Abstract
Common mycorrhizal networks (CMNs) link multiple plants together. We hypothesized that CMNs can serve as an underground conduit for transferring herbivore-induced defence signals. We established CMN between two tomato plants in pots with mycorrhizal fungus Funneliformis mosseae, challenged a ‘donor' plant with caterpillar Spodoptera litura, and investigated defence responses and insect resistance in neighbouring CMN-connected ‘receiver' plants. After CMN establishment caterpillar infestation on ‘donor' plant led to increased insect resistance and activities of putative defensive enzymes, induction of defence-related genes and activation of jasmonate (JA) pathway in the ‘receiver' plant. However, use of a JA biosynthesis defective mutant spr2 as ‘donor' plants resulted in no induction of defence responses and no change in insect resistance in ‘receiver' plants, suggesting that JA signalling is required for CMN-mediated interplant communication. These results indicate that plants are able to hijack CMNs for herbivore-induced defence signal transfer and interplant defence communication.
To adapt to natural enemies including pathogens and herbivores plants have evolved various mechanisms to perceive enemies and to respond to their attacks by rapid and dynamic alterations in morphology, physiology and biochemistry. In response to insect herbivores plants use both constitutive defence that is constantly presented, and induced defence that is only activated upon attack1,2,3. The induced defence is often associated with the production of defensive compounds such as secondary metabolites that are toxic, repellent, or antinutritive for herbivores2,3,4,5, proteinase inhibitors that affect insect feeding1, or volatiles that either repel herbivores or recruit parasitoids and/or predators of the herbivores feeding on the plant6,7,8.
Plant defensive traits are costly, and induced defences minimize the cost by eliciting defence only when necessary. Although the induced defences allow plants to avoid the defence costs in the absence of enemies, plants may suffer considerable damage during the time required to mount defences once an insect attack occurs9. To compensate for this vulnerability, some plants have evolved ability to ‘eavesdrop' on plant defence signals from neighbouring plants, a phenomenon called plant-to-plant or interplant communication. These plants perceive the induced volatiles from neighbours that are being attacked by herbivores and hence increase their defence accordingly4,7,8,10,11. However, almost all interplant communication studies are focusing on aboveground air transfer of signals from a sender to a receiver whilst studies on belowground interplant communication are scarce.
Arbuscular mycorrhizas (AMs), symbiotic associations between AM fungi (AMF) and roots of ~80% terrestrial plants, play key roles in enhancing plant nutrient acquisition and tolerance to abiotic or biotic stresses12. AMs also enhance plant's defences against the attack from insect herbivores13,14. Furthermore, the same or different mycorrhizal hyphae are able to form belowground common mycorrhizal networks (CMNs) interconnecting roots of different plant individuals, species, genera and families in a plant community15,16,17. Mycorrhizal networks are able to transfer carbon, nitrogen and phosphorus from one plant to another18,19,20, which may have a significant influence on plant performance and resource distribution within plant communities17,19. Resource sharing among neighbouring plants through CMNs raises intriguing possibilities that CMNs may serve as a conduit for defence signal transfer among plants in a community21. Our previous study showed that disease resistance and induced defence signals could be transferred between the healthy and pathogen-infected neighbouring plants22. Recently Barto et al. showed that CMNs facilitated transport of allelochemicals released from a plant to target plants, thereby affecting allelopathic interactions23. However, whether or not CMNs could transfer herbivore-induced defence signals is unknown. Jasmonate (JA) signalling pathway plays an essential role in plant responses to chewing insects24,25 and AM symbiosis26, but its role in interplant communication is undiscovered.
Here we test the hypothesis that plants hijack CMNs for herbivore-induced defence signal transfer between insect-attacked and un-attacked plants. In this study Funneliformis mosseae (syn. Glomus mosseae) was used to establish CMN between tomato plants (Lycopersicon esculentum Mill.), and a leaf-chewing caterpillar common cutworm (Spodoptera litura Fabricius) (Lepidoptera: Noctuidae) was used to attack tomato plants. Transgenic tomato plants deficient in jasmonate biosynthesis and perception were analyzed to determine the potential involvement of jasmonate signalling in CMN-mediated interplant communication.
Results
Root colonization and CMN establishment by F. mosseae
No AM fungal colonization was observed in non-inoculated roots of ‘donor' and ‘receiver' tomatoes in treatment B (Table 1). Thirty-five days after F. mosseae inoculation and before insect attack root mycorrhizal colonization was 59.7, 55.4 and 46.4% in the ‘donor' plants in treatments A, C and D, and 38.1, 41.5 and 38.2% in the ‘receiver' plants in treatments A, C and D, respectively (Table 1). In general, mycorrhizal colonization was higher in the ‘donor' than in the ‘receiver' plants. Nested PCR confirmed that both ‘donor' and ‘receiver' tomato plants in treatments A and D were colonized by F. mosseae (Fig. S2), indicating successful CMN establishment between the ‘donor' and ‘receiver' plants.
Table 1. Percentage of root mycorrhizal colonization in ‘donor' and CMN-connected ‘receiver' tomato plants, and weight gain of Spodoptera litura fed on the leaves of these plants.
| Mycorrhizal cononization (%) | Larval weight gain (mg) | |||||
|---|---|---|---|---|---|---|
| Treatment | Inoculation on ‘donor' | CMN | ‘Donor' | ‘Receiver' | ‘Donor' | ‘Receiver' |
| A | Fm + SL | Yes | 59.7 ± 0.8 a | 38.1 ± 3.5 a | 43.6 ± 4.9 b | 48.5 ± 4.9 c |
| B | SL | No | 0 c | 0 b | 72.2 ± 7.2 a | 95.7 ± 7.7 a |
| C | Fm + SL | No | 55.4 ± 1.3 a | 41.5 ± 1.6 a | 55.6 ± 5.8 ab | 61.3 ± 5.2 bc |
| D | Fm | Yes | 46.4 ± 1.7 b | 38.2 ± 1.4 a | Un-infested | 73.0 ± 5.7 b |
Funneliformis mosseae was used to establish common mycorrhizal networks (CMNs) between ‘donor' and ‘receiver' tomato plants. Third instar larvae of S. litura (SL) were used to attack tomato plants. In bioassays the ‘receiver' plants were inoculated with larvae 24 h later than ‘donor' plants. Four treatments included: A) a ‘receiver' plant was connected with a S. litura attacked ‘donor' plant through CMNs; B) a ‘receiver' plant was grown near a SL-attacked ‘donor' plant without mycorrhizal inoculation; C) a mycorrhizal ‘receiver' plant was grown near a SL-attacked mycorrhizal ‘donor' plant but the two tomato plants were separated by water proof membrane; D) a ‘receiver' plant was connected with a neighboring plant by CMNs without insect infestation. Four sets of bioassays were independently carried out and three pots per treatment were set up for each set of bioassays. Values are means ± standard error. Significant differences (P < 0.05 using Tukey post-hoc test) among treatments in the same column are indicated by different letters. Results of ANOVA analyses are presented in the Supporting Information (Table S2).
Effects of CMNs on tomato plant resistance against SL
To test whether CMN-mediated interplant communication can enhance tomato resistance against insect attack, the ‘receiver' plants were inoculated with SL after the CMN establishment and 24 h after the ‘donor plant' had been inoculated with SL. Mycorrhizal colonization significantly enhanced tomato resistance of both ‘donor' and ‘receiver' plants to SL (Table 1), consistent with that AMF colonization improved plant resistance to generalist chewing insects9,10. The SL-larvae fed on mycorrhizal ‘receiver' plants in treatment A, C and D had 48.5, 61.3 and 73.0 mg weight gain, respectively, while those on non-mycorrhizal ‘receiver' plants in treatment B had 95.7 mg weight gain. More interestingly, larvae fed on ‘receiver' plants in treatment A gained significant less weight relative to those in treatment D, suggesting that herbivore attack on ‘donor' plants induces resistance of CMN-connected neighbouring ‘receiver' plants.
Induction of defence-related enzymes in ‘receiver' plants
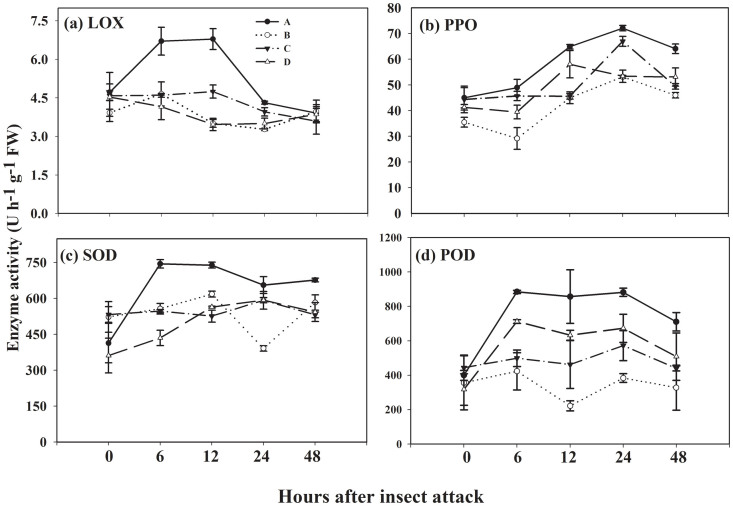
As expected the activity of four leaf defence-related enzymes (LOX, lipoxygenase; POD, peroxidase; PPO, polyphenol oxidase; and SOD, superoxide dismutase) were similar among four treatments at the beginning time point of insect inoculation (Fig. 1). Thereafter, as a general rule, the activity of all four tested enzymes was significantly higher in the ‘receiver' plants of treatment A than that in other three treatments B, C and D. Six hours after insect inoculation, the LOX activity in ‘receiver' tomato plants in treatment A showed a general increase compared with other three treatments (Fig. 1a). The LOX activity displayed increases of 42.6 (P = 0.043), 45.7 (P = 0.038) and 59.5% (P = 0.014), and increases of 94.3 (P < 0.001), 41.7 (P = 0.004) and 94.3% (P < 0.001) compared to that in treatment B, C and D at 6 and 12 h after insect inoculation, respectively. In the other treatment conditions (B, C and D), however, due to the absence of F. mosseae/CMN the activity of LOX was not significantly different.
Figure 1. Levels of four defence-related enzymes in leaves of ‘receiver' tomato plants in response to common mycorrhizal networks (CMNs) connected with insect-infested neighbouring tomatoes.
Funneliformis mosseae was used to establish CMNs between ‘donor' and ‘receiver' tomatoes. Third instar larvae of Spodoptera litura were used to attack tomatoes. Four defence-related enzymes are (a) lipoxygenase (LOX), (b) polyphenol oxidase (PPO), (c) superoxide dismutase (SOD) and (d) peroxidase (POD). Four treatments (A, B, C, D) were set up as described in Table 1. Enzyme activities were analyzed 24 h after insect infestation on ‘donor' plants. Values are means ± standard error from three sets of independent experiments with three pots per treatment for each set of experiments. Significant differences among treatments were tested at P = 0.05 by Tukey post-hoc test. Results of ANOVA analysis are presented in the Supporting Information (Table S3).
PPO activity in ‘receiver' plants of treatment A increased by 39.1 (P = 0.001), 30.6 (P = 0.005) and 20.5% (P = 0.026) at 48 h after insect attack on ‘donor' plants relative to that in treatment B, C and D, respectively (Fig. 1b). The SOD activity in the healthy ‘receiver' plants of treatment A displayed increases of 19.6 (P = 0.005), 40.6 (P < 0.001) and 31.2% (P < 0.001) at 12 h after insect attack compared to those in treatment B, C and D (Fig. 1c). Remarkably higher POD activity was observed in treatment A, which was increased by 286.0 (P = 0.002), 84.3 (P = 0.032) and 35.5% (P = 0.159) 24 h after insect inoculation compared with treatment B, C and D (Fig. 1d). Although mycorrhization in healthy ‘receiver' plants led to some increase in POD in treatment C and D, POD induction was more pronounced in the presence of F. mosseae/CMN connection with insect-attacked plants.
Induction of defence-related genes in ‘receiver' plants
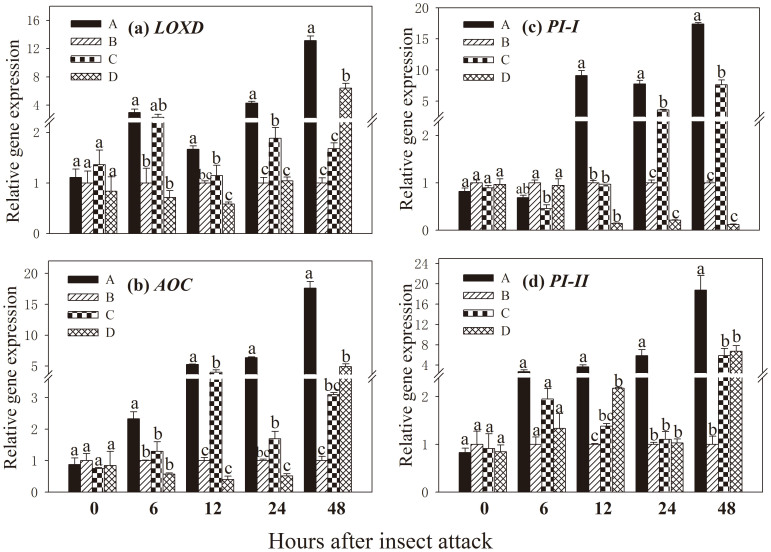
In the leaves of ‘receiver' plants quantitative real time RT-PCR was used to detect the transcripts of four defence genes: genes encoding lipoxygenase D (LOXD) and allene oxide cyclase (AOC), which are two key enzymes of the jasmonic acid biosynthesis pathway; two wound-response genes encoding serine protease inhibitors (PI-I and PI-II). No significant difference in transcript levels of four defence-related genes was observed among four treatments at the beginning time point of insect inoculation (Fig. 2). However, 6 h after insect inoculation on ‘donor' plant transcript levels of LOXD, AOC and PI-II in ‘receiver' plants of treatment A were significantly higher relative to those in the other three treatments, and 12 h after the insect attack transcript levels of all four genes in ‘receiver' plants of treatment A were significantly higher. This effect was particularly evident at 24–48 h post insect inoculation, for example LOXD transcript levels increased by 4.3-, 2.3- and 4.1-fold at 24 h after the insect attack, and by 13.1-, 7.8- and 2.0-fold relative to those in treatment B, C and D, respectively (Fig. 2a).
Figure 2. Expression of four defence-related genes in leaves of ‘receiver' tomato plants in response to common mycorrhizal networks (CMNs) connected with Spodoptera litura-infested neighbouring tomatoes.
Funneliformis mosseae was used to establish CMNs between ‘donor' and ‘receiver' tomatoes. Third instar larvae of Spodoptera litura were used to attack tomatoes. Quantitative real time RT-PCR was used to detect the transcripts of five defence-related genes encoding (a) lipoxygenase D (LOXD), (b) allene oxide cyclase (AOC), (c) proteinase inhibitor I (PI-I) and (d) proteinase inhibitor II (PI-II). Four treatments (A, B, C, D) were set up as described in Table 1. Values are means + standard error from three sets of independent experiments with three pots per treatment for each set of experiments. For each time point, letters above bars indicate significant difference among treatments (P < 0.05 according to Tukey's multiple range test). Results of ANOVA analysis are presented in the Supporting Information (Table S4).
In the ‘receiver' plants of treatment A, the expression levels of AOC were up-regulated by 6.4-, 3.8- and 12.4-fold at 24 h after the insect attack, and by 17.6-, 5.7- and 3.6-fold at 48 h compared with those in treatment B, C and D, respectively (Fig. 2b). A similar and significant increases in PI-I and PI-II transcript levels were observed in ‘receiver' plants of treatment A, where PI-I expression levels were found to increase by 7.7-, 2.2- and 37.0-fold at 24 h after the insect attack, and by 17.4-, 2.3- and 147.7-fold at 48 h relative to those in treatment B, C and D, respectively (Fig. 2c). PI-I expression levels were induced 5.9-, 5.3- and 5.7-fold at 24 h after the insect attack, and by 18.8-, 3.2- and 2.9-fold at 48 h relative to those in treatment B, C and D, respectively (Fig. 2d). The expression levels of defence-related genes in the ‘receiver' plants in treatment C and D increased to some extent, which may have resulted from root infection by the mycorrhizal fungus, but they were not as high as those in treatment A in which those ‘receiver' and ‘donor' plants were linked by CMN.
JA accumulation and perception
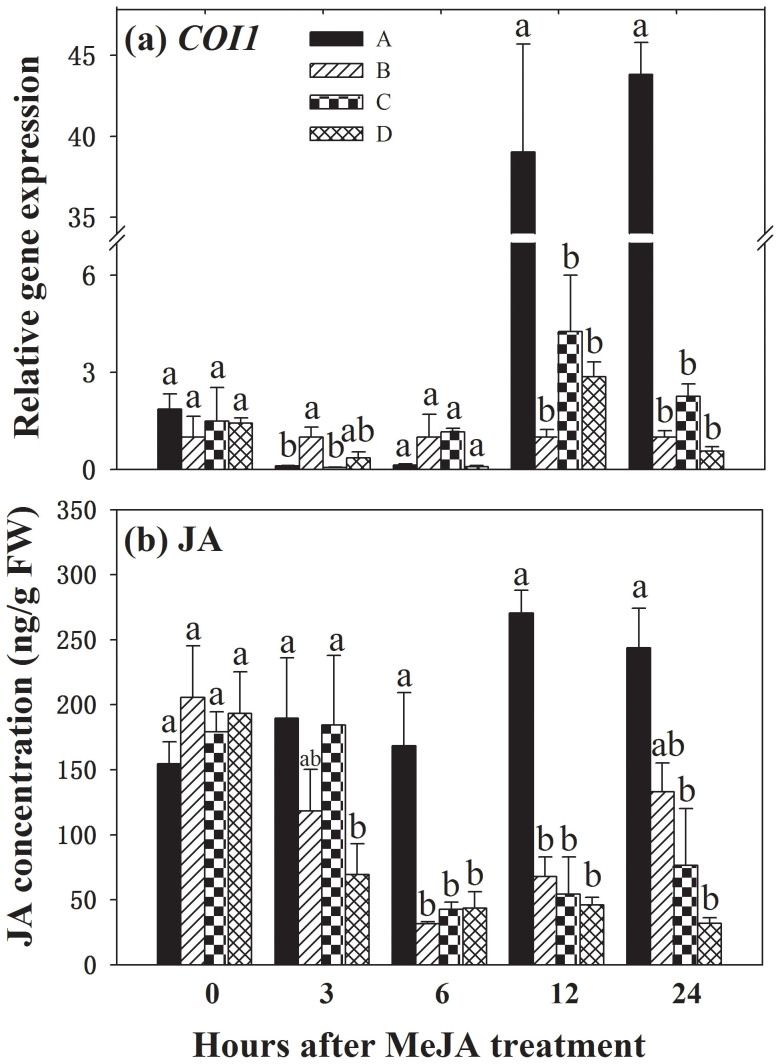
To determine involvement of the JA signalling in CMN-mediated plant communication, after methyl jasmonate (MeJA) was applied on ‘donor' plants, JA levels and JA signalling perception response in CMN-connected ‘receiver' plants were investigated. It was found that 6 h after MeJA application on ‘donor' plants and thereafter, JA level in CMN-connected ‘receiver' plants in treatment A was significantly higher than that in other three treatments (Fig. 3a). JA levels were increased by 284.1, 487.2 and 661.0% at 6, 12 and 24 h after MeJA application, respectively relative to those in treatment D. Similarly, transcripts of COI1, a JA signalling perception gene, in the leaves of CMN-connected ‘receiver' plant in treatment A were induced 13.7- and 77.0-fold 12 h and 24 h after MeJA application, respectively (Fig. 3b).
Figure 3. Levels of COI1 transcripts (a) and jasmonic acid (JA, b) in leaves of ‘receiver' tomato plants in response to common mycorrhizal networks (CMNs) connected with MeJA-treated neighbouring tomatoes.
Funneliformis mosseae was used to establish CMNs between ‘donor' and ‘receiver' tomatoes. Leaves of each ‘donor' plant were sprayed with 1 ml MeJA (1 mM) or buffer (treatment D). Quantitative real time RT-PCR was used to detect the transcripts of COI1. Four treatments (A, B, C, D) were set up as described in Table 1. Values are means + standard error from three sets of independent experiments with three pots per treatment for each set of experiments. For each time point, letters above bars indicate significant difference among treatments (P < 0.05 according to Tukey's multiple range test). Results of ANOVA analysis are presented in the Supporting Information (Table S6).
‘Mute donor' and ‘deaf receiver'
A further bioassay with transgenic tomato plants showed that if ‘donor' plants were spr2 tomato that were defective in JA biosyntheses, larvae fed on WT ‘receiver' plants in treatment E gained significantly more weight relative to those in treatment A, though these ‘receiver' plants were WT and connected with insect-attacked ‘donor' plants via CMN (Table 2). There was no significant difference in weight gain between treatment E and treatment B, C, F and G. In treatment H and I, although ‘donor' plants were attacked by the insect and connected with ‘receiver' plants via CMN, these ‘receiver' plants that were defective in either JA production or JA perception showed highly susceptible to SL herbivory.
Table 2. Percentage of root mycorrhizal colonization in wild-type (WT), JA biosynthesis mutant spr2 and JA signaling mutant jai1 of ‘donor' tomato plants inoculated with Funneliformis mosseae and ‘receiver' plants, and weight gain of Spodoptera litura fed on the leaves of these plants.
| Mycorrhizal colonization (%) | Weight gain (mg) | |||||
|---|---|---|---|---|---|---|
| Treatment | Inoculation on ‘donor' | CMN | ‘Donor' (genotype) | ‘Receiver' (genotype) | ‘Donor' (genotype) | ‘Receiver' (genotype) |
| A | Fm + SL | Yes | 47.8 ± 7.5 a (WT) | 32.9 ± 5.3 a (WT) | 57.3 ± 2.7 c (WT) | 39.4 ± 1.3 e (WT) |
| B | SL | No | 0 ± 0 b (WT) | 0 ± 0 b (WT) | 82.8 ± 2.5 b (WT) | 82.1 ± 3.2 bcd (WT) |
| C | Control | No | 0 ± 0 b (WT) | 0 ± 0 b (WT) | Un-infested (WT) | 87.0 ± 4.6 bc (WT) |
| D | Fm | Yes | 52.4 ± 3.3 a (WT) | 43.1 ± 6.8 a (WT) | Un-infested (WT) | 71.1 ± 3.8 d (WT) |
| E | Fm + SL | Yes | 8.4 ± 0.6 b (spr2) | 6.2 ± 0.8 b (WT) | 140.2 ± 2.6 a (spr2) | 87.5 ± 4.0 bc (WT) |
| F | Fm | Yes | 6.9 ± 1.2 b (spr2) | 6.0 ± 1.0 b (WT) | Un-infested (spr2) | 76.5 ± 2.4 cd (WT) |
| G | SL | No | 0 ± 0 b (spr2) | 0 ± 0 b (WT) | 132.1 ± 1.7 a (spr2) | 94.7 ± 3.3 b (WT) |
| H | Fm + SL | Yes | 51.6 ± 1.5 a (WT) | 10.2 ± 1.6 b (spr2) | 56.9 ± 2.8 c (WT) | 145.8 ± 3.3 a (spr2) |
| I | Fm + SL | Yes | 46.4 ± 8.2 a (WT) | 33.3 ± 10 a (jai1) | 63.7 ± 0.6 c (WT) | 139.9 ± 3.1 a (jai1) |
F. mosseae (Fm) was used to established common mycorrhizal network (CMN) between ‘donor' and ‘receiver' tomato plants. Third instar larvae of S. litura (SL) were used to attack tomato plants. JA biosynthesis mutant spr2, JA signaling mutant jai1, as well as their corresponsive wild-type (WT, cv Castlemart) tomato were used for comparison. In bioassays the ‘receiver' plants were inoculated with larvae 24 h later than the ‘donor' plants. Nine treatments included: A) a ‘receiver' WT plant was connected with a SL-attacked ‘donor' WT plant through CMN; B) a ‘receiver' WT plant was grown near a SL-attacked ‘donor' WT plant without mycorrhizal inoculation; C) a ‘receiver' WT plant was grown near a ‘donor' WT plant without AMF and insect applied. D) a ‘receiver' WT plant was connected with a no-infested WT plant by CMN; E) a ‘receiver' WT plant was connected with a neighboring SL-attacked ‘donor' spr2 plant through CMN; F) a ‘receiver' WT plant was grown near a SL-attacked ‘donor' spr2 plant without mycorrhizal inoculation; G) a ‘receiver' WT plant was connected with a no-infested spr2 plant by CMN; H) a ‘receiver' spr2 plant was connected with a SL-attacked tomato ‘donor' WT plant through CMN; I) a ‘receiver' jai1 plant was connected with a SL-attacked tomato ‘donor' WT plant through CMN. Four sets of bioassays were independently carried out and three pots per treatment were set up for each set of bioassays. Values are means ± standard error. Significant differences (P < 0.05 using Tukey post-hoc test) among treatments in the same column are indicated by different letters. Results of ANOVA analysis and each set of bioassays are presented in the Supporting Information (Table S5).
The mutant spr2 had significantly lower mycorrhizal colonization relative to WT plants (Table 2). In treatment E and F mycorrhizal colonization rates were less than 8.4% in both ‘donor' and ‘receiver' plants. However, mutant jai1 in treatment I had similar mycorrhizal colonization with WT ‘receiver' plants in treatment A.
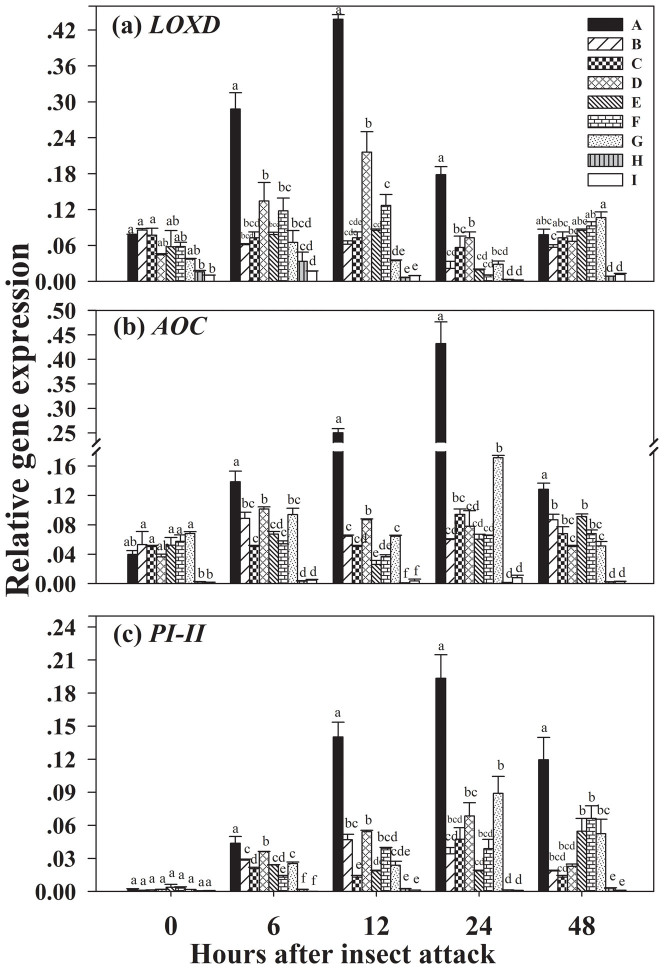
Real time RT-PCR analyses showed that transcripts of LOXD, AOC and PI-II in the leaves of CMN-connected WT ‘receiver' plant in treatment A were induced 2.0-, 2.9- and 2.6-fold 12 h after insect inoculation on ‘donor' plant, and 2.4-, 5.5- and 2.8-fold 24 h after insect inoculation, respectively relative to treatment D with similar mycorrhizal colonization (Fig. 4). However, in treatment E no induction of transcripts of LOXD, AOC and PI-II was found in WT ‘receiver' plant that connected mutant jai1 ‘donor' plant via CMN. No induction was either detected in treatment I with well-established CMNs between ‘donor' and ‘receiver' plant (Fig. 4).
Figure 4. Expression of three defence-related genes in leaves of ‘receiver' tomato plants in response to common mycorrhizal networks (CMNs) connected with Spodoptera litura-infested neighbouring tomatoes.
Funneliformis mosseae was used to establish CMNs between ‘donor' and ‘receiver' tomatoes. Third instar larvae of Spodoptera litura were used to attack tomatoes. Quantitative real time RT-PCR was used to detect the transcripts of three defence genes encoding (a) lipoxygenase (LOXD), (b) allene oxide cyclase (AOC) and (c) proteinase inhibitor II (PI-II). Nine treatments were set up as described in Table 2. Values are means + standard error from three sets of independent experiments with three pots per treatment for each set of experiments. For each time point, letters above bars indicate significant difference among treatments (P < 0.05 according to Tukey's multiple range test). Results of ANOVA analysis are presented in the Supporting Information (Table S7).
Discussion
Ecological significance of plant-to-plant carbon and other nutrient movement through CMNs has been addressed15,17,18,27. Our previous study showed that CMNs can transfer disease defence signals between tomato plants22. Barto et al. reported that CMNs facilitate transport of allelochemicals from supplier to target plants and expand the bioactive zones of allelochemicals in natural environments28. Our results in this study show that tomato plants can hijack the CMNs as an underground communication conduit for herbivore-induced defence signal transfer between insect-attacked and intact neighboring tomato plants. Significant increases in defence-related enzymatic activities, gene expression and insect resistance in the neighboring intact ‘receiver' tomato plants in the treatment A (Table 1, Fig. 1 & 2), which were connected by CMNs with insect-attacked ‘donor' plants, indicated that CMNs could enhance the defence response of neighboring plants and thereby increased their insect resistance.
Two possible mechanisms involving induced defences in neighbouring intact plants have been proposed: i) aboveground communications mediated by herbivore-induced volatiles from ‘donor' plants29,30,31; ii) belowground communications mediated by root exudates and fungal mycelia32,33,34. In our experiment the airborne volatile communication was controlled by covering the SL-infected ‘donors' with air-tight plastic bags after the insect inoculation. Many filamentous soil organisms, such as bacteria and saprobic or parasitic fungi, can form mycelial networks in soil35, but these have not been reported to connect plants23. Our results from treatment B in this study would exclude the second possibility. If root exudates and mycelial networks of the other fungi had mediated plant-plant communication, defence responses in ‘receiver' plants of treatment B should have been induced. The possible mycorrhization effects on enhanced defence of ‘receiver' plants were eliminated by treatments C and D. In treatment C both ‘donor' and ‘receiver' plants were inoculated by F. mosseae, but they were separated by a water-proof membrane to prevent interplant CMN formation (Fig. S1). Significantly higher defence-related enzymatic activities and gene transcripts in the ‘receiver' plants of treatment A than those in treatment C or D suggested that defence signals had been transferred from herbivore-infested ‘donor' plants to CMN-connected ‘receiver' plants in treatment A. Thus, CMNs do mediate belowground communications between herbivore-infected ‘donor' and intact ‘receiver' plants.
Babikova et al. showed that mycorrhizal mycelia can serve as a conduit for signaling between plants of broad bean (Vicia faba), acting as an early warning system for aphid attack36. However, the signal that may be released from the ‘donor' and transferred to the ‘receiver' remains unknown. The JA signalling plays a vitally key role in plant defence response to insect herbivory25. To determine whether JA signalling in involved in CMN-mediated interplant communication we detected JA accumulation and JA-response genes transcripts in ‘receiver' plants. Six hours after insect attack on the leaves of ‘donor' plants three genes including LOXD and AOC in jasmonic acid (JA) pathway and as well as JA-response gene PI-II, were up-regulated in the leaves of ‘receiver' plants (Fig. 2a, b, d & Fig. 4a, b, c), suggesting that at this time point the ‘receiver' plants had already triggered their JA-related defence responses. The fact that exogenous application of MeJA to ‘donor' plants increased accumulation of JA level and COI1 transcripts in the leaves of ‘receiver' plants (Fig. 3) also indicated that JA might be the signal sending from ‘donor' to ‘receiver' plants. Use of a JA biosynthesis defective mutant spr2 as ‘donor' plant in replace of WT plant led to no induction of defence responses and no change in plant resistance in ‘receiver' plants although two plants were connected by CMNs and the ‘donor' plant was attacked by the insect (Fig. 4 & Table 2). Furthermore, no induction of defence responses was found either upon use of both spr2 and jai1 as ‘receiver' plants in replace of WT plants. These results suggest that JA signalling was required for the CMN-mediated interplant communication.
Labelling studies in tobacco showed that both JA and MeJA could be transported from leaves to roots and to induce defence systemically37,38. Heil & Ton suggested that JA and its derivatives were optimal candidates for long-distance signals39. Giovannetti et al. found that a number of particles (e.g., vacuoles, mitochondria, nuclei, and fat droplets) moved at the speed of 1.8 μm/s (approximately 15.5 cm/d) in both directions within the CMNs of Glomus caledonium40. It seems highly probable that jasmonate signals moved from roots of one plant to another via CMNs and systemically induced defence of neighbouring plants.
Based on these results that the receiver plants connected with insect-attacked donor plants by CMNs in treatment A had less insect damage, higher levels of defence-related enzymatic activities and gene expression than controls without insect-attacked neighbouring donor plants (treatment D) or without CMN linkages (treatment C), as well as JA biosynthesis deficient mutant spr2 donor plant failed to induce defence responses and insect resistance in CMN-connected ‘receiver' plant, we propose that tomato plants can hijack CMNs for herbivore-induced defence signal transfer and interplant defence communication to activate defence responses more rapidly and aggressively upon insect attack and to increase their insect resistance.
Since most land plants are colonized by mycorrhizal fungi and CMNs widely distribute in plant communities, CMN-mediated interplant communications may have an important ecological role in plant defence at the community level. Plants in a community may join together through CMNs to form a functional ‘guild of mutual aid'41. CMNs not only function as nutrient and carbon allocation networks to influence patterns of interplant competition, plant diversity and plant community dynamics15,17,18, but also could act as herbivore defence networks in plant communities. This interplant underground communication through common symbionts may operate more reliably and over greater distance than air-borne communications. Continuous tillage and agrochemical application in agricultural ecosystems may disrupt mycorrhizal networks and affect self defence of crops. Maintenance of mycorrhizal networks is of particular importance in crop protection. Documenting this communication will promote our understanding of ecological functions of CMNs and community-level systemic defence in natural ecosystems.
Methods
Plant, fungus and insect
Tomato seeds (S. lycopersicum cv. Jinbao) were surface-sterilized by 10% H2O2 for 5 min and germinated in autoclaved quartz sand after rinsing with sterilized distilled water. The 10-day-old seedlings were transplanted to pots for further experimentations. F. mosseae (Nicol. & Gerd, FM) Gerdemann & Trappe BEG 167, kindly provided by Dr. Runjin Liu at Qingdao Agricultural University (China), was used to establish CMNs between two tomato seedlings in the same pot. Mycorrhizal inocula were reproduced in pots with corn (Zea mays cv. Gaoyou-115) in autoclaved sand media42 and consisted of rhizospheric sand with root segments and hypahe (35 infective propagules/gram).
A leaf-chewing caterpillar common cutworm (S. litura, SL) was used to attack tomato plants. SL larvae were reared on a semisynthetic diet containing wheat germ43 and maintained in an insectary at 23–26°C, 16 h/8 h (day/night) and 65–70% relative humidity. Homogenously 12-hour-molted third instar larvae were used to attack tomato plants.
Chemicals
TRIzol reagent, M-MLV reverse transcriptase, Taq polymerase, RNase inhibitor and dNTPs were from TaKaRa (Shuzo Co. Ltd., Shiga, Japan), 4-morpholine-propanesulfonic acid (MOPS) and diethylpyrocarbonate (DEPC) were from AMRESCO (Solon, OH), SYBR Green Real Time PCR Master Mix was from Toyobo Life Science (TOYOBO Co. Ltd, OSAKA, Japan), and methyl jasmonate (MeJA) was from Sigma-Aldrich (St. Louis, MO, USA), respectively.
Experimental design and growth conditions
A rectangular plastic pot measuring 29 × 13 × 11 cm (length × height × width) was separated by two fine stainless steel screens (25 μm, TWP Inc. Berkeley, CA, USA) into two equal compartments (Compartment I and II), which prevent direct root contacts but allowing fungal mycelia to get through screens to establish CMNs between plants. One tomato plant in the compartment I or ΙΙ was denoted as ‘donor' and ‘receiver' plant, respectively. To determine the direct effects of CMNs whilst excluding possible root contacts and mycorrhization, four treatments were designed (Table 1 & Fig. S1)22: A) a ‘receiver' plant was connected with a SL-attacked ‘donor' plant through a CMN; B) a ‘receiver' plant was grown near a SL-attacked ‘donor' plant without AMF inoculation or CMNs connection between plants; C) a mycorrhizal ‘receiver' plant was grown near a SL-attacked mycorrhizal ‘donor' plant but being separated by a water-proof membrane to prevent root and mycelial contacts between them, and D) a ‘receiver' plant was connected with the neighbouring ‘donor' plant by a CMN without SL feeding. For enzymatic and molecular analysis all ‘receiver' plants in these four treatments did not receive any insect attack. In bioassays all ‘receiver' plants were infested with SL larvae.
Each compartment was filled with 1.5 kg autoclaved soil/sand mixture (2:1). The brown loam soil was obtained from the university campus containing 2.37% organic matter, 0.21% total N, 56.2 mg/kg available P with a pH of 5.64. For mycorrhizal inoculation 100 g F. mosseae inocula were applied to the compartment I in treatments A and D before sowing, but 50 g to each compartment in treatment C. In treatment B 100 g above-mentioned corn growth media without mycorrhizal inoculation were applied to the compartment I.
Plants were grown in the same conditions as Song et al.22. Under the microscopic observation CMNs between plants over the fine steel mesh were established after 35 d of transplanting and this was also confirmed at harvest (40 days after transplanting) (Table 1). Leaves of each ‘donor' tomato were infested with five newly-molted third instar SL larvae after 40 d of transplanting. After insect infestation both ‘receivers' and insect-attacked ‘donors' were covered by air-tight plastic bags for 5 d to eliminate possible volatile signal contact between plants. Leaves of ‘receivers' were collected at 3, 6, 12, 24 and 48 h after insect feeding on ‘donors' for real-time RT-PCR and enzymatic analyses. Root AM colonization was measured according to Mukerji et al.42.
Involvement of the JA signalling pathway
Jasmonic acid (JA) signalling pathway plays an essential role in plant responses to chewing insects24,25. Both JA biosynthesis defective mutant suppressor of prosystemin-mediated responses2 (spr2) and JA signalling defective mutant jasmonic acid–insensitive1 (jai1) tomato, which were derived from a tomato wild-type (WT, Solanum lycopersicum Castlemart) parent44, were used to determine the role of JA signalling in CMNs mediated plant communication. The experiment design was similar to that described above, but spr2 was used as a ‘donor' in treatments E, F and G to determine if a ‘receiver' WT tomato could ‘eavesdrop' on defence signals from its insect-attacked JA signal defective ‘donor' through CMNs (Table 2). The treatment C in this experiment was different from the above one since the ‘donor' did not receive both insect and AMF inoculation, but still separated by two stainless steel screens (25 μm). The spr2 and jai1 were ‘receivers' in treatment H and I, respectively, but the ‘donors' were the WT tomatoes receiving both insect and AMF inoculation (Table 2).
Bioassay
To test if induced defence by CMNs communication could enhance insect resistance, bioassays were conducted to compare the weight gain of larvae fed on both ‘receiver' and ‘donor' tomatoes. Both CMN establishment and insect feeding on the ‘donor' plants were the same as in the above experimental design. However, in bioassays all ‘receiver' plants were infested with third instar SL larvae 24 h after insect feeding on the ‘donor' plants. The larval weight fed on ‘receiver' and ‘donor' plants were recorded 72 h after insect inoculation. Before feeding treatment all larvae had been starved for 2 h and then weighed. Four sets of bioassays were independently carried out and four pots per treatment were set up for each set of bioassays.
Enzyme assays
Four defence-related enzymes including peroxidase (POD), polyphenol oxidase (PPO), superoxide dismutases (SOD) and lipoxygenase (LOX) were analysed. Leaf samples (0.2 g fresh weight) were ground in liquid nitrogen and homogenized in 2.0 ml ice cold 0.05 M phosphate buffer (pH 7.2 for POD, but 7.8 for PPO and SOD) containing 1% (w/v) polyvinylpyrrolidone (PVP). The supernatant after 12,000 g centrifugation for 15 min at 4°C was used for enzyme assays (Song et al., 2010). Activities of LOX, POD, PPO and SOD were spectrophotometrically determined according to Rodríguez-Rosales et al.45, Kraus & Fletcher46, Zauberman et al.47 and McCord & Fridovich48, respectively. Leaf samples for enzyme analyses were harvested from ‘receiver' plants of three sets of independent experiments with three pots per treatment for each set of experiments.
Analysis and perception of JA
Four treatments (A, B, C and D) as described above were used to determine if an application of methyl jasmonate (MeJA) on ‘donor' plants could increase JA production and JA signalling perception in CMN-connected ‘receiver' plants. Forty days after transplanting ‘donor' plants in treatment A, B and C were sprayed with 1.0 ml 1.0 mM MeJA (pH 8.0, dissolved in 50 mM sodium phosphate buffer containing 0.01% Tween 20 and adjusted by 1.0 M citric acid) or in treatment D with 1.0 ml buffer lacking MeJA. Leaves from ‘receiver' plants were then harvested for JA analysis and RNA extraction at 0, 3, 6, 12 and 24 h after the MeJA application on ‘donor' plants. For each treatment at each time point, four plants were sampled. JA levels were determined by gas chromatography (GC) as described by Ye et al.49. CORONATINE-INSENSITIVE1 (COI1) encoding an F-box protein that is required for JA signalling perception50 was used to detect the transfer of JA signalling from ‘donor' plants to ‘receiver' plants.
Real-time RT-PCR analysis
Differential expression of selected genes was verified by real-time RT-PCR using the RNA samples isolated from tomato leaves obtained from all ‘receiver' plants. The Ubi3 gene was used as a reference gene. Total RNA from tomato leaves was extracted and isolated according to Kiefer et al.51 including a DNase (Promega, Madison, USA) treatment. First strand cDNA was synthesized from 1 μg total RNA using ImProm-II™ Reverse transcription system (Promega, Madison, USA) according to the manufacturer's instructions.
The primers for target's genes LOXD, AOC, COI1, PI-1 and PI-11 were designed by Primer 3.0 software (Applied Biosystems, http://fokker.wi.mit.edu/primer3/input.htm) based on tomato mRNA sequences deposited in the GenBank. The gene-specific primer sequences used are listed in Table S1. Real-time PCR reactions were carried out with 0.2 μl (0.15 μM) of each specific primers, 1 μl cDNA, 12.5 μl of the SYBR green master mix (Quanti Tech SYBR Green kit, Qiagen, Gmbh Hilden, Germany) and the final volume made up to 25 μl with RNase-free water. In the negative control cDNA was replaced by RNase free water. The reactions were performed on a DNA Engine Opticon 2 Continuous Fluorescence Detection System (MJ Research Inc., Waltham, MA, USA). The programme used for real-time PCR was 3 min initial denaturation at 95°C, followed by 35 cycles of denaturation for 20 s at 95°C, annealing for 20 s (LOXD: 56.9°C;AOC: 56.5°C;PI-1: 47.6°C;PI-11: 55°C;COI1: °C; Ubi3: 51.5°C and extension for 20 s at 72°C. The fluorescence signal was measured immediately after incubation for 2 s at 75°C following the extension step, which eliminated possible primer dimer detection. At the end of the cycles, melting temperatures of the PCR products was determined between 65°C and 95°C. Amplicon specificity was verified by the melting curve analysis and agarose gel electrophoresis. Transcripts of targeted genes were calculated by the Double-standard Curves method. Three biological replicates were independently carried out and three pots per treatment were set up for each biological replicate. Each leaf sample for RNA extract was collected from tomato leaves of the ‘receiver' plants in each pot.
Statistical analysis
For each treatment three replicates were maintained in a completely randomized design. SAS 8.0 (SAS Institute, Cary, North Carolina) package for windows was used for statistical analysis. The data for root AM colonization, larva weight, enzymatic activity, JA concentrations and gene expression levels were analyzed with a one-way analysis of variance with the significant differences among means identified by the Turkey multiple range test at P < 0.05.
Author Contributions
Y.Y.S., M.Y. and R.S.Z. designed research; Y.Y.S., M.Y., R.L.W. and Y.J.S. performed research; C.Y.L. constructed transgenic plants; Y.Y.S., X.H.H., K.Z., S.M.L. and R.S.Z. analyzed data; and Y.Y.S., X.H.H. and R.S.Z. wrote the paper.
Supplementary Material
Supplementary Information
Acknowledgments
This research was supported by the National 973 Program of China (2011CB100400), National Natural Science Foundation of China (31070388, 31028018, 31100286), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2010), Guangdong Natural Science Foundation of China (S2011040004336), and Ph.D. Foundation of the Ministry of Education of China (20104404110004). We thank Dr. Ron A. Salzman (Stoller Enterprises) for language improvement.
Footnotes
The authors have no competing interests, or other interests that might be perceived to influence the results and / or discussion reported in this article.
References
- Green T. R. & Ryan C. A. Wound-induced proteinase inhibitor in plant leaves: a possible defense mechanism against insects. Science 175, 776–777 (1972). [DOI] [PubMed] [Google Scholar]
- Zangerl A. R. Furanocoumarin induction in wild parsnip: evidence for an induced defense against herbivores. Ecology 71, 1926–1932 (1990). [Google Scholar]
- Agrawal A. A. Induced responses to herbivory and increased plant performance. Science 279, 1201–1202 (1998). [DOI] [PubMed] [Google Scholar]
- Baldwin I. T. & Schultz J. C. Rapid changes in tree leaf chemistry induced by damage: evidence for communication between plants. Science 221, 277–279 (1983). [DOI] [PubMed] [Google Scholar]
- Mithöfer A. & Boland W. Plant defense against herbivores, chemical aspects. Annu. Rev. Plant Biol. 63, 431–450 (2012). [DOI] [PubMed] [Google Scholar]
- Hare J. D. Ecological role of volatiles produced by plants in response to damage by herbivorous insects. Annu. Rev. Entom. 56, 161–180 (2011). [DOI] [PubMed] [Google Scholar]
- Farmer E. E. & Ryan C. A. Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc. Natl. Acad. Sci. U. S. A. 87, 7713–7716 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura G. et al. Herbivory-induced volatiles elicit defence genes in lima bean leaves. Nature 406, 512–515 (2000). [DOI] [PubMed] [Google Scholar]
- Frost C. J., Mescher M. C., Carlson J. E. & De Moraes C. M. Plant defense priming against herbivores: getting ready for a different battle. Plant Physiol. 146, 818–824 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruin J., Sabelis M. W. & Dicke M. Do plants tap SOS signals from their infested neighbours? Trends Ecol. Evol. 10, 167–170 (1995). [DOI] [PubMed] [Google Scholar]
- Baldwin I. T. et al. Volatile signaling in plant-plant interactions: “talking trees” in the genomics era. Science 311, 812–815 (2006). [DOI] [PubMed] [Google Scholar]
- Smith S. E. & Read D. J. Mycorrhizal symbiosis, 3rd edn. London, UK, Academic Press (2008).
- Rabin L. B. & Pacovsky R. S. Reduced larva growth of two Lepidoptera (Noctuidae) on excised leaves of soybean infected with a mycorrhizal fungus. J. Econ. Entomol. 78, 1358–1363 (1985). [Google Scholar]
- Hartley S. E. & Gange A. C. Impacts of plant symbiotic fungi on insect herbivores, Mutualism in a multitrophic context. Annu. Rev. Entom. 54, 323–342 (2009). [DOI] [PubMed] [Google Scholar]
- Simard S. W. et al. Mycorrhizal networks: mechanisms, ecology and modelling. Fungal Biol. Rev. 26, 39–60 (2012). [Google Scholar]
- Giovannetti M. et al. At the root of the wood wide web. Self recognition and non-self incompatibility in mycorrhizal networks. Plant Signal. Behav. 1, 1–5 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selosse M. A., Richard F., He X. & Simard S. W. Mycorrhizal networks, des liaisons dangereuses? Trends Ecol. Evol. 21, 621–628 (2006). [DOI] [PubMed] [Google Scholar]
- Leake J. et al. Networks of power and influence, the role of mycorrhizal mycelium in controlling plant communities and agroecosystem functioning. Can. J. Bot. 82, 1016–1045 (2004). [Google Scholar]
- He X. H., Xu M. G., Qiu G. Y. & Zhou J. B. Use of 15N stable isotope to quantify nitrogen transfer between mycorrhizal plants. J. Plant Ecol. 2, 107–118 (2009). [Google Scholar]
- Ren L. X. et al. Role of arbuscular mycorrhizal network in carbon and phosphorus transfer between plants. Biol. Fertil. Soils 49, 3–11 (2013). [Google Scholar]
- van der Heijden M. G. A. & Horton T. R. Socialism in soil? The importance of mycorrhizal fungal networks for facilitation in natural ecosystems. J. Ecol. 97, 1139–1150 (2009). [Google Scholar]
- Song Y. Y. et al. 2010. Interplant communication of tomato plants through underground common mycorrhizal networks. PLoS ONE 5(10), e13324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barto K. E., Weidenhamer J. D., Cipollini D. & Rillig M. C. Fungal superhighways: do common mycorrhizal networks enhance below ground communication? Trends Plant Sci. 17, 633–637 (2012). [DOI] [PubMed] [Google Scholar]
- Browse J. & Howe G. A. New weapons and a rapid response against insect attack. Plant Physiol. 146, 832–838 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G. A. & Jander G. Plant immunity to herbivores. Annu. Rev. Plant Biol. 59, 41–66 (2008). [DOI] [PubMed] [Google Scholar]
- Hause B. & Schaarschmidt S. The role of jasmonates in mutualistic symbioses between plants and soil-born microorganisms. Phytochemistry 70, 1589–1599 (2009). [DOI] [PubMed] [Google Scholar]
- He X. H. et al. Rapid nitrogen transfer from ectomycorrhizal pines to adjacent ectomycorrhizal and arbuscular mycorrhizal plants in a California oak woodland. New Phytol. 170, 143–51 (2006). [DOI] [PubMed] [Google Scholar]
- Barto E. K. et al. The Fungal Fast lane: Common mycorrhizal networks extend bioactive zones of allelochemicals in soils. PLoS ONE 6(11), e27195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberth J., Alborn H. T., Schmelz E. A. & Tumlinson J. H. Airborne signals prime plants against insect herbivore attack. Proc. Natl. Acad. Sci. USA 101, 1781–1785 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U. et al. Priming: getting ready for battle. Mol. Plant Microbe Interact. 19, 1062–1071 (2006). [DOI] [PubMed] [Google Scholar]
- Kessler A., Halitschke R., Diezel C. & Baldwin I. T. Priming of plant defense responses in nature by airborne signaling between Artemisia tridentata and Nicotiana attenuata. Oecologia 148, 280–292 (2006). [DOI] [PubMed] [Google Scholar]
- Dicke M. & Dijkman H. Within-plant circulation of systemic elicitor of induced defence and release from roots of elicitor that affects neighbouring plants. Biochem. System. Ecol. 29, 1075–1087 (2001). [Google Scholar]
- Guerrieri E. et al. Plant-to-plant communication mediating in-flight orientation of Aphidius ervi. J. Chem. Ecol. 28, 1703–1715 (2002). [DOI] [PubMed] [Google Scholar]
- Rasmann S. et al. Recruitment of entomopathogenic nematodes by insect-damaged maize roots. Nature 434, 732–737 (2005). [DOI] [PubMed] [Google Scholar]
- Thorn R. G. & Lynch M. D. J. [Fungi and eukaryotic algae]. Soil microbiology, ecology, and biochemistry [Paul, E. A. (ed., 3rd edn)] [145–162] (Elsevier, Oxford, UK, 2007). [Google Scholar]
- Babikova Z. et al. Underground signals carried through common mycelial networks warn neighbouring plants of aphid attack. Ecol. Lett. 16, 835–843 (2013). [DOI] [PubMed] [Google Scholar]
- Thorpe M. R., Ferrieri A. P., Herth M. M. & Ferrieri R. A. C-11-imaging, methyl jasmonate moves in both phloem and xylem, promotes transport of jasmonate, and of photoassimilate even after proton transport is decoupled. Planta 226, 541–551 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang Z. P. & Baldwin I. T. Transport of [2–14C]jasmonic acid from leaves to roots mimics wound-induced changes in endogenous jasmonic acid pools in Nicotiana sylvestris. Planta 203, 436–441 (1997). [Google Scholar]
- Heil M. & Ton J. Long-distance signalling in plant defence. Trends Plant Sci. 13, 264–272 (2008). [DOI] [PubMed] [Google Scholar]
- Giovannetti M., Azzolini D. & Citernesi A. S. Anastomosis formation and nuclear and protoplasmic exchange in arbuscular mycorrhizal fungi. Appl. Environ. Microbiol. 65, 5571–5575 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry D. A. et al. Ectomycorrhizal mediation of competition between coniferous tree species. New Phytol. 112, 501–511 (1989). [DOI] [PubMed] [Google Scholar]
- Mukerji K. G., Manoharachary C. & Chamola B. P. Techniques in mycorrhizal studies. (Kluwer Academic Publishers, Dordrecht, The Netherlands, 2002). [Google Scholar]
- Waldbauer G. P., Cohen R. W. & Riedman S. An improved procedure for laboratory rearing of the corn earworm, Heliothis zea (Lepidoptera, Noctuidae). Great Lakes Entomol. 17, 113–118 (1984). [Google Scholar]
- Li C. Y. et al. The tomato Supperssor of prosystemin-mediated responses2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell 15, 1646–1661 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rosales M. P., Kerkeb L., Ferrol N. & Donaire J. P. Lipoxygenase activity and lipid composition of cotyledons and oil bodies of two sunflower hybrids. Plant Physiol. Biochem. 36, 285–291 (1998). [Google Scholar]
- Kraus T. E. & Fletcher R. A. Paclobutrazol protects wheat seedlings from heat and paraquat injury. Is detoxification of active oxygen involved? Plant Cell Physiol. 35, 45–52 (1994). [Google Scholar]
- Zauberman G. et al. Post-harvest retention of the red colour of litchi fruit pericarp. Sci. Horticul. 47, 89–97 (1991). [Google Scholar]
- McCord J. M. & Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244, 6049–6055 (1969). [PubMed] [Google Scholar]
- Ye M. et al. Silencing COI1 in rice increases susceptibility to chewing insects and impairs inducible defense. PLOS ONE 7, e36214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J. B. et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer E., Heller W. & Ernst D. A simple and efficient protocol for isolation of functional RNA from plant tissues rich in secondary metabolites. Plant Mol. Biol. Rep. 18, 33–39 (2000). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information