Human and simian immunodeficiency viruses (HIV and SIVs) contain several auxiliary genes not found in other retroviruses. These genes are thought to be functionally important for optimal viral replication and persistence in infected individuals. Primate lentiviruses can be classified by the composition of these accessory genes. While viruses of the HIV type1 (HIV-1) group have vif, vpr, vpu, and nef genes, those of the HIV-2 group carry vif, vpx, vpr, and nef genes (Fujita et al., 2010). Vpx protein encoded by the vpx gene is unique to non-HIV-1 viruses, and is essential for viral replication in macrophages in contrast to its structural paralog Vpr (Fujita et al., 2010). The most outstanding sequence feature to distinguish Vpx from Vpr is the presence of poly-proline motif (PPM) at its C-terminal region. We have recently shown, by in vitro and in vivo assay systems, that the PPM in HIV-2 Vpx is critical for its efficient translation (Miyake et al., 2014).
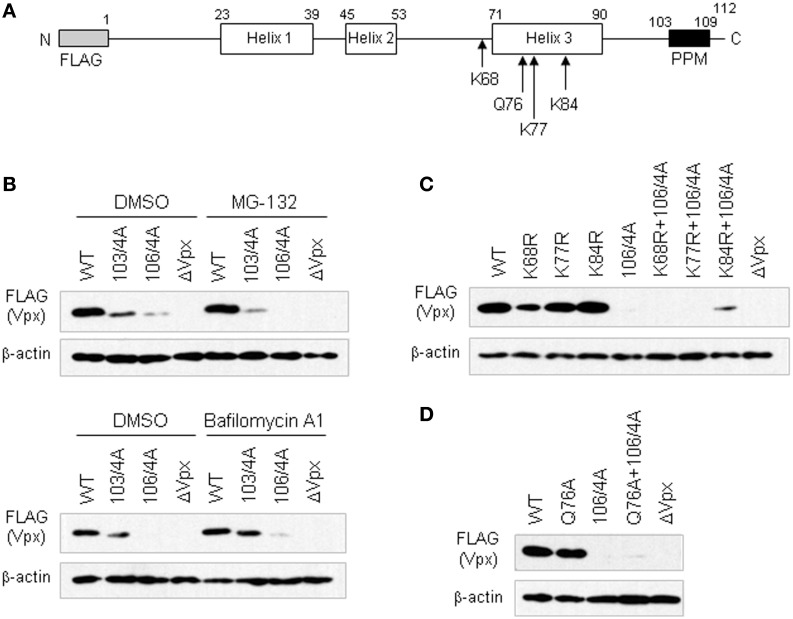
Although PPM consisting of seven consecutive prolines has been demonstrated to be required for efficient HIV-2 Vpx translation, thereby acquiring viral infectivity in macrophages, the effects of PPM mutations on the degradation of Vpx in cells was not formally analyzed as yet (Fujita et al., 2008; Miyake et al., 2014). Therefore, in this study, we asked whether the PPM plays a role in keeping away from proteasomal and/or lysosomal degradation (Figure 1). In order to assess this, we used various expression plasmids for HIV-2 Vpx (pEF-Fvpx series) described in a previous study (Miyake et al., 2014): wild-type (WT) plasmid has the vpx gene derived from HIV-2 GL-AN clone (Kawamura et al., 1994); mutants 103/4A and 106/4A have four consecutive alanine-substitutions at the site of P103-P106 and P106-P109, respectively, and have been shown to express a low/minimum level of mutant Vpx proteins in cells (Figure 1A); a negative control is a frame-shift mutant pEF-FxSt that lacks Vpx expression (ΔVpx).
Figure 1.
Steady-state levels of various Vpx-PPM mutants in cells as monitored by Western blotting. (A) Structure of the FLAG-tagged HIV-2 GL-AN Vpx construct. Numerals above the schema represent amino acid numbers of the Vpx protein. Positions of lysine and glutamine residues mutated are indicated. (B) Expression of Vpx-PPM mutants in the presence of a proteasome inhibitor MG-132 or a lysosome inhibitor Bafilomycin A1. (C) Expression of lysine-mutants with or without 106/4A mutation. (D) Expression of Q76A mutants with or without 106/4A mutation. For (B) to (D) experiments, 293T cells were transfected with the plasmids indicated, and harvested for Western blotting 24 h later. To examine lysosomal and proteasomal degradation processes (B), 100 nM of BafilomycinA1 (Yoshimori et al., 1991) and 7.5 μM of MG-132 (McCulley and Ratner, 2012) were added at 5 and 16 h post-transfection, respectively. WT, pEF-Fvpx; ΔVpx, pEF-FxSt.
Various expression plasmids were transfected into human 293T cells (Lebkowski et al., 1985) as described before (Adachi et al., 1986), and the amounts of WT and mutant Vpx proteins produced in cells in the absence or presence of a proteasome inhibitor MG-132 (Fujita et al., 2004; McCulley and Ratner, 2012) were comparatively examined by Western blotting (Miyake et al., 2014). A drastic reduction in Vpx expression was observed for mutants 103/4A and 106/4A, 106/4A in particular, both in the absence and presence of MG-132 (Figure 1B). These results showed that neither of these mutants could be rescued with MG-132, suggesting no involvement of the PPM in the proteasome-mediated degradation. Similarly, a lysosome inhibitor Bafilomycin A1 (Yoshimori et al., 1991) did not affect much the level of 103/4A and 106/4A in transfected 293T cells, although a small increase was observed for both mutants (Figure 1B). These results suggested that the low expression level of these PPM mutants may not be attributable to the lysosomal degradation.
Proteasomal degradation is generally triggered by the polyubiquitin modification of lysine residues in a protein. There are three lysines in the Vpx of HIV-2 GL-AN clone (Khamsri et al., 2006) (Figure 1A). We generated several clones carrying mutations in these residues. Furthermore, we focused on the 76th glutamine residue (Figure 1A). This amino acid has been reported to interact with DCAF1 for formation of Cullin4-based E3 ubiquitin ligase complex to degrade an anti-HIV restriction factor SAMHD1 (Hrecka et al., 2011; Laguette et al., 2011) by proteasome (Le Rouzic et al., 2007; Srivastava et al., 2008). Mutants K68R, K77R, K84R, and Q76A with or without the 106/4A mutation were constructed as described previously (Miyake et al., 2014) (Figure 1A), and examined for their expression in transfected cells (Figures 1C,D). As shown in Figure 1C, only one clone with K84R and 106/4A mutations showed a slight enhancement in agreement with a previous report (Srivastava et al., 2008). Moreover, no significant effect was observed for a mutant carrying Q76A and 106/4A mutations (Figure 1D). These results also suggested that PPM may not be associated with the proteasome-mediated degradation.
In total, proteasomal or lysosomal degradation does not account for the extremely low expression level of Vpx exhibited by the PPM mutants. This is consistent with our previous conclusion that PPM is critical for efficient translation of Vpx (Miyake et al., 2014). Molecular mechanism by which PPM enhances Vpx translation to a remarkable extent needs to be determined.
Acknowledgments
This study was supported in part by a grant from the Ministry of Health, Labour and Welfare of Japan (Research on HIV/AIDS project no. H24-005).
References
- Adachi A., Gendelman H. E., Koenig S., Folks T., Willey R., Rabson A., et al. (1986). Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Akari H., Sakurai A., Yoshida A., Chiba T., Tanaka K., et al. (2004). Expression of HIV-1 accessory protein Vif is controlled uniquely to be low and optimal by proteasome degradation. Microbes Infect. 6, 791–798 10.1016/j.micinf.2004.04.011 [DOI] [PubMed] [Google Scholar]
- Fujita M., Otsuka M., Nomaguchi M., Adachi A. (2008). Functional region mapping of HIV-2 Vpx protein. Microbes Infect. 10, 1387–1392 10.1016/j.micinf.2008.08.005 [DOI] [PubMed] [Google Scholar]
- Fujita M., Otsuka M., Nomaguchi M., Adachi A. (2010). Multifaceted activity of HIV Vpr/Vpx proteins: the current view of their virological functions. Rev. Med. Virol. 20, 68–76 10.1002/rmv.636 [DOI] [PubMed] [Google Scholar]
- Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., et al. (2011). Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 10.1038/nature10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura M., Sakai H., Adachi A. (1994). Human immunodeficiency virus Vpx is required for the early phase of replication in peripheral blood mononuclear cells. Microbiol. Immunol. 38, 871–878 10.1111/j.1348-0421.1994.tb02140.x [DOI] [PubMed] [Google Scholar]
- Khamsri B., Murao F., Yoshida A., Sakurai A., Uchiyama T., Shirai H., et al. (2006). Comparative study on the structure and cytopathogenic activity of HIV Vpr/Vpx proteins. Microbes Infect. 8, 10–15 10.1016/j.micinf.2005.05.020 [DOI] [PubMed] [Google Scholar]
- Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Segeral E., et al. (2011). SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 10.1038/nature10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebkowski J. S., Clancy S., Calos M. P. (1985). Simian virus 40 replication in adeno-virus-transformed human cells antagonizes gene expression. Nature 317, 169–171 10.1038/317169a0 [DOI] [PubMed] [Google Scholar]
- Le Rouzic E., Belaidouni N., Estrabaud E., Morel M., Rain J. C., Transy C., et al. (2007). HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6, 182–188 10.4161/cc.6.2.3732 [DOI] [PubMed] [Google Scholar]
- McCulley A., Ratner L. (2012). HIV-2 viral protein X (Vpx) ubiquitination is dispensable for ubiquitin ligase interaction and effects on macrophage infection. Virology 427, 67–75 10.1016/j.virol.2012.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake A., Fujita M., Fujino H., Koga R., Kawamura S., Otsuka M., et al. (2014). Poly-proline motif in HIV-2 Vpx is critical for its efficient translation. J. Gen. Virol. 95, 179–189 10.1099/vir.0.057364-0 [DOI] [PubMed] [Google Scholar]
- Srivastava S., Swanson S. K., Manel N., Florens L., Washburn M. P., Skowronski J. (2008). Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 4:e1000059 10.1371/journal.ppat.1000059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T., Yamamoto A., Moriyama Y., Futai M., Tashiro Y. (1991). Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 266, 17707–17712 [PubMed] [Google Scholar]