Abstract
Background
Our interest in Candida albicans mitochondria began with the identification of GOA1. We demonstrated its role in cell energy production, cross-talk among mitochondria and peroxisomes, non-glucose energy metabolism, maintenance of stationary phase growth, and prevention of premature apoptosis. Its absence results in avirulence. However, what regulated transcription of GOA1 was unknown.
Results
To identify transcriptional regulators (TRs) of GOA1, we screened a C. albicans TF knockout library (TRKO) and identified Rbf1p, Hfl1p, and Dpb4p as positive TRs of GOA1. The phenotypes of each mutant (reduced respiration, inability to grow on glycerol, reduced ETC CI and CIV activities) are reasonable evidence for their required roles especially in mitochondrial functions. While the integration of mitochondria with cell metabolic activities is presumed to occur, there is minimal information on this subject at the genome level. Therefore, microarray analysis was used to provide this information for each TR mutant. Transcriptional profiles of Rbf1p and Hfl1p are more similar than that of Dpn4p. Our data demonstrate common and also gene-specific regulatory functions for each TR. We establish their roles in carbon metabolism, stress adaptation, cell wall synthesis, transporter efflux, peroxisomal metabolism, phospholipid synthesis, rRNA processing, and nuclear/mtDNA replication.
Conclusions
The TRs regulate a number of common genes but each also regulates specific gene transcription. These data for the first time create a genome roadmap that can be used to integrate mitochondria with other cell processes. Of interest, the TRs are fungal-specific, warranting consideration as antifungal drug targets.
Keywords: Transcription factor, Non-glucose carbon metabolism, Mitochondria, Lipid oxidation, Metabolic regulation, Candida albicans
Background
Fungal invasive infections of humans are now referred to as “hidden killers” [1]. More than 90% of these infections are caused by species of Candida, Cryptococcus, Aspergillus, and Pneumocystis[1]. Blood-borne, nosocomial candidiasis is ranked 4th in frequency in the USA with a crude and attributable mortality of 49 and 27% (USA), similar to other developed countries [1,2]. The incidence of candidiasis has increased sharply over the past few decades primarily due to cancer chemotherapy, organ/bone marrow transplantation, surgical intervention, and the AIDS pandemic [3,4]. Treatment of these infections costs ~ $2.0-2.6 billion per year [5-8]. Global cryptococcal meningitis (usually caused by C. neoformans) in HIV/AIDS patients and others with immunosuppression therapies is estimated at 1 million cases per year; 620,000 deaths alone are in Sub-Saharan Africa [1]. Cryptococcus gattii is an emerging pathogen of apparently healthy people, signifying its potential as an even more dangerous invasive fungus. Death from the top 10 invasive fungi (1-1.5 million) is equal to or more than that of tuberculosis (1.4 million) or malaria (971,000) [1]. The enormity of fungal infections is magnified by the non-invasive (superficial) infections such as nail and scalp infections (1.9 billion), vaginal infections of women during child-bearing years (frequency of 50-75%), and oral and esophageal candidiasis in HIV/AIDS patients (12 million). In part, the rising costs are associated with inappropriate therapy, defined as delayed intervention, inadequate dosage, or administration of an antifungal to which an isolate was considered drug resistant [6].
C. albicans remains as the most common cause of candidiasis among all Candidia species. Virulence of this organism is commonly attributed to factors that initiate colonization of host cells (the ALS gene family and others), cause invasion (secreted lipases and proteases), regulate morphogenesis (the yeast hyphal transition), and biofilm formation [1]. In vivo virulence of these factors has been established in animal models fulfilling the paradigm of “Molecular Koch’s postulates”. Aside from the construction of single mutants to verify a role in pathogenesis, another useful approach to understanding virulence is to characterize global gene differences between a pathogen (C. albicans) and a non-pathogen (Saccharomyces cerevisiae, model yeast) or between two pathogens, one with a much lower incidence of causing candidiasis (C. dublinensis) [9]. Both types of data suggest interpretations of the gene repertoire needed by a pathogen. One of the major differences between C. albicans and model yeast is a rewiring of transcriptional regulation [10]. For C. albicans, enzymes of alternative carbon metabolism (non-glucose substrates) are stabilized even in the presence of glucose, compared to model yeast of which these same enzymes are regulated by glucose-repressible events [11]. Speculation is that C. albicans maintains a backup source for energy and carbon conservation to respire when confronted with low levels of host glucose. Model yeast when grown aerobically uses glucose via glycolysis and is referred to as Crabtree-positive. Oppositely, C. albicans respires oxidatively in the presence of glucose and is Crabtree-negative [12]. These observations are not surprising, given the differences in their environmental niches. In the case of C. albicans, low blood levels of glucose cause the utilization of alternative carbon sources as mentioned above and described in other labs [13-16]. Some peroxisomal activities in C. albicans are critical to the pathogenesis of candidiasis, since these organelles house alternative carbon metabolic pathways (such as the glyoxylate cycle) that are critical to survival of the organisms in macrophages [15].
Our interest in mitochondria of C. albicans began with the identification of GOA1[16]. Functional annotation was developed based upon phenotypic assays of a goa1Δ null mutant. Goa1p translocates to mitochondria during stress and in the presence of non-glucose substrates such as glycerol. The protein regulates complex I (CI) of the electron transport chain and also interacts with peroxisomes, organelles that house alternative carbon metabolic pathways. The loss of GOA1 causes a major reduction in mitochondrial membrane potential and a concomitant reduction in the formation of ATP. We have shown that a dysfunctional CI causes an increase in reactive oxidant species (ROS), triggering apoptosis and an associated shortened chronological aging in vitro[16-18]. We demonstrated that cross-talking between mitochondria and peroxisomes in the presence of either glucose or non-glucose substrates requires Goa1p [17,19]. Importantly, there are several defects in the mutant in regard to virulence and host cell interactions [20]. Compared to parental and gene-reconstituted strains, goa1Δ is avirulent in a murine model of blood-borne candidiasis, killed readily by human neutrophils ex vivo, and hypersusceptible to triazoles. Microarray analyses of goa1Δ indicate a major down regulation of genes associated with peroxisomal functions and carbon metabolism [17,19]. But, what regulates GOA1? Herein, we report on the identification of three transcription regulator (TR) Rbf1p, Hfl1p, and Dpb4p, each of which positively regulates GOA1. Importantly, we assign functions to each of these transcription factors in the regulation of cellular processes. Two of these TRs have not been functionally annotated, and the third (Rbf1p) is a repressor of filamentation [21,22]. Using microarray analysis, we show that there is both TR-specific gene regulation as well as regulation of a set of common genes.
Results
Identification of transcription regulators of GOA1
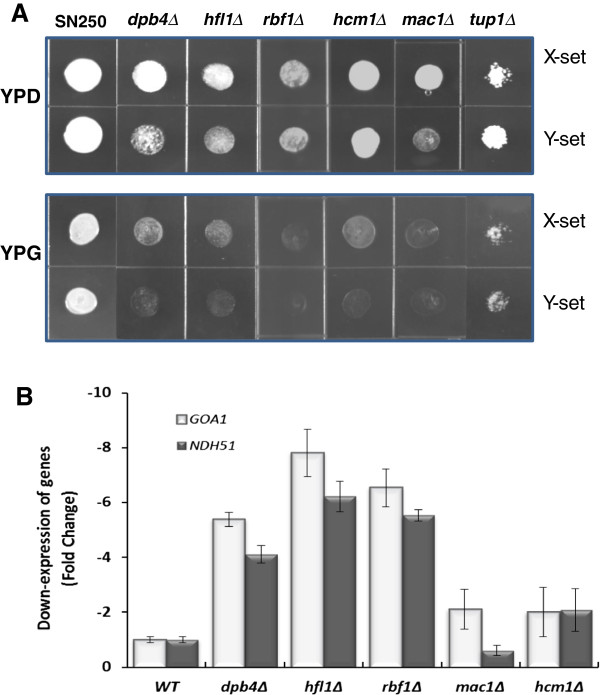
To initially identify transcription factors that regulate GOA1, we screened a transcription regulator knockout (TRKO) library of C. albicans[21]. The library was maintained in 96-well microtiter plates at -80°C. The initial screen of 163 TR mutants was done in 36-well plates containing YP-dextrose (YPD) or YP-glycerol (YPG), since glycerol is only a substrate for mitochondrial oxidation. Cultures of each mutant were grown overnight and inoculated in each medium. We identified 6 mutants that were either unable to grow or grew poorly only on YP-glycerol (YPG) (Figure 1A and Table 1). Two independently generated mutants of each gene were used in assays for growth on both YPD and YPG (Figure 1A). Each independent gene mutant is identified as X and Y.
Figure 1.
Glycerol utilization and transcriptional regulation of GOA1 and NDH51 by the TRKOs. A. Drop plate assays of 6 TRKO mutants in YP-dextrose (YPD) or YP-glycerol (YPG). Each mutant was spot inoculated with 3 μl of stock cells and grown at 30°C for 24 h. X- and Y-sets refer to two independently constructed mutants lacking each gene. The reduction or lack of growth on YPG indicates a mitochondrial respiratory dysfunction. B. Each of the mutants shown in (A) was measured for transcription of GOA1 by Real-time PCR. Data are indicated as a down regulation of either GOA1 or NDH51. The latter encodes a Complex I subunit of the mitochondrial electron transport chain. Dpb4p, Hfl1p and Rbf1p are positive regulators of both GOA1 and NDH51.
Table 1.
TRKOs those are unable to grow on YP-Glycerol and their mutant phenotypes
| ORF | Gene | Description | Mutant phenotype |
|---|---|---|---|
| orf19.4853 |
HCM1 |
Has a forkhead domain; involved in hyphal morphogenesis; similar to the S.cerevisiae ortholog that regulates transcription during S-phase of the mitotic cell cycle |
Vegetative growth decreased in YPG |
| orf19.7068 |
MAC1 |
Regulon of CaCTR1, a high affinity copper transporter gene; role in ion homeostasis |
Vegetative growth decreased in YPG |
| orf19.2088 |
DPB4 |
Putative DNA polymerase epsilon subunit D; Ortholog of S.cerevisiae involved in DNA-dependent DNA replication |
Viable, slow-growing and displays abnormal invasive growth |
| orf19.3063 |
HFL1 |
HAP5-like; phosphorylated protein |
Vegetative growth decreased in YPG |
| orf19.5558 |
RBF1 |
Glutamine-rich activation domain; binds RPG-box DNA sequences; antigenic during human oral infection |
Vegetative growth decreased in YPG |
| orf19.6109 | TUP1 | Co-repressor; represses filamentous growth; regulates switching | Abnormal colony appearance on YPD and YPG |
The mutants that did not grow on YPG were assayed for transcription of GOA1 using real-time PCR (Figure 1B). Of the six mutants, we observed significant down regulation of GOA1 in mutants lacking HFL1, RBF1, or DPB4. Transcription levels for both GOA1 and NDH51 (Ndh51p is a complex I subunit protein) were decreased 4-6 fold in RBF1, HFL1, and DPB4 TRKO strains. Orf19.2088 is named DPB4 which corresponds to the S. cerevisiae ortholog. The MAC1 and HCM1 mutants had much smaller changes than the RBF1, HFL1, or DPB4 TRKO strains vs. wild type (WT) cells (Figure 1B). The knockout strain for TUP1 was excluded from further studies because of its poor viability. HCM1 and MAC1 are conserved transcriptional regulators in C. albicans and S. cerevisiae[21]. Mac1p and Tup1p are thought to regulate copper and iron uptake [23]. Our data indicate that we have identified three TRs that regulate GOA1 and NDH51 both of which are required for mitochondrial activity in C. albicans. Of importance, their role as regulators of cell metabolism has not been described. The remaining sections reflect our studies of the 3 TR mutants whose transcription of GOA1 was down regulated.
Morphology and growth of hfl1Δ, rbf1Δ, and dpb4Δ
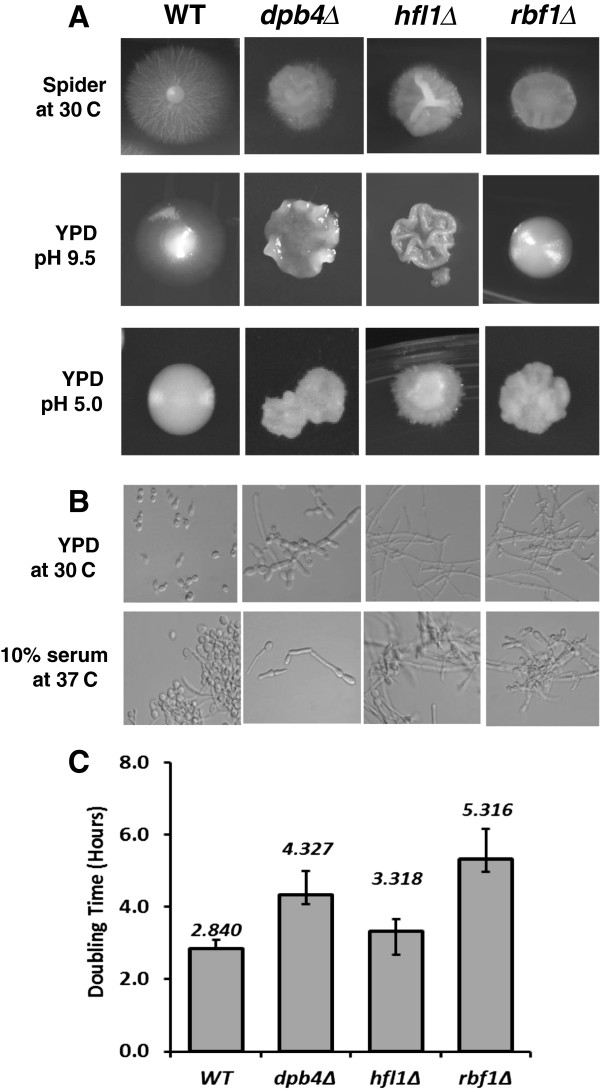
A comparison of morphology as well as doubling times was carried out with the RBF1, HFL1 and DPB4 TRKO strains (Figure 2,A-C and Table 1). Abnormal colony phenotypes (Figure 2A) and filamentous microscopic growth (Figure 2B) were observed in most growth conditions compared to the parental strain. Each of the three TRKO strains exhibited smaller colonies and a near absence of filamentous rings at the edges of colonies compared to WT colonies on Spider medium (Figure 2A). The surface of colonies from mutant strains was extensively wrinkled by day 7 on both Spider and YPD (pH 9.5) agar media. Invasive growth on YPD-2% glucose, pH 9.5, also was diminished in the three TRKO strains. All mutants were constitutively filamentous in YPD at 30°C. However, dpb4∆ was less filamentous compared to the other two mutants (50% of cells), and those were mostly pseudohyphae. Rbf1Δ and hfl1Δ were similar to WT cells in 10% serum at 37°C, as reported previously for rbf1Δ [21]. The growth phenotypes of each mutant are summarized in Table 1 and also described at the Candida CGD database (http://www.candidagenome.org). Doubling times varied according to the specific mutant from 3.32 to 5.32 hr compared to WT cells (2.84 hours) (Figure 2C).
Figure 2.
The yeast to hyphal transition, generation time, and transporter activity are altered in each of the TRKO mutants. A. Growth on Spider agar, YPD pH 9.5 and YPD pH 5.0. On Spider agar, all mutants had reduced colony growth. On YPD (below), colony diameter was again reduced and each mutant had a wrinkled colony appearance with a lack of filamentation at pH 9.5. B. Microscopic growth of each mutant is shown. In YPD (30°C), each of the TRKO mutants was constitutively filamentous compared to the yeast growth of WT cells (SN250) as reported previously [21,22]. In 10% serum, all mutants and WT strains are filamentous. C. Doubling times are shown for WT (SN250) as well as each TRKO strain. Strains dpb4∆ and rbf1∆ were significantly higher than WT cells. Strain hfl1∆ was not significantly different than WT cells. Doubling time was determined by cell mass measurements.
Antifungal susceptibilities of the TR mutants
Since we showed previously that deletions of GOA1 and NDH51 resulted in hypersensitivity to triazole antifungals [19], we compared the susceptibilities of each TR mutant to the antifungals fluconazole, amphotericin B (AmB), and caspofungin (Table 2). Since trailing has been reported, we measured both MIC50 and MIC100 for fluconazole. Similar to our published data on goa1Δ and ndh51Δ, rbf1Δ and hfl1Δ are hypersusceptible to 1-2.0 μg/ml fluconazole (MIC100), while dpb4Δ was similarly susceptible as parental cells. The susceptibilities of all mutants to AmpB and caspofungin were not statistically different compared to parental cells.
Table 2.
Antifungal drug susceptibilities to cell membrane and cell wall inhibitors*
| Gene KO |
Fluconazole(μg/ml) |
AmB (μg/ml) |
Caspofungin (μg/ml) |
|
|---|---|---|---|---|
| MIC 100 | MIC 50 | MIC 100 | MIC 100 | |
|
WT |
256 |
0.5 |
0.25 |
0.25 |
|
GOA1
|
1.0 |
1.0 |
1.0 |
0.0625 |
|
DPB4
|
256 |
1.0 |
1.0 |
0.5 |
|
HFL1
|
2.0 |
0.25 |
0.5 |
0.125 |
| RBF1 | 2.0 | 0.25 | 0.25 | 0.0625 |
*Data reflect averages of 3 experiments.
The TF mutants are hypersusceptible to inhibitors of cell wall formation
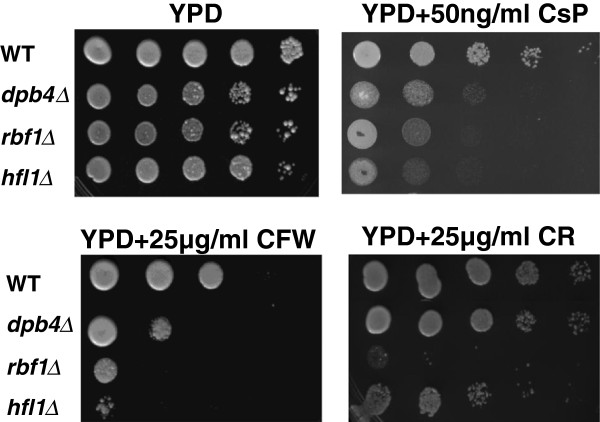
Surprisingly, all mutants were hypersusceptible to calcofluor white (CFW) and caspofungin on drop plate assays, but rbf1Δand hfl1Δ were more so than dpb4Δ (Figure 3). As for susceptibility to Congo red (a β-1,3-glucan inhibitor), only the rbf1Δ and to a lesser extent hfl1Δ were more susceptible than WT cells. Thus, rbf1Δ was affected most by cell wall formation inhibitors. The differences between the caspofungin MICs (described above) and cell wall inhibitor drop plate assays suggest that the regulation of cell wall integrity among the three TRs is different. However, the disparities of both assays could also be an explanation as MIC determinations were done using RPMI medium at 37°C while drop plate assays were done in YPD agar at 30°C.
Figure 3.
The TRKO mutants (indicated on the left) are susceptible to the cell wall inhibitors calcofluor white, Congo red and caspofungin. Cell wall inhibitors are YPD only (control), YPD + caspofungin (CsP) first row; YPD + calcofluor white (CFW) or + Congo Red (CR) in the second row. Compared to growth on YPD only, each mutant was hypersensitive to CFW and caspofungin, while only rbf1∆ was hypersensitive to CR. The hfl1∆ displayed a slight sensitivity to CR.
Mitochondrial defects in rbf1Δ, hfl1Δ and dpb4Δ
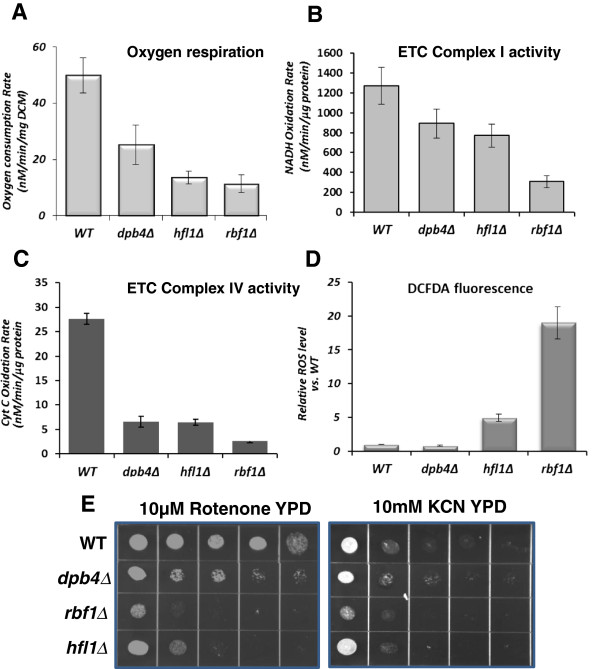
The inability of each mutant to assimilate a non-fermentable carbon source (glycerol) indicates defects in mitochondrial respiration. Further, we were interested in comparing the functions of each of the TRKO strains to GOA1 in energy production and carbon metabolism. To determine the mitochondrial status of the TRKOs, we first measured oxygen consumption among mutants and parental cells. The oxygen consumption rates were decreased by 2.2-fold for dpb4Δ, and about 5-fold in hfl1Δ and rbf1Δ compared to WT cells (Figure 4A). For these experiments, total oxygen consumption was determined from equal masses of cells (per mg dry cell mass, DCM).
Figure 4.
The TRKO mutants are deficient in respiratory functions. A. respiration; B. ETC CI activity; C. ETC CIV activity, and D. ROS production.dpb4∆, hfl1∆, and rbf1∆ each respired less (2.2-5-fold reduction) than WT cells. CI activity decreased in each mutant proportionally to their decrease in respiration. CIV activity was 5.5 fold lower in the TRKO mutants. In D, ROS production compared to WT cells was highest in the rbf1∆, although also significantly increased in hfl1∆. The dpb4∆ produced ROS equal to WT cells. E. ETC CI and CIV inhibitors: growth of mutants and WT cells on YPD containing either rotenone (C1 inhibitor) or KCN (CIV inhibitor) is shown. rbf1∆ and hfl1∆ are hypersensitive to both inhibitors while dpb4∆ was less so.
The ETC CI and CIV activities (Figure 4B, C), reactive oxidant levels (ROS) (Figure 4D, and susceptibilities to ETC CI and CIV inhibitors (Figure 4E) were also evaluated in rbf1Δ, hfl1Δ and dpb4Δ compared to WT cells. ETC CI and CIV enzyme activities for the rbf1Δ mutant were significantly reduced by 4-fold and 14-fold, respectively. Corresponding to the decrease in CI enzyme activity was an increase in sensitivity to rotenone, a CI inhibitor and KCN (CIV inhibitor) in rbf1Δ. For hfl1Δ, CI activity was less affected than rbf1Δ, but CIV activity was reduced similarly to rbf1Δ. CI enzyme activity in dpb4Δ was similar to that of hfl1Δ. Sensitivity of the dpb4∆ to rotenone was less than that of the other 2 mutants but the same as hfl1∆ in regard to KCN sensitivity. These data indicate that each of the TR mutants have altered CI and especially CIV enzyme activity although correlates with complex inhibitors are not absolute.
One of the striking features of mitochondria with dysfunctional CI and CIV activities of the ETC is an increase in mitochondrial ROS [17,18]. In this regard, ROS levels were nearly 20-fold higher in rbf1Δ and ~5-fold higher in hfl1Δ; however, ROS production in dpb4Δ was similar to that of parental cells (Figure 4D), indicating that the ROS scavenging system was less functional in hfl1Δ and rbf1Δ but not affected in dpb4Δ. Microarray data indicated that genes associated with ROS detoxification such as SOD3, GPX1, GPX2, in each mutant were increased slightly, but a down regulation in SOD6 and GRX1 occurred in both hfl1Δ and rbf1Δ (Additional file 1: Table S1, Additional file 2: Table S2 and Additional file 3: Table S3). The decrease in SOD6 and GRX1 transcription may partially explain the high ROS levels in hfl1Δ and rbf1Δ.
Global transcriptional profiling in rbf1Δ, hfl1Δ, and dpb4Δ
Based upon our published data on transcriptional profiling of the goa1Δ [19] and the functions of the RBF1, HFL1, and DPB4 as positive regulators of GOA1, we expected common gene pools as well as TR-specific gene changes. To obtain data to support this premise, we compared array data from each TR mutant to goa1Δ versus their own parental strains. A 2-fold increase/decrease in transcription was used to determine if significant changes occurred.
General observations of transcriptional changes for each TR mutant
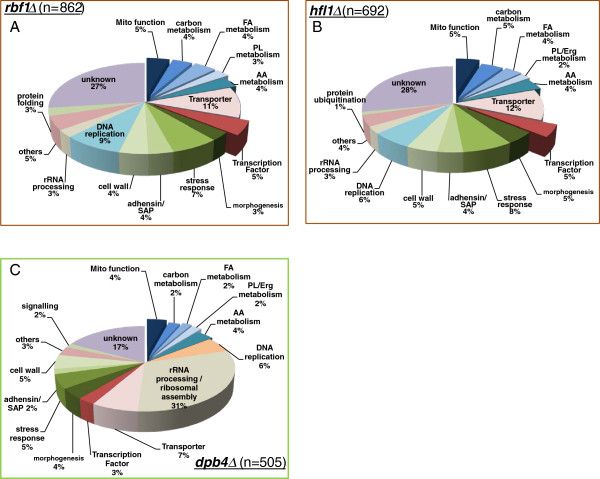
The total number of genes whose transcription changed significantly compared to SN250 was 862 (rbf1Δ), 692 (hfl1Δ) and 505 (dpb4Δ) (Figure 5). The genes with up/down changes in expression vs. the parental strain were grouped for each TR mutant based on their functional classification. The assignment of functional categories is based on the information provided by the C. albicans CGD and S. cerevisiae databases. The% of uncharacterized genes in hfl1Δ and rbf1Δ was 27-28%, while that of dpb4∆ was 17%. We found that 20% of the genes from rbf1Δ and hfl1Δ were associated with mitochondria, carbon, lipid and amino acid metabolic process. For dpb4Δ, the genes of metabolic processes accounted for only 14% (Figure 5).
Figure 5.
Total gene changes in the TRKO mutants of C. albicans rbf1∆ (A), hfl1∆ (B) dpb4∆ (C). Genes with more than a 2-fold or less than 2-fold change in expression levels are included in the diagram. Functional classification of each category is based on the C. albicans genome database (http://www.candidagenome.org/) and curated manually with S. cerevisiae genome database (http://www.yeastgenome.org/) when needed.
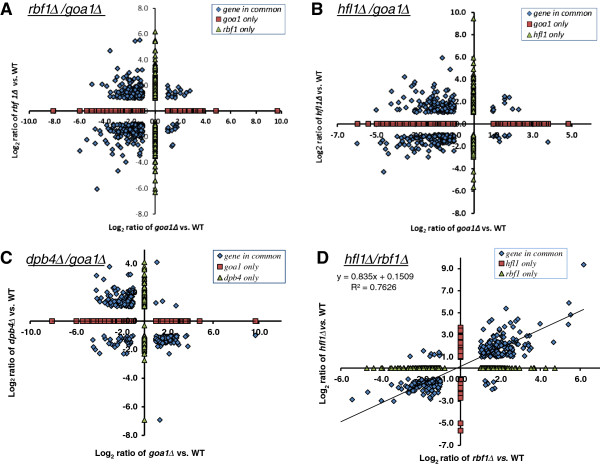
To determine differences among the three TRs in metabolic genes, we compared each TR transcriptome with the goa1Δ transcription profile. We found that a significant overlap of common genes was observed between rbf1Δ and goa1Δ (334 in total) and between hfl1Δ and goa1Δ (302 in total). By contrast, the number of genes common to both dpb4Δ and goa1Δ was less (247 in total). Gene data are represented in separate sections in Figure 6A-C for each TR mutant in comparison to goa1Δ data published previously [19], and Figure 6D compares rbf1Δ and hfl1Δ. Common down regulated genes encoding putative metabolism functions cluster in the lower left quadrants for the three TR mutants (Figure 6A-C), respectively. Common cell wall, morphology switching, and stress upregulated genes cluster in the upper left quadrants. The group of genes in the lower right quadrant in dpb4Δ (Figure 6C) is related to ribosomal /mtRNA processing and DNA/mtDNA replication or maintenance, which is down regulated in dpb4Δ. The upper right quadrants for each TR mutant represent a number of transcriptionally altered but non-functionally clustered genes. Green triangles indicate TR-specific genes and red squares indicate GOA1-specific genes (Figure 6A-C). The down regulated cluster of genes in dpb4∆ only (Y axis of Figure 6C) includes 5 mtDNA genes that encode the ETC CI subunit. The similarity of gene transcriptome changes between rbf1Δ and hfl1Δ reaches a maximum in the co-linearized rate (R2 = 0.76) that is derived from common gene pools in a total of 326 genes (Figure 6D).
Figure 6.
A comparison of the transcriptomes reveal differences among the 3 TRKO mutants. Data are derived from RBF1 (A), HFL1 (B), DPB4 (C) TRKO strains vs. the GOA1 mutant, and rbf1 vs. hfl1 mutants (D). Genes common to both the goa1∆ and either rbf1∆, hfl1∆, or dpb4∆ are depicted as blue diamonds. The goa1∆-specific genes are indicated as red squares, and TRKO-specific genes as green triangles. Upregulated genes for each TR mutant are show above the horizontal axis, down regulated genes below the horizontal axis; while the right side and left side of vertical axis represent upregulated and down regulated genes for goa1Δ. Each quadrant reflects a cluster of similar genes between goa1∆ and TR mutants. Upper left: cell wall, stress adaptation, and morphological switching; lower left: metabolism; lower right: different rewiring responses for common genes, in Figure 6C reflecting DNA replication or maintenance and rRNA processing between dpb4∆ and goa1∆; Upper right: upregulated genes of different functional annotation.
There are several points of interest from these data. First, down regulated metabolic genes (left lower quadrants) are much greater in number from rbf1Δ (Figure 6A) and hfl1Δ (Figure 6B) than in the dpb4Δ (Figure 6C). Second, dpb4Δ has a much greater number of down regulated genes that are associated with mitochondria DNA and protein synthesis machinery, such as mtDNA encoding genes and mitochondrial rRNA ribosomal assembly (lower right quadrant) compared to the other TRs (Figure 6C, Table 3). As shown in Figure 5, the total% of gene changes in rRNA processing and ribosomal assembly was 31% in dpb4∆ compared to 3% in rbf1Δ and hfl1Δ. Third, the cluster size of upregulated genes associated with cell wall integrity, stress responses, morphological switching, and adhesion are reasonably similar for all TRs but opposite with goa1Δ (upper left quadrant). These data demonstrate that the number of genes associated with metabolic and other cellular bioprocesses varies among the TR mutants, suggesting differences in their regulatory roles.
Table 3.
The transcription profiles of mitochondrial functions of TRKOs
| Biological processes |
rbf1Δ
|
hfl1Δ
|
dpb4Δ
|
|||
|---|---|---|---|---|---|---|
| down | up | down | up | down | up | |
| ETC subunits & assembly, alternative oxidase |
orf19.1691 |
NAD5, AOX2 |
NDE1 |
NAD6, MRF1 |
NAD2, NAD3 |
MRF1 |
|
ATP18, CYT1 |
orf19.3353 |
COX2 |
NAD4L, NAD4 |
|||
|
QCR8, QCR9 |
orf19.6374 |
orf19.230 |
NAD5, ATP19 |
|||
|
RIP1, orf19.2249 |
MRF1 |
orf19.3353 |
orf19.4273 |
|||
| Mitochondrial genome maintenance |
SUV3 |
PRP13,OGG1 |
SUV3,CPA1 |
|
SUV3, RPP1 |
|
|
RFC4 |
orf19.3062,RAD32 |
|||||
| Mitochondrial tRNA processing & ribosome assembly |
orf19.1201 |
MRPL6 |
orf19.1356 |
|
orf19.2309, orf19.2384 |
|
|
orf19.1356 | ||||||
|
orf19.5161 |
orf19.3303, orf19.4160 |
|||||
|
orf19.414, TRM1 | ||||||
| Mitochondrial carbon metabolism |
GCV2, ALD6 |
orf19.850 |
UCF1,CTN1 |
orf19.3043 |
IDH2 |
|
|
orf19.2184, ALD6 |
GDB1,MAL2 |
|||||
|
ILV6,DLD1 DLD2,PDK2 | ||||||
| Mitochondrial transporters |
orf19.4583 |
PAM17, ERV1 |
ATM1,orf19.28 |
|
RIM2, PET9 |
|
|
orf19.5129 |
orf19.6532 |
orf19.5628 |
||||
|
orf19.6555 |
orf19.6555 orf19.4966 |
orf19.28 |
||||
|
MDL2,FRP5 | ||||||
|
orf19.3455 | ||||||
| Others |
ISA2 |
orf19.2401 |
orf19.2175 |
PST2 |
|
|
|
orf19.2749 |
orf19.2544 |
orf19.6736 |
||||
|
orf19.6550 |
orf19.2825, orf19.4523 |
GTT12 |
||||
| orf19.5270 | orf19.4612, orf19.6283 | |||||
Functional clustering of gene changes in rbf1Δ, hfl1Δ, and dpb4Δ
Mitochondrial respiration
As shown in Figure 4A, oxygen consumption was reduced by 5-fold of WT levels in the hfl1Δ and rbf1Δ and by 2.2-fold in dpb4Δ. This observation is consistent with their increased doubling times in vitro in Figure 2C, while also indicating some degree of dysfunctional mitochondria. To better understand the reasons for the reduction in respiration, transcriptional profiling was performed. Statistically significant changes in mitochondria genes are summarized among the three mutants (Table 3).
Although hfl1∆ and rbf1∆ displayed the same reduction in oxygen consumption, mechanistically the decrease in respiration was not identical. For example, the genes that are required for assembly of ETC CIV (orf19.1691) and CV (ATP18, orf19.2249) were down regulated only in rbf1∆ (Table 3). Other respiratory–related genes such as the ubiquinol cytochrome c reductase (QCR8 and QCR9) were down-regulated in rbf1∆ as well. Candida species are known to have at least 3 respiratory pathways in addition to the classical ETC chain [18,24]. The other pathways (AOX and PAR) are utilized when the classical pathway is not functioning well. Thus, in rbf1∆, an impairment of mitochondrial ETC chain results in subtle increases of NAD5, orf19.3353 and a significant increase of AOX2 (11.5-fold). However, these apparent compensatory changes in the ETC and alternative respiratory route are not sufficient to overcome the mitochondrial dysfunctions in rbf1∆. This is especially true for AOX2, which is a hallmark gene of a mitochondrial stress response rather than its role in respiration [25].
The transcription profile of genes related to mitochondrial respiration in hfl1∆ was different. Instead of a direct impact on ETC complex subunits, we found that the down-regulated genes in this mutant were more related to mitochondrial carbon metabolism such as pyruvate (ALD6, PDK2), lactate (DLD1, DLD2) and amino acids (orf19.2184, ILV6) (Table 3); also four mitochondrial mtDNA maintenance genes (orf19.3062, SUV3, CPA1, RAD32) were transcriptionally reduced in hfl1∆. At same time, eight mitochondrial transporters of amino acid/protein, ammonium, FAD, and magnesium are down regulated in hfl1∆. Similar to the rbf1∆ mutant, a possible compensatory response of mitochondrial respiration in hfl1∆ was reflected by up-regulation of CI (NAD6), CIV (COX4) and others (Table 3). For this same mutant, the only down regulated gene with a respiratory function was NDE1 (NADH dehydrogenase), which functions as an ETC CI subunit in S. cerevisiae.
In contrast, the absence of DPB4 suppressed expression of NAD2, NAD3, NAD4L, NAD4, and NAD5 more than 2-fold; these five genes comprise the mitochondrial respiration chain CI which are encoded by mitochondrial DNA. Also the mitochondrial nucleotide transporter genes (orf19.28, and RIM2) are down regulated in dpb4Δ (Table 3). Meanwhile, the house-keeping genes for mtDNA maintenance and mitochondrial RNA processing (a total of 8 genes) were transcriptionally reduced (Table 3). Obviously, Dpb4p regulates respiration in a different manner than the other two TRs.
Fatty acid catabolism is correlated transcriptionally with decreases in phospholipid (PL) biosynthetic encoding genes
Similar to mammalian cells, in C. albicans lipids provide a source for energy generation via catabolism as well as phospholipid biosynthesis via anabolic pathways [26]. Regulation of both catabolic and anabolic pathways is critical to cell growth [27]. After comparing the transcriptome of lipid metabolism with goa1∆, differences are seen among the three TR mutants of C. albicans. The absence of DPB4 resulted in an upregulation of β-oxidation (lipid catabolism) and genes of the peroxisomal glyoxylate cycle (Table 4). But its PL biosynthesis may be compromised since INO4 (PL biogenesis activator) was down regulated by 100-fold vs. WT cells. The other TRKO strains (rbf1∆ and hfl1∆) resembled goa1∆, and each other, with significant down regulation in lipid oxidation, lipase, the glyoxylate cycle, and peroxisomal importing systems such as the peroxins. In addition, genes for PL biosynthesis including sphingolipid (SL) biosynthesis were down regulated while genes for PL catabolic processes were up regulated. In contrast to the DPB4 mutant that may regulate PL biosynthetic process, decreased gene expression for lipid catabolism and PL biosynthesis in the other two mutants indicate that RBF1 and HFL1 positively regulate both lipid catabolism and PL biosynthesis.
Table 4.
The transcription profiles of alternative carbon utilization and phenotype-related genes among TRKOs
| Biological processes | rbf1Δ | hfl1Δ | dpb4Δ |
|---|---|---|---|
| Lipid metabolism |
(n = 62)
a
|
(n = 52) |
(n = 19) |
| Dw-Peroxins (4/4)
b
|
Dw-Peroxins (5/5) |
|
|
| Dw-lipid catabolism(29/31) |
Dw-lipid catabolism(14/17) |
Up-lipid catabolism (6/9) |
|
| & glyoxylate cycle (2/2) |
& glyoxylate cycle(2/2) |
& glyoxylate cycle(2/2) |
|
| Dw-PL biosynthesis (10/12) |
Dw-PL biosynthesis (15/17) |
Dw-PL biosynthesis (4/4) |
|
|
Up-PL catabolism (3/4) |
Up-PL catabolism (2/2) |
Up-PL catabolism (3/3) |
|
| Dw-SL biosynthesis (3/4) |
Dw-SL biosynthesis (2/2) |
|
|
| Dw-ERG biosynthesis (2/4) |
Dw-ERG biosynthesis (3/4) |
Up-ERG biosynthesis (2/2) |
|
| Non-glucose and glucose utilization |
(n = 31) |
(n = 32) |
(n = 12) |
| Dw-carbon utilization (26) |
Dw-carbon utilization (23) |
Up-carbon utilization (9) |
|
| GAL1, GAL10 |
GAL1, 10,102,7 |
|
|
|
Up-fermentation |
Up-fermentation |
Up-fermentation |
|
| glycolysis |
glycolysis |
glycolysis |
|
| glycogen |
glycogen |
glycogen |
|
| glucose utilization |
glucose utilization |
glucose utilization |
|
| xylose |
trehalose |
xylose |
|
| Amino acid metabolism |
(n = 31) |
(n = 28) |
(n = 19) |
| Dw-aa biosynthesis (8) |
Dw-aa biosynthesis (9) |
Dw-aa biosynthesis (8) |
|
|
Up-aa biosynthesis(3) |
MET2,3,10,15 |
MET2,3,6,10,13,14 |
|
| Dw-aa catabolism (5) |
Dw-aa catabolism (5) |
Dw-aa catabolism (5) |
|
| |
ARO3, ARO8 |
|
|
|
Up-aa catabolism(9) |
Up-aa catabolism(8) |
Up-aa catabolism (6) |
|
|
ARO9,ARO10 |
ARO9,ARO10 |
|
|
|
Up-sulfur/nitrogen assimilation (6) |
Dw-sulfur/nitrogen assimilation (6) |
|
|
| Morphogenesis |
(n = 27) |
(n = 33) |
(n = 17) |
|
Up-hyphal formation (13) |
Up-hyphal formation (12) |
Up-hyphal formation (8) |
|
|
ECE,1 HWP1,DEF1, HGC1,FGR43 |
ECE1, HWP1, FGR18 , HGC1 |
FGR6-1,3,4,10, RBR1, IHD2 |
|
|
RBR1, IHD2,FGR6-1,4,10 |
FGR43, RBR1,IHD2 |
||
| Transporters |
(n = 101) |
(n = 80) |
(n = 37) |
| Dw: sugar, amino acid, MSF |
Dw: sugar, amino acid,MSF |
Dw: lactate, polyamine |
|
| sterol/PL, nucleosides, |
sterol/PL, nicotinamide, |
|
|
| choline, nicotinamide, |
CDRs efflux pump, urea |
|
|
| ion (K+, NH4+, Ca+2, P-, Cl-) |
ion (S-, NH4+, Zn+2, P-) |
|
|
|
Up: urea, allantoate |
Up:spermidine/polyamine |
Up: glucose, acetate, MSF |
|
| spermidine/polyamine |
cation (H+, Ca+2,Cu+2, Fe+3) |
fatty acid, aa, |
|
| cation (H+, Cu+2, Fe+3) |
ions (H+, Cu+2, Fe+3 , S-) |
||
a: Total number of genes in this group;
b: x/y indicates “x” number of genes are down (Dw) or up (Up) regulated among total of “Y” number of genes in this metabolic process.
Alternative carbon source metabolism is also regulated by each TR
The biological implications for the assimilation of non-glucose carbon sources even when glucose is not limiting for C. albicans has been described [12,28-30]. We observed that numerous genes, required for non-glucose utilization in both rbf1∆ (26 of a total of 31 genes) and hfl1∆ (23 of 32), were down regulated along with mitochondrial defects. Notably, the GAL gene cluster was significantly reduced by 4.6-6.4 fold in hfl1∆ (GAL1, 7, 10, 102) and 2.9-3.0-fold in rbf1∆ (GAL1, 10) (Table 4). On the other hand, most of the genes for alternative carbon consumption in dpb4∆ increased transcriptionally (9 of 12 in total), including genes for fermentation (IFD6), glycogen catabolism, and the xylose catabolic gene XYL2. The genes of these three metabolic processes also were upregulated in RBF1 and HFL1 mutants. Therefore, we assume that the growth defects of RBF1 and HFL1 mutants were also contributed by their reduced ability to use non-glucose carbon sources including lipids mentioned above. However, gene transcription of glycolysis and fermentation was upregulated in each mutant.
Amino acid metabolism is regulated by each TR
Regarding genes of amino acid biosynthesis, more genes were downregulated than upregulated for each of the TRKO mutants (Table 4). However, for the hfl1∆ and dpb4∆, down regulation of methionine synthesis genes were particularly common. Interestingly, transcription of the aromatic amino acid catabolic genes ARO9 and ARO10 were up-regulated only in rbf1∆ and hfl1∆ (Table 4). Both gene products are aromatic transaminases [31]. Their functions are associated with providing an alternative, energy efficient means for NADH regeneration, nitrogen assimilation, and pseudohyphal growth [31]. As stated above, down regulation of the MET genes was observed in hfl1∆ and dpb4∆. Methionine, as a constituent of proteins, is also critical to biochemical pathways, including the “methyl cycle” which generates the key metabolite S-adnosylmethioinine (AdoMet) [32]. As the main donor of methyl groups in methylation reactions, AdoMet plays a vital role in de novo phosphatidylcholine (PC) synthesis that requires three AdoMet-dependent methylation steps [33].
Morphogenesis and cell wall responses are regulated by each TF
The repressive activity of RBF1 on filamentous growth in C. albicans was first noted by Aoki et al[22]. In Table 4, we list the most common genes that are related to filamentous growth and their expression level in each mutant. We show that the production of hyphae was associated with the upregulation of genes, such as RBR1, HWP1 and ECE1 in rbf1Δ and hfl1Δ mutants, but much less so in dpb4Δ. Transcriptional changes were not noted in the transcription factors CPH1 and EFG1. These partial transcriptional profiles mostly correspond to the hyphal phenotypes of the rbf1Δ and hfl1Δ mentioned above.
Microarray data support a general increase of genes encoding cell wall β-glucan biosynthesis among three mutants, such as EXG2, PHR1, PHR2, GSC1 and KRE1. Up or down regulation of genes associated with the regulation of mannosylation are noted in the hfl1Δ and rbf1Δ (Additional file 1: Table S1 and Additional file 2: Table S3). In addition to the cell wall glucan biosynthesis genes, those of the cell wall integrity and MAPK pathways were up-regulated, including the CHK1 histidine kinase and the CEK1 MAP kinase. Both genes are known to regulate cell wall polysaccharide synthesis [34,35].
Regulation of metabolic flux transporters
The regulatory roles of the three TRs on transporter activity have been noted (Tables 3 and 4). The major changes in both rbf1∆ and hfl1∆ mutants were down-regulation of transporters for sugar, lipid, amino acids, as well as the MFS transporter family (major facilitating superfamily). Quantitatively, 101 transporters were downregulated in rbf1∆, 80 in hfl1∆, and 37 in dpb4∆, of which the mitochondrial transporters and inter organelle transporters are not included. Certainly, the circuits for nutrient import from extracellular environment or intracellular translocation between compartments are regulated by all TRs but less so by DPB4. In dpb4∆, gene expression for MFS, sugar, lipid and amino acid importers are increased.
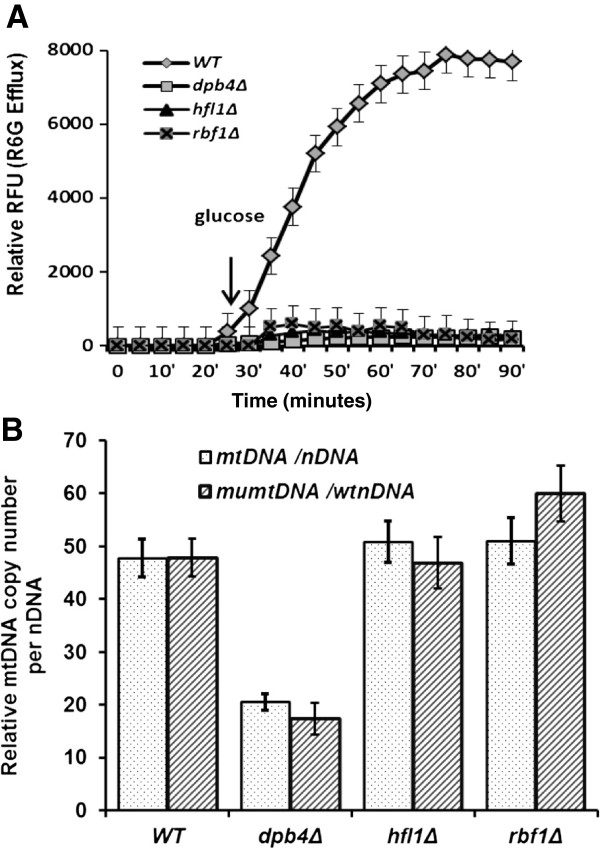
The measurement of intracellular accumulation of R6G is a useful method to reflect the activity of the CDR drug efflux pumps. The extracellular release of R6G in C. albicans was inversely correlated with the level of this group of efflux exporters [36]. Similar to goa1∆, the CDR genes (CDR2, CDR4 and CDR11) are down regulated in hfl1∆, which may explain its poor extracellular efflux rate of R6G shown in Figure 7 and hypersusceptibility to fluconazole (Table 2). However, these CDR genes were not changed in rbf1∆ and dpb4∆ although they displayed a similar rate of R6G efflux as hfl1∆. Rbf1p, Hfl1p, or Dpb4 may regulate efflux by a different mechanism. Because R6G has a permanent positive charge, its cellular accumulation relies on a plasma membrane potential that is localized mainly in the mitochondria [37].
Figure 7.
Membrane transport of R6G is reduced in each TF mutant and relative mtDNA copy number is less in dpb4∆ mutant (7B). (A) Parental (SN250) and each mutant were assayed for transport of R6G. Relative RFU, relative fluorescent units over a 90 min time interval were determined. Cells were starved in buffer for 20 min then glucose was added to each culture. Compared to parental cells, all mutants had little transporter activity. (B) The ratio of mtDNA copy number to that of nuclear DNA (nDNA) is calculated by ΔCt with matched pairs of mtDNA/nDNA primers. The relative copy number of mtDNA is averaged from three biological replicates (mtDNA/nDNA). Compared to the parental strain, dpb4Δ has less mtDNA copies compared to its own nDNA or parental nDNA. However mutants (rbf1Δ and hfl1Δ) have a similar mtDNA copy number as the parental strain. (mumtDNA: mutant mtDNA; wtnDNA: wild type strain nDNA).
The spermidine transporter was only upregulated in rbf1∆ and hfl1Δ. These data may illustrate that these mutants have a high demand for sustaining intracellular pH and membrane potential since the spermidine transporter synchronizes Ca2+, Na+, K+ -ATPase in plant cells [38]. However, transporters of metal cations were upregulated in each of the TRKO mutants. The significance of uptake of Fe3+ and Cu2+ uptake is related to mitochondrial respiration since electron transfer among ETC complexes is carried out by reduced metal ions [39]. The high demand for metal ion uptake in the TRKO mutants again suggests their defective mitochondria.
Dpb4p is required for mitochondrial genome maintenance in C. albicans
Giving the fact that a few complex I genes are down-regulated transcriptionally in dpb4Δ described above (Figure 4), we performed real-time PCRs to determine if mtDNA maintenance is affected in this mutant with four sets of primers: two sets of mtDNA encoded genes NAD1 (complex I subunit) and COX1 (complex IV subunit), and two sets of nDNA genes (18S rRNA and SOD1). The average number of copies of mtDNA per nDNA for DPB4 TRKO strain is less than half the levels of WT and other two mutants tested in Figure 7B. Since nDNA replication is also extensively affected in dpb4Δ mutant microarray data, we also normalized the mtDNA copy number by comparing the mtDNA Ct of dpb4Δ with nDNA Ct of WT cells. Again we see the reduction of mtDNA replication rate in this mutant.
The TRs regulate other TRs
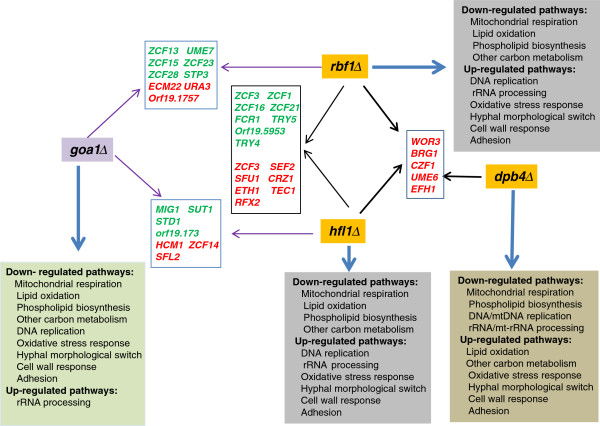
In eukaryotes, ~3-5% of their protein repertoire is transcription factors [40]. In our previous studies of GOA1, we found ~100 transcription factors that were down regulated in the gene-deleted mutant, including reduction of a large group of Zn2-Cys6 cluster TRs. Presumably, the TRs regulate metabolic pathways. However, of 77 such genes, most are poorly characterized, yet they are fungal-specific [41]. We compared the regulation of other TRs by RBF1, HFL1 and DPB4. The TRs regulated by RBF1 and HFL1 are closely related. Fifteen of the TR genes were either up or down regulated (Figure 8 and see Discussion). In this group, some genes shared between RBF1 and HFL1 mutants were also changed in the GOA1 mutant, such as ZCF1, ZCF5, ZCF16, ZCF21, FCR1, TRY4 and RFX2. The likely involvement of these 7 TRs in the regulation of metabolic process needs to be determined, although the last three transcriptional regulators have been reported to process filamentous growth and azole drug resistance [42,43].
Figure 8.
TRKOs and their influence on transcription of other TR genes. Each TRKO mutant and goa1∆ is shown with arrows that connect common TR genes (rectangular boxes) affected in each null mutant. The TR genes are indicated in red (upregulated) or green (down regulated). Thus for dpb4∆, 5 upregulated genes are shown (connected by a right-facing black arrow) that are common to both rbf1∆ and hfl1∆. Common TR genes of rbf1∆ and hfl1∆ are similarly inked by black arrows. The number of TR genes common to both rbf1∆ and hfl1∆ is much greater than those common to dpb4∆. Also shown are comparisons of presumed up or down regulated pathways for each mutant including goa1∆ (square boxes of various colors).
Only 5 common gene changes were noted in the three TRKOs (Figure 8). Interestingly, the predicated functions of UME6, WOR3, BRG1, CZF1 and EFH1 are all associated with hyphal formation or the white-opaque switch that actually matches their hyphal phenotypes. However, these genes were down-regulated in goa1Δ which correlates with the inability of that strain to form hyphae under hyphal inducing conditions. In conclusion, RBF1, HFL1 and DPB4 are repressors of the five genes mentioned above for morphologic switching. Overall, the regulatory network for DPB4 mutant is very different, since only 14 of TRs were changed in dpb4Δ compared to 44 for rbf1Δ and 31 for hfl1Δ, including the five common TRs just mentioned above and two other TRs all of which relate to morphogenesis.
Discussion
Goa1p is required (directly or indirectly) for a number of cell processes [16-20]. Those processes include homeostasis of stationary phase cells, morphogenesis, prevention of early apoptosis, ATP production through its regulation of the ETC CI, and communication between peroxisomes and mitochondria. The latter function ensures that both organelles coordinate pathways of energy production, including aerobic respiration, β-oxidation, gluconeogenesis, and carbon conservation via the glyoxylate cycle. We believe these are the core functions of Goa1p. Deletion of GOA1, achieved by constructing a null mutant, causes loss of chronological aging, constitutive yeast growth, lethal levels of ROS, apoptosis, heightened susceptibility to triazole antifungal drugs, and reduced energy production from both mitochondria and peroxisomes. The host-pathogen outcomes from a lack of Goa1p are avirulence, a failure to colonize kidney tissues, and increased killing by human neutrophils and macrophages [16,20].
The availability of a transcription regulator knockout (TRKO) library proved suitable for facile screening based upon the ability of each mutant to grow or not on YP medium containing glycerol as the sole carbon source. Six such mutants were then reduced to three whose gene absence caused a down regulation of GOA1 indicating that each TR is a positive regulator of GOA1. However, we have not as yet identified gene targets of each TF. Thus, our supposition of direct or indirect regulation by each TF awaits confirmation. Our ongoing studies focus on chromatin immunoprecipitation (ChIP) to identify gene targets of each TF.
We have not completed phenotyping profiles of the TRKOs except for antifungal susceptibilities and respiratory competency. In this regard, Rbf1p and Hfl1p are similar to Goa1p in that they each are required for maintenance of WT levels of fluconazole susceptibility. Each of the three TRs is required for respiratory activities but to varying degrees. For example, Rbf1p is much more critical to the maintenance of CI activity and, consequently, low levels of ROS. Each of the TRs is a negative regulator of morphogenesis (yeast-hyphal transition), yet Goa1p would appear to be part of a positive regulatory circuit. Further, Goa1p is critical to host cell recognition. Loss of GOA1 results in a downregulation of genes encoding this function (17-19). Oppositely, the TR mutants display upregulation of these genes. This difference needs to be correlated with the binding of the TRKOs to epithelial and innate immune cells. The cell wall inhibitor studies may point to a role of each TF in compensatory changes in response to cell wall defects caused by inhibitors. A last look at functional annotation is that of transporter activity, of which each of the TRs and Goa1p is essential (Figure 7A).
An important advantage of the TRKO mutants is to identify genes that are common to specific biological processes. For example, SUV3 is the only common down-regulated gene of mitochondrial genome maintenance for all TR mutants (Table 3). SUV3 is an RNA helicase that is essential for mitochondrial RNA catabolism and respiratory growth. Oppositely, for the filamentous morphogenesis circuitry, three genes among several are commonly upregulated in each TR [RBR1 and IHD2 (Table 4)].
To visualize the overall roles of each TR and Goa1p, we aligned genes that were up or down regulated in each mutant and also shared among the TRKOs. Also, for each TR, we have summarized the gene responses as presumed functional activities (Figure 8). The three TRs share similar roles in metabolic pathways, stress responses, cell wall regulation, fluconazole susceptibility, and morphogenesis, but transcriptional changes are different. As stated previously, there was a higher number of genes committed to metabolic processes in Rbf1p and Hfl1p than in Dbp4p regarding alternative carbon utilization of non-favorable carbon, lipid and amino acid catabolism. Oppositely, metabolic regulation performed by Dbp4p is likely because of its role in the maintenance of mtDNA genome (Figure 7B). Thus, when mtDNA replication rate in dpb4Δ is compromised, the cell metabolism and respiration are also then affected. While Rbf1p and Hfl1p regulated lipid oxidation and other peroxisomal functions such as the glyoxylate pathway, Dbp4p is less critical to these processes. Likely, phospholipid homeostasis in C. albicans is also contributed by each TR but in different ways. In fact, a more than 100-fold reduction of INO4 suggests that Dpb4p provides a positive regulation of PL biosynthesis. Ino4p is the PL biosynthetic activator that forms a heterodimer with Ino2p to regulate UASINO-containing genes [44]. When PA levels drop in ER, the Opi1p repressor disassociates from the nuclear/ER membrane, then translocates to nuclei to attenuate the Ino2p-Ino4p complex activity [45].
Oxidative stress caused by ROS is usually associated with aberrations of mitochondrial metabolism that can threaten cell survival [46]. In this regard, the levels of cell ROS are quite different among the TRKO mutants. ROS production by rbf1∆ was much higher than in hfl1∆, while that of dpb4∆ was similar to WT cells. However, most stress response genes in rbf1∆ and hfl1∆ were similarly represented. Perhaps the higher level of ROS in rbf1∆ is due to the combined loss of CI and CIV activities. CI accounts for ~ 50% of the oxidoreduction activity of the ETC, and if dysfunctional, one could expect much more ROS as superoxide. Another major functional difference in the three TRs as compared to Goa1p is that they negatively regulate genes associated with cell wall and cell integrity while Goa1p positively regulates this gene cluster including the signaling transduction pathway MAPK CEK1 and CHK1[24,25]. Again, direct versus indirect regulatory effects need to be sorted out. Also, as stated above, Goa1p is required for the yeast to hypha phase transition along with cell surface GPI-anchored adhesins, while each of the three TRs repress these growth requirements. Finally, we note again that Dbp4p positively regulates nuclear DNA/mtDNA replication or maintenance as well as rRNA/mt-rRNA processing. The assignment of genes in these groups quantitatively is much greater in Dbp4p than the other two TRs.
Translational applications of these data are apparent. The TFs are fungal-specific, and therefore are of reasonable purpose as drug targets for antifungal drug discovery. Inhibitors likely will have severe consequences on cell metabolism and growth. High throughput screens for such inhibitors might utilize heterozygote libraries of TRs. Hypersensitivity of strains to compounds compared to a parental strain indicates a haploinsufficiency response (decreased fitness) suggesting a drug target. Or, screens of the TRKO mutants with a compound library may be useful. In this case, a resistance phenotype likely indicates the target of the compound.
Conclusions
Our data are the first to integrate the regulation of metabolic processes by the transcription regulators Rbf1p, Hfl1p and Dpb4p. That these TRs regulate mitochondria and peroxisomal metabolism also is new to fungal pathogens and speaks to their regulation of organelle cross-talking. Equally important, each TR was previously unstudied except for two reports on Rbf1p and its role as a negative regulator of morphogenesis in C. albicans. Of particular interest is that there is little overlap in genes of mitochondrial functions regulated by the 3 TRs, yet each is functionally important to this organelle (Table 3). In regard to other cell metabolic activities, such as lipid metabolism, non-glucose utilization, and amino acid metabolism, Rbf1p and Hfl1p are more similar in their gene profiles compared to Dpb4p (Table 4). Two of the major differences in gene transcription among the TRs are transporter genes, of which Rbf1p and Hfl1p clearly are of broader scope than Dpb4p. Second, Dpb4p is a regulator for mtDNA genome maintenance. These observations provide the beginning of an integrative view of global cell functions by transcriptional regulators of fungi.
Methods
Strains and media
The homozygous mutants (TRKO) of a C. albicans transcription factor rbf1∆, hfl1∆ and dpb4∆ (orf19.2088) were obtained from transcription factor (TR) library provided by Dr. Alexander Johnson’s lab [21]. All assays with these mutants include the parental strain C. albicans SN250 [21]. All strains were grown at 30°C in 2% YPD, 2% glycerol YPG medium (glycerol used as carbon source), minimal medium (0.67% yeast nitrogen base) containing 2% glucose (SD), or Spider agar plates and YPD at pH 9.5.
TR deletion library screens
The entire TRKO mutant library was initially screened in parallel YPD and YPG plates. For these experiments, TR mutants with mitochondrial defects were identified by their lack of growth on YP-glycerol. 3 μl of cells was added to YPD or YP-2% glycerol (YPG) plates. Plates were kept at 30°C incubator for 24 hours and those with growth defects in YPG only were identified and verified by a second screening with a second deletion strain of the same gene. Subsequent experiments were done only with those mutants that had defective growth in YPG.
Morphology and generation times
All strains were grown overnight in YPD at 30°C for all experiments described below. Cells were washed, diluted to a cell suspension of 1 × 105/ul, and streaked on YPD, pH 9.5 or Spider agar media and incubated at 30°C. Plates were observed on day 7 and photographed. The morphologic switch from yeast to filamentous forms in 10% serum at 37°C for all strains was done with the same growth conditions. Mutants were compared to SN250. Generation times for rbf1∆, hfl1∆, and dpb4∆ strains were evaluated as described [16]. All strains were grown in YPD media at 30°C for 20 hours and cell suspensions were adjusted to an initial cell concentration of OD = 0.1. Also, since the mutants were constitutively filamentous, 50 ml of each culture was centrifuged, and cell pellets were dried, and weighed every 2 hours. Doubling time was determined based on the biomass for each strain in duplicate cultures.
Functional mitochondrial assays
The measurement of oxygen consumption, reactive oxidant species (ROS) levels, and mitochondrial enzymatic activities of each mutant (rbf1, hfl1 and dpb4) and SN250 were done as described [21]. In brief, for oxygen consumption experiments, each strain was inoculated into 100 ml of YPD (2% glucose) broth until exponential growth was achieved. Cells were washed twice with PBS and suspended into fresh YPD at a cell concentration of OD = 0.3. 1 ml of cells was then loaded immediately into the sealed respirometer chamber (Hansatech Instruments Ltd., Norfolk, England). Oxygen consumption was measured over 10 min and polarographically recorded using Oxygraph Plus software. The remaining cultures were centrifuged to determine cell biomass. Oxygen consumption is presented as nmol per min per mg cell dry weight. Data from three experiments were averaged.
Intracellular ROS levels for each strain were evaluated by staining cells using the ROS sensitive fluorescent dye DCFDA (2′,7′-dichlorofluorescein diacetate; Sigma). Since growth was filamentous, the final step in ROS measurement was performed using a fluorescence microplate reader in 96-well black plates (Dynex Technologies Inc., Chantilly, VA, USA) at λex: 485 nm and λem: 530 nm. Cell suspensions were kept in the dark to minimize loss of fluorescent signal during the assay. Cell cultures for each strain were prepared in 20 ml of YPD using an inoculum of 5 × 104/ml; cells were grown overnight at 30°C, in shake culture (200 rpm). The cell pellets from 1 ml of cultures were washed once with PBS and suspended to 1 ml of PBS with 50 μM DCFDA for 30 min at 30°C, 100 rpm. Cells were washed twice with PBS, and 200 μl from each strain was introduced into a 96-well microtiter plate. Cell fluorescence in the absence of DCFDA was used to verify that background fluorescence was similar per strain. ROS data was obtained from duplicate cultures, and all experiments were repeated a total of 3 times.
Enzyme activities of the mitochondrial electron transport chain (ETC) CI and CIV were measured spectrophotometrically following procedures described previously [17,18]. CI (NADH:ubiquinone oxidoreductase) and CIV (cytochrome c oxidase) activities are plotted from duplicated samples for each strain as nmol per min per μg of mitochondrial protein.
Antifungal susceptibility tests
The susceptibility (MIC50 and MIC100) for all strains to fluconazole, amphotericin B (AmB) and caspofungin was determined using the broth microdilution method according to CLSI guidelines M27-A3. The range of drugs tested was 0.25-256 μg/ml for fluconazole; 0.03-32 μg/ml for AmB; and 0.016-16 μg/ml for caspofungin. Exponentially grown cultures for each tested strain were diluted in RPMI-1640 to a density of 1 × 104 CFU/ml and 100 μl was added to each well of 96-well plate containing 100 μl RPMI-1640 with different concentration of drug. All plates were incubated for 48 h at 37°C. The MIC100 was determined as the concentration resulting in complete growth inhibition, and MIC50 for fluconazole corresponded as an inhibition of at least 50% of fungal growth.
Cell wall and ETC CI and CIV inhibitor assays
Overnight cultures of all strains were collected and washed twice with PBS. The cell suspension, adjusted to 5 × 105 to 5 × 101 in 10 μl PBS, was spotted onto YPD agar with or without inhibitors. For identifying the cell wall defects, 25 μg/ml of calcofluor white (CFW) or Congo red (CR) was added to YPD plates. CI and CIV inhibitors were used at concentrations of 10 μM rotenone and 10 mM KCN in YPD agar. Cultures were incubated at 30°C for 24 h and photographed.
Rhodamine 6G (R6G) efflux
These experiments were performed using a modified procedure of our earlier published data [19] using 96-well microtiter plates. In brief, cells were initially seeded into 10 ml of fresh YPD after an overnight culture. Exponentially growing cells were washed twice with PBS (pH 7.0, without glucose), and suspended in glucose-free PBS to 108/ml for 2 hours incubation to deplete glucose. Rhodamine 6G was then added at a final concentration of 10 μM for 20 min. Again, cells were washed and suspended in glucose-free PBS before introducing 2% glucose. At every 10 min base, 0.2 ml of cells were removed and energy-dependent efflux of R6G was measured by monitoring the absorption at 527 nm in that were transferred into a black 96-well plate in triplicate, glucose-free controls were included in all experiment.
Quantitative PCR analysis of Mitochondrial DNA (mtDNA) replication rate
The total DNAs were isolated from SN250 strain and mutants using RNase to remove RNA followed by standard phenol/chloroform extraction and ethanol precipitation. The concentration of DNAs was determined by a nano-spectrophotometer. The primers for analysis of mtDNA are NAD1F (5′-TAGGTTGTGTTGCTGAATGTGC) and NAD1R (5′-CCAGTACCACCACCCATAAATAAG), COX1F (5′-GGTGAATTACGTCTAGCTGTTCC) and COX1R (GCACCATCTAATAGCCCTACTCA). Two sets of nuclear DNA (nDNA) gene are 18SrRNAF (5′-CGCAAGGCTGAAACTTAAAGG) and 18SrRNAR (5′-AGCAGACAAATCACTCCACC), SOD1F(5′-GCTCCAACCACAATTTCCTG) and SOD1R (5′-TGGATTGAAATGAGGACCAGC). The 20 μL PCR reaction contains 1× iQSyBR green supermix (Bio-Rad), 0.25 μM of each primer, and approximately 5 ng of total genomic DNA for each strain. PCR conditions are 2 min at 95°C, followed by 40 cycles of 15 s of denaturation at 95°C and 30 s of annealing at 55°C and 30s of extension at 60°C. The relative copy number of mitochondrial DNA over the nuclear DNA was averaged from the threshold cycle number (Ct) difference for each pairs of mtDNA/nDNA [47,48]. The individual ratio was determined from each sets of mtDNA/nDNA pairs use the calculation equation N = 2ΔCt where ΔCt = CtnDNA1 -CtmDNA1 or ΔCt = CtnDNA2 -CtmDNA2. Statistical analysis of data was conducted by the t test.
RNA and microarray analyses
For transcriptional profiling, RNA was obtained from the TRKO mutants and SN250 grown in 20-ml of 2% SD medium at 30°C for 5 h as previously described [17,19]. RNA was quantified using an RNA 6000 Nano device, and RNA integrity was assessed using an Agilent 2100 bioanalyzer. For real time PCR measurement of GOA1 and NDH51 transcription, overnight cultures in YPD were seeded into 20 ml of fresh SD medium containing 2% glucose. When exponential growth was achieved for all strains, cells were collected and washed, then suspended in YPG medium for one hour before RNA was extracted. Approximately 800 ng of RNA was used to prepare cDNA. Quantitative real-time PCR was carried out in 20 μl of 1x iQ SyBR green Supermix (Bio-Rad) containing 0.25 μM concentration of each primer. The experiment was performed in triplicate using Bio-Rad iQ5, and the transcription level of each gene was normalized to C. albicans 18S rRNA levels. The 2–ΔΔCT method of analysis was used to determine the fold change in gene transcription [17,18].
One-color microarray-based gene expression analysis was done using the Agilent low input Quick Amp Labing kit. The RNAs for each strain were prepared from exponential cells cultured in 20-ml of SC medium containing 2% glucose. cDNA was synthesized from 100 ng total RNA for each strain according to the manufacturer’s instructions. Hybridization was completed in a Agilent SureHyb hybridization chamber and scan processed with an Agilent SCAN G2505C Microarray Scanner System. The array used in this study was provided by Agilent Technologies (eArray, ID 037557). The total of 6101 genes (including 12 mitochondrial genes) was done in duplicate.
The image files were first analyzed by Agilent Feature Extraction Software and cyanine 3 intensities were then logarithmically transformed and statistically normalized. The fold change for each gene was calculated by comparing to wild type. In this study, we adopted the cut off for the parametric p-value <0.05 and fold change >2 to determine the significance. The entire significant genes list for rbf1Δ, hfl1Δ and dpb4Δ are available in the supplemental material (Additional file 1: Table S1, Additional file 3: Table S2 and Additional file 2: Table S3).
Availability of supporting data
The microarray data of three TRKO strains and wild type SN250 have been deposited to the GEO database with accession number [GEO:GSE54057]. The microarray data of each mutant with gene changes more than 2- fold are included in this manuscript as additional files indicated below.
Abbreviations
AA: Amino acid; ALS: Agglutinin-like sequence; CFW: Calcofluor white; CLSI: The Clinical and Laboratory Standards Institute; CR: Congo red; ERG: Ergosterol; ETC: Electron transport chain; MIC: Minimum inhibitory concentration; PA: Phosphatidic acid; PL: Phospholipid; R6G: Rhodamine 6G; ROS: Reactive oxidant species; SL: Sphingolipid; TR: Transcription regulator; TRKO: Transcriptional regulator knockout library; UASINO: Upstream activation sequence INO; WT: Wild type.
Competing interests
The authors declare no competing interests exist.
Authors’ contributions
P.S performed the TRKO library screening and partial functional study; K.K and D.L performed the most of functional studies, morphological studies, Q-PCR and microarray data analysis; N.S and D.L performed microarray assay; R.C and D. L provided the theoretical framework and guidance for this study and wrote the manuscript. All authors read and approved the final manuscripts.
Supplementary Material
Up and down-regulated genes list in rbf1Δ mutant.
Up and down-regulated genes list in dpb41Δ mutant.
Up and down-regulated genes list in hfl1Δ mutant.
Contributor Information
Kasra Khamooshi, Email: kk493@georgetown.edu.
Patricia Sikorski, Email: patrycjasikorski@gmail.com.
Nuo Sun, Email: nuosun@gmail.com.
Richard Calderone, Email: calderor@georgetown.edu.
Dongmei Li, Email: dl33@georgetown.edu.
Acknowledgements
The experiments were supported by a grant from the NIH-NIAID (AI09029). The authors also wish to thank the Biomedical Graduate Research Organization of the Georgetown University Medical Center for funds.
References
- Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;15:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- Calderone R, Gay-Andrieu F, Li D, Alex D, Sun N. In: Antimicrobial Drug Discovery. Tegos G, Mylonakis E, editor. UK: CABI; 2012. Antifungals and antifungal discovery, chapter 17. [Google Scholar]
- Arnold HM, Micek ST, Shorr AF, Zilberberg MD, Labelle AJ, Kothari S, Kollef MH. Hospital resource utilization and costs of inappropriate treatment of candidemia. Pharmacotherapy. 2010;15:361–368. doi: 10.1592/phco.30.4.361. [DOI] [PubMed] [Google Scholar]
- Gauwerky K, Borelli C, Korting HC. Targeting virulence. A new paradigm for antifungals. Drug Discov Today. 2009;15:214–222. doi: 10.1016/j.drudis.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Pfaller M, Neofytos D, Diekema D, Azie N, Meier-Kriesche HU, Quan SP, Horn D. Epidemiology and outcomes of candidemia in 3648 patients: data from the Prospective Antifungal Therapy (PATH Alliance®) registry, 2004-2008. Diagn Microbiol Infect Dis. 2012;15:323–31. doi: 10.1016/j.diagmicrobio.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Blumberg HM, Jarvis WR, Soucie JM, Edwards JE, Patterson JE, Pfaller MA, Rangel-Frausto MS, Rinaldi MG, Saiman L, Wiblin RT, Wenzel RP. National Epidemiology of Mycoses Survey(NEMIS) Study Group. Risk factors for Candida blood stream infections in surgical intensive care unit patients. Clin Infect Dis. 2001;15:177–186. doi: 10.1086/321811. [DOI] [PubMed] [Google Scholar]
- Thompson GR 3rd, Patel PK, Kirkpatrick WR, Westbrook SD, Berg D, Erlandsen J, Redding SW, Patterson TF. Oropharyngeal candidiasis in the era of anti-retroviral therapy. Oral Surg Oral Med Oral Pathol Oral Radiol Endo. 2010;15:488–495. doi: 10.1016/j.tripleo.2009.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J, Goldfarb N. Candidemia in the in-patient setting: treatment options and economics. Expert Opin Pharmacother. 2007;15:1643–1650. doi: 10.1517/14656566.8.11.1643. [DOI] [PubMed] [Google Scholar]
- Grumaz C, Lorenz S, Stevens P, Lindemann E, Schöck U, Retey J, Rupp S, Sohn K. Species and condition specific adaptation of the transcriptional landscapes in Candida albicans and Candida dubliniensis. BMC Genomics. 2013;15:212. doi: 10.1186/1471-2164-14-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahi P, Moye-Rowley WS. Coordinate control of lipid composition and drug transport activities is required for normal multidrug resistance in fungi. Biochim Biophys Acta. 2009;15:852–859. doi: 10.1016/j.bbapap.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandai D, Yin Z, Selway L, Stead D, Walker J, Leach MD, Bohovych I, Ene IV, Kastora S, Budge S, Munro CA, Odds FC, Gow NA, Brown AJ. The evolutionary rewiring of ubiquitination targets has reprogrammed the regulation of carbon assimilation in the pathogenic yeast Candida albicans. MBio. 2012;15:e00495-12. doi: 10.1128/mBio.00495-12. doi: 10.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi M, Kamiyama A, Tokunaga M. Respiration of medically important Candida species and Saccharomyces cerevisiae in relation to glucose effect. J Med Vet Mycol. 1988;15:195–198. doi: 10.1080/02681218880000271. [DOI] [PubMed] [Google Scholar]
- Ramírez MA, Lorenz MC. Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes. Eukaryot Cell. 2007;15:280–290. doi: 10.1128/EC.00372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barelle CJ, Priest CL, Maccallum DM, Gow NA, Odds FC, Brown AJ. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 2006;15:961–971. doi: 10.1111/j.1462-5822.2005.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M, Bender J, Fink G. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;15:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambach A, Fernandes MP, Ghosh A, Kruppa M, Alex D, Li D, Fonzi WA, Chauhan N, Sun N, Agrellos OA, Vercesi AE, Rolfes RJ, Calderone R. Goa1p of Candida albicans localizes to the mitochondria during stress and is required for mitochondrial function and virulence. Eukaryot Cell. 2009;15:1706–1720. doi: 10.1128/EC.00066-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Calderone R, Sun N, Wang Y, Li D. Caloric restriction restores the chronological life span of the Goa1 null mutant of Candida albicans in spite of high cell levels of ROS. Fungal Genet Biol. 2012;15:1023–32. doi: 10.1016/j.fgb.2012.09.007. [DOI] [PubMed] [Google Scholar]
- Li D, Chen H, Florentino A, Alex D, Sikorski P, Fonzi WA, Calderone R. Enzymatic dysfunction of mitochondrial complex I of the Candida albicans goa1 mutant is associated with increased reactive oxidants and cell death. Eukaryot Cell. 2011;15:672–682. doi: 10.1128/EC.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Fonzi W, Chen H, She X, Zhang L, Zhang L, Calderone R. Azole susceptibility and transcriptome profiling in Candida albicans mitochondrial electron transport chain complex I mutants. Antimicrob Agents Chemother. 2013;15:532–542. doi: 10.1128/AAC.01520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Zhang L, Chen H, Calderone R, Li D. Cell surface changes in the Candida albicans mitochondrial mutant goa1Δ are associated with reduced recognition by innate immune cells. Cell Microbiol. 2013;15 doi: 10.1111/cmi.12135. doi:10.1111/cmi.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;15:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Ishii N, Watanabe M, Yoshihara F, Arisawa M. Rbf1 (RPG-box binding factor), a transcription factor involved in yeast-hyphal transition of Candida albicans. Nippon Ishinkin Gakkai Zasshi. 1998;15:67–71. doi: 10.3314/jjmm.39.67. [DOI] [PubMed] [Google Scholar]
- Levitin A, Whiteway M. The effect of prostaglandin E2 on transcriptional responses of Candida albicans. Microbiol Res. 2007;15:201–210. doi: 10.1016/j.micres.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Chamilos G, Lewis RE, Kontoyiannis DP. Inhibition of Candida parapsilosis mitochondrial respiratory pathways enhances susceptibility to caspofungin. Antimicrob Agents Chemother. 2006;15:744–747. doi: 10.1128/AAC.50.2.744-747.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph-Horne T, Hollomon DW, Wood PM. Fungal respiration: a fusion of standard and alternative components. Biochim Biophys Acta. 2001;15:179–195. doi: 10.1016/S0005-2728(00)00251-6. [DOI] [PubMed] [Google Scholar]
- Nohturfft A, Zhang SC. Coordination of lipid metabolism in membrane biogenesis. Annu Rev Cell Dev Biol. 2009;15:539–566. doi: 10.1146/annurev.cellbio.24.110707.175344. [DOI] [PubMed] [Google Scholar]
- Shingu-Vazquez M, Traven A. Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryot Cell. 2011;15:1376–1383. doi: 10.1128/EC.05184-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lorenz M. Carnitine acetyltransferases are required for growth on non-fermentable carbon sources but not for pathogenesis in Candida albicans. Microbiology. 2008;15:500–509. doi: 10.1099/mic.0.2007/014555-0. [DOI] [PubMed] [Google Scholar]
- Carman A, Vylkova S, Lorenz M. Role of acetyl coenzyme A synthesis and breakdown in alternative carbon source utilization in Candida albicans. Eukaryot Cell. 2008;15:1733–1741. doi: 10.1128/EC.00253-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M, Fink G. The glyoxylate cycle is required for fungal virulence. Nature. 2001;15:83–86. doi: 10.1038/35083594. [DOI] [PubMed] [Google Scholar]
- Hazelwood LA, Daran JM, van Maris AJ, Pronk JT, Dickinson JR. The ehulich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol. 2008;15:2259–2266. doi: 10.1128/AEM.02625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barz T, Ackermann K, Pyerin W. Control of methionine biosynthesis genes by protein kinase CK2-mediated phosphorylation of Cdc34. Cell Mol Life Sci. 2006;15:2183–2190. doi: 10.1007/s00018-006-6213-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanovic N, Streith I, Wolinski H, Rechberger G, Kohlwein SD, Tehlivets O. S-adenosyl-L-homocysteine hydrolase, key enzyme of methylation metabolism, regulates phosphatidylcholine synthesis and triacylglycerol homeostasis in yeast: implications for homocysteine as a risk factor of atherosclerosis. J Biol Chem. 2008;15:23989–2399. doi: 10.1074/jbc.M800830200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisman B, Alonso-Monge R, Román E, Arana D, Nombela C, Pla J. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot Cell. 2006;15:347–358. doi: 10.1128/EC.5.2.347-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Williams D, Lowman D, Monteiro MA, Tan X, Kruppa M, Fonzi W, Roman E, Pla J, Calderone R. The Candida albicans histidine kinase Chk1p: signaling and cell wall mannan. Fungal Genet Biol. 2009;15:731–41. doi: 10.1016/j.fgb.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maesaki S, Marichal P, Vanden Bossche H, Sanglard D, Kohno S. Rhodamine 6G efflux for the detection of CDR1-overexpressing azole-resistant Candida albicans strains. J Antimicrob Chemother. 1999;15:27–31. doi: 10.1093/jac/44.1.27. [DOI] [PubMed] [Google Scholar]
- Saengkhae C, Loetchutinat C, Garnier-Suillerot A. Kinetic analysis of rhodamines efflux mediated by the multidrug resistance protein (MRP1) Biophys J. 2003;15:2006–2014. doi: 10.1016/S0006-3495(03)74628-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Song CP, He J, Zhu H. Polyamines improve K+/Na+ homeostasis in barley seedlings by regulating root Ion channel activities. Plant Physiol. 2007;15:1061–1072. doi: 10.1104/pp.107.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt ME, Vickery LE. Charge pair interactions stabilizing ferredoxin-ferredoxin reductase complexes. Identification by complementary site-specific mutations. J Biol Chem. 1993;15:17126–30. [PubMed] [Google Scholar]
- van Steensel B. Mapping of genetic and epigenetic regulatory networks using microarrays. Nat Genet. 2005;15(Suppl):S18–24. doi: 10.1038/ng1559. [DOI] [PubMed] [Google Scholar]
- Vandeputte P, Ischer F, Sanglard D, Coste AT. In vivo systematic analysis of Candida albicans Zn2-Cys6 transcription factors mutants for mice organ colonization. PLoS ONE. 2011;15:e26962. doi: 10.1371/journal.pone.0026962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B, Clancy CJ, Cheng S, Raman SB, Iczkowski KA, Nguyen MH. Candida albicans RFX2 encodes a DNA binding protein involved in DNA damage responses, morphogenesis, and virulence. Eukaryot Cell. 2009;15:627–639. doi: 10.1128/EC.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talibi D, Raymond M. Isolation of a putative Candida albicans transcriptional regulator involved in pleiotropic drug resistance by functional complementation of a pdr1 pdr3 mutation in Saccharomyces cerevisiae. J Bacteriol. 1999;15:231–240. doi: 10.1128/jb.181.1.231-240.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumme J, Dietz M, Wagner C, Schüller HJ. Dimerization of yeast transcription factors Ino2 and Ino4 is regulated by precursors of phospholipid biosynthesis mediated by Opi1 repressor. Curr Genet. 2008;15:35–45. doi: 10.1007/s00294-008-0197-7. [DOI] [PubMed] [Google Scholar]
- Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;15:317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, King MS, Pryde KR. The production of reactive oxygen species by complex I. Biochem Soc Trans. 2008;15:976–80. doi: 10.1042/BST0360976. [DOI] [PubMed] [Google Scholar]
- Bai RK, Perng CL, Hsu CH, Wong LJC. Quantitative PCR analysis of mitochondrial DNA content in patients with mitochondrial disease. Ann N Y Acad Sci. 2004;15:304–309. doi: 10.1007/978-3-662-41088-2_29. [DOI] [PubMed] [Google Scholar]
- Kelly RDW, Mahmud A, McKenzie M, Trounce IA, St Joh JC. Mitochondrial DNA copy number is regulated in a tissue specific manner by DNA methylation of the nuclear-encoded DNA polymerase gamma A. Nucleic Acids Res. 2012;15:10124–10138. doi: 10.1093/nar/gks770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Up and down-regulated genes list in rbf1Δ mutant.
Up and down-regulated genes list in dpb41Δ mutant.
Up and down-regulated genes list in hfl1Δ mutant.