Q, Falk KI, Orem J, Kironde F, Wahlgren M, Bejarano MT 2007. A molecular link between malaria and Epstein-Barr virus reactivation. PLoS Pathog 3: e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM 1982. Human c-myc oncogene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci 79: 7824–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalldorf G, Linsell CA, Barnhart FE, Martyn R 1964. An epidemiologic approach to the lymphomas of African children and Burkitt’s Sarcoma of the jaws. Perspect Biol Med 7: 435–449 [DOI] [PubMed] [Google Scholar]
- Dang CV 2012. MYC on the path to cancer. Cell 149: 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave SS, Fu K, Wright GW, Lam LT, Kluin P, Boerma EJ, Greiner TC, Weisenburger DD, Rosenwald A, Ott G, et al. 2006. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med 354: 2431–2442 [DOI] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Zindy F, Sherr CJ 1997. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin–proteasome pathway. Genes Dev 11: 957–972 [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Victora GD, Ying CY, Phan RT, Saito M, Nussenzweig MC, Dalla-Favera R 2012. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol 13: 1083–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati D, Espmark E, Kironde F, Mbidde EK, Kamya M, Lundkvist A, Wahlgren M, Bejarano MT, Falk KI 2006. Clearance of circulating Epstein-Barr virus DNA in children with acute malaria after antimalaria treatment. J Infect Dis 193: 971–977 [DOI] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC 1992. Induction of apoptosis in fibroblasts by c-Myc protein. Cell 69: 119–128 [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, Cleveland JL, Tansey WP, Lowe SW 2005. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature 436: 807–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, Bernd HW, Cogliatti SB, Dierlamm J, Feller AC, et al. 2006. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med 354: 2419–2430 [DOI] [PubMed] [Google Scholar]
- Illingworth J, Butler NS, Roetynck S, Mwacharo J, Pierce SK, Bejon P, Crompton PD, Marsh K, Ndungu FM 2013. Chronic exposure to Plasmodium falciparum is associated with phenotypic evidence of B and T cell exhaustion. J Immunol 190: 1038–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MS, Soni V, Bronson R, Kieff E 2008. Epstein-Barr virus nuclear antigen 1 does not cause lymphoma in C57BL/6 J mice. J Virol 82: 4180–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee BL 2009. E and ID proteins branch out. Nat Rev Immunol 9: 175–184 [DOI] [PubMed] [Google Scholar]
- Klapproth K, Sander S, Marinkovic D, Baumann B, Wirth T 2009. The IKK2/NF-κB pathway suppresses MYC-induced lymphomagenesis. Blood 114: 2448–2458 [DOI] [PubMed] [Google Scholar]
- Küppers R 2005. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer 5: 251–262 [DOI] [PubMed] [Google Scholar]
- Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, Busslinger M 2008. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 28: 751–762 [DOI] [PubMed] [Google Scholar]
- *.Levens D 2013. Cellular MYCro economics: Balancing MYC function with MYC expression. Cold Spring Harb Perspect Med 3: a014233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, et al. 2012. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet 44: 1321–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan IC 1994. Germinal centers. Annu Rev Immunol 12: 117–139 [DOI] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ III 2011. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci 108: 16669–16674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N, Kim SS, Penn LZ 2006. The Oscar-worthy role of Myc in apoptosis. Semin Cancer Biol 16: 275–287 [DOI] [PubMed] [Google Scholar]
- Morrow RH, Kisuule A, Pike MC, Smith PG 1976. Burkitt’s lymphoma in the Mengo Districts of Uganda: Epidemiologic features and their relationship to malaria. J Natl Cancer Inst 56: 479–483 [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56: 777–783 [DOI] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151: 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L 2009. miR-19 is a key oncogenic component of mir-17–92. Genes Dev 23: 2839–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orem J, Mbidde EK, Weiderpass E 2008. Current investigations and treatment of Burkitt’s lymphoma in Africa. Trop Doct 38: 7–11 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Guglielmino R, Houldsworth J, Mohr J, Aoufouchi S, Polakiewicz R, Chaganti RS, Dalla-Favera R 2004. Expression of the AID protein in normal and neoplastic B cells. Blood 104: 3318–3325 [DOI] [PubMed] [Google Scholar]
- Peled JU, Yu JJ, Venkatesh J, Bi E, Ding BB, Krupski-Downs M, Shaknovich R, Sicinski P, Diamond B, Scharff MD, et al. 2010. Requirement for cyclin D3 in germinal center formation and function. Cell Res 20: 631–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Rahl PB, Young RA 2014. MYC and transcription elongation. Cold Spring Harb Perspect Med 10.1101/cshperspect.a020990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC 2004. AID is required for c-myc/IgH chromosome translocations in vivo. Cell 118: 431–438 [DOI] [PubMed] [Google Scholar]
- Refaeli Y, Young RM, Turner BC, Duda J, Field KA, Bishop JM 2008. The B cell antigen receptor and overexpression of MYC can cooperate in the genesis of B cell lymphomas. PLoS Biol 6: e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J, Schlesner M, Hoffmann S, Kreuz M, Leich E, Burkhardt B, Rosolowski M, Ammerpohl O, Wagener R, Bernhart SH, et al. 2012. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet 44: 1316–1320 [DOI] [PubMed] [Google Scholar]
- Rowe M, Kelly GL, Bell AI, Rickinson AB 2009. Burkitt’s lymphoma: The Rosetta Stone deciphering Epstein-Barr virus biology. Semin Cancer Biol 19: 377–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L, Schmitz R, Ceribelli M, Staudt LM 2011. Malignant pirates of the immune system. Nat Immunol 12: 933–940 [DOI] [PubMed] [Google Scholar]
- Sander S, Calado DP, Srinivasan L, Kochert K, Zhang B, Rosolowski M, Rodig SJ, Holzmann K, Stilgenbauer S, Siebert R, et al. 2012. Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell 22: 167–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, Wright G, Shaffer AL, Hodson DJ, Buras E, et al. 2012. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 490: 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, Aaronson S, Leder P 1982. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci 79: 7837–7841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorley-Lawson DA 2001. Epstein-Barr virus: Exploiting the immune system. Nat Rev Immunol 1: 75–82 [DOI] [PubMed] [Google Scholar]
- Vereide DT, Sugden B 2011. Lymphomas differ in their dependence on Epstein-Barr virus. Blood 117: 1977–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC 2012. Germinal centers. Annu Rev Immunol 30: 429–457 [DOI] [PubMed] [Google Scholar]
- Victora GD, Dominguez-Sola D, Holmes AB, Deroubaix S, Dalla-Favera R, Nussenzweig MC 2012. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood 120: 2240–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JB, Bell JL, Levine AJ 1996. Expression of Epstein-Barr virus nuclear antigen-1 induces B cell neoplasia in transgenic mice. EMBO J 15: 3117–3126 [PMC free article] [PubMed] [Google Scholar]
- Wright GW, Simon RM 2003. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 19: 2448–2455 [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K 2008. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat Immunol 9: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RM, Staudt LM 2013. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat Rev Drug Discov 12: 229–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yustein JT, Dang CV 2007. Biology and treatment of Burkitt’s lymphoma. Curr Opin Hematol 14: 375–381 [DOI] [PubMed] [Google Scholar]
- Zech L, Haglund U, Nilsson K, Klein G 1976. Characteristic chromosomal abnormalities in biopsies and lymphoid-cell lines from patients with Burkitt and non-Burkitt lymphomas. Int J Cancer 17: 47–56 [DOI] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]