Abstract
As a critical regulator of drug metabolism and inflammation, Pregnane X Receptor (PXR), plays an important role in disease pathophysiology linking metabolism and inflammation (e.g. hepatic steatosis)1,2. There has been much progress in the identification of agonist ligands for PXR, however, there are limited descriptions of drug-like antagonists and their binding sites on PXR3,4,5. A critical barrier has been the inability to efficiently purify full-length protein for structural studies with antagonists despite the fact that PXR was cloned and characterized in 1998. Our laboratory developed a novel high throughput yeast based two-hybrid assay to define an antagonist, ketoconazole's, binding residues on PXR6. Our method involves creating mutational libraries that would rescue the effect of single mutations on the AF-2 surface of PXR expected to interact with ketoconazole. Rescue or "gain-of-function" second mutations can be made such that conclusions regarding the genetic interaction of ketoconazole and the surface residue(s) on PXR are feasible. Thus, we developed a high throughput two-hybrid yeast screen of PXR mutants interacting with its coactivator, SRC-1. Using this approach, in which the yeast was modified to accommodate the study of the antifungal drug, ketoconazole, we could demonstrate specific mutations on PXR enriched in clones unable to bind to ketoconazole. By reverse logic, we conclude that the original residues are direct interaction residues with ketoconazole. This assay represents a novel, tractable genetic assay to screen for antagonist binding sites on nuclear receptor surfaces. This assay could be applied to any drug regardless of its cytotoxic potential to yeast as well as to cellular protein(s) that cannot be studied using standard structural biology or proteomic based methods. Potential pitfalls include interpretation of data (complementary methods useful), reliance on single Y2H method, expertise in handling yeast or performing yeast two-hybrid assays, and assay optimization.
Keywords: Biochemistry, Issue 81, Orphan nuclear receptor, ketoconazole, yeast two-hybrid, Pregnane X Receptor, ligand, antatogist, coactivators SRC-1 (steroid receptor coactivator 1), drug-receptor interaction
Introduction
The yeast two-hybrid (Y2H) assay is widely used to discover protein-protein interactions and more recently for discovery of novel small molecules that disrupt protein-protein interaction complexes 7, 8, 9, 10, 11. However, the conventional approaches of this assay, used for drug discovery or "hits", do not allow for detection of allosteric interaction residues of chemicals compounds within protein-protein surfaces, that when altered still interact and allow for interrogation of the altered residues11. Indeed, such a method(s), if feasible to develop, would enable a tractable yeast system for high throughput assessment of allosteric interaction residues critical for protein-protein interaction disruption. In the context of drug discovery, the most direct way to establish interaction of compounds with proteins would involve structural determination (e.g. crystalization of protein-inhibitor complex). These methods are cumbersome, use elaborate resources and it is not technically feasible to perform structural studies on every protein.
Tractable genetic drug screening systems have been established in bacteria1, 2 and other model systems like mammalian two-hybrid. However, these systems need optimization and alternative systems like Y2H are still the most tested in drug discovery. There are limitations that include poor sensitivity and reliability of interactions using singular methods13 , however, a single Y2H assay can be modified to answer specific questions regarding interaction residues. In the field of nuclear receptor research, Y2H has been used to define interacting proteins14, however, these protein interactions have rarely been used to define the nature in which ligands/antagonists interact with nuclear receptor-protein complexes. Thus, our laboratory focused efforts on defining a method, especially for receptor proteins that are not readily amenable to proteomic based methods, that would unearth novel ligand/antagonist interacting residues using a reverse Y2H based discovery platform.
Based on our previous finding that ketoconazole disrupts PXR and its activator SRC-1, we developed a novel reverse Y2H system that enable us to define and interrogate ketoconazole interacting residues on PXR6. Our method is based on the properties of the yeast GAL4 protein that consists of separable domains responsible for DNA-binding and transcriptional activation. The PXR LBD protein is expressed as a fusion to the LexA DNA-binding domain (DNA-BD), while the full length co-activators SRC-1 (steroid receptor coactivator 1) proteins are expressed as fusions to the GAL4 activation domain (AD). Interaction between PXR and SRC-1 fusion proteins leads to the transcriptional activation of GAL4-binding sites containing reporter gene β-LacZ that is integrated into the yeast genome. Ketoconazole, a PXR antagonist, disrupts PXR and SRC-1 interaction 15, 16, 17 and we can detect the interaction of PXR and SRC-1 in the presence or absence of ketoconazole after staining colonies on filters for X-gal activity. The principle of Y2H is illustrated in Figure 1 and the experimental procedure is summarized in Figure 2.
Protocol
1. Construction of PXR and SRC-1 Fusions in Yeast Vectors
- PCR amplification of Human PXR LBD (107-434 amino acids) and Human SRC-1 full length (1-1401 amino acids).
- Use pSG5-hPXR plasmid8 as PXR LBD template, use pCMX-SRC1 plasmid as SRC-1 templates.
- Thaw PCR SuperMix, DNA templates and primers (see Materials), keep them on ice.
- Add 0.25 μg/μl DNA template 1 μl, 10 mM primer pairs 2 μl each, PCR SuperMix 45 μl and add H2O to bring total volume to 50 μl.
- Set PCR at one cycle of 94 °C for 2 min, 20 cycles of 94 °C for 30 sec, 55 °C for 30 sec and 72 °C for 2 min, then one cycle of 72 °C for 10 min for extension. Check PCR products by running on 1% agarose gel.
- PCR products cloning into Y2H vectors.
- Purify PCR products from 1% agarose gel. Digest PXR, LBD, PCR, and Y2H vector pSH2-1 with BamHI and SalI; digest SRC-1 PCR and Y2H vector pGADNOT with NotΙ and SalΙ. To do this, add 1 μg DNA , 3 μl 10x reaction buffer, 1 μl restriction enzymes, and finally H2O to bring total volume to 30 μl.
- Put digestion samples in 37 °C water bath for 1 hr. Check digestion by running on 1% agarose gel.
- Purify digestion samples from 1% agarose gel, ligate digested PCR products and Y2H vectors and transform to DH5α competent cells.
- Pick the colonies and isolate the plasmid DNA.
Identify pSH2-1-PXR plasmid DNA by BamHI/SalI digestion, identify pGADNOT-SRC-1 plasmid DNA by NotI/SalI digestion. Verify all positive plasmid DNA by sequencing.
2. Yeast Two-hybrid Assays
Inoculate the yeast strain into 5 ml YPAD and incubate over night by constant agitation (220 rpm at 30 °C). Note: The Saccharomyces cerevisiae strain was CTY10-5d (MATa ade2 trp1-901 leu2-3,112 his3-200 ga l4-gal80- URA3::lexA-lacZ) that contains an integrated GAL1-lacZ gene with lexA operator10. To obtain viable yeast cells resistant to ketoconazole, but those that did not carry transporter alterations as a cause of azole resistance18-20, novel strains of CTY10-5d yeast were obtained by first deleting ERG3 ( erg3Δ)and then introducing additional deletion in ERG11 ( erg3Δ/erg11Δ) genes by homologous recombination. The construction details have been described in a previously published manuscript6 .
The next day (in the morning), inoculate yeast (1:20) in 5 ml YAPD and incubate to obtain an OD600 ~0.2 by constant agitation (220 rpm at 30 °C).
Pellet the cells at 2,000 rpm in a microcentrifuge for 2 min and discard the supernatant. Then wash the pellet twice with autoclaved water and once with 0.1 M LiAc.
After the final wash, discard supernatant and add the following reagents to the cell pellet: 240 μl PEG 3500 50% w/v, 36 μl 1 M LiAc, 50 μl 2 mg/ml boiled salmon sperm ssDNA, H2O 34 μl. Vortex to mix, add 1 μg pSH-PXR and 1 μg pGADNOT-SRC-1 DNA and mix.
Transfer the mix to polystyrene round-bottom tube, agitate the transformation, mix (220 rpm at 30 °C) for 30 min. Move the tube to 42 °C water bath and incubate for an additional 30 min.
Centrifuge the tube at 2,000 rpm for 2 min and remove the supernatant. Wash pellet once with autoclaved water.
Pipette 100 μl sterile water into the tube, re-suspend the cells by gentle finger tapping and pipetting, then plate onto plates of -His/-Leu Dropout Medium (Formulation per One Liter -His/-Leu Dropout Medium: 20 g dextrose, 1.7 g YNB, 0.67 g CSM-His/-Leu, 5 g ammonium sulfate, 1.5% agar) and incubate at 30 °C for 3-4 days to isolate transformants.
3. X-gal Filter Assay
Cut nitrocellulose membrane to the size of part of a Petri dish. Label the nitrocellulose membrane to mark the orientation.
Place premarked nitrocellulose membrane on yeast colonies, make sure that the membrane is in contact with the surface of the plate medium.
Remove the membrane, place it colony side up on a 3 mm filter paper, and place the membrane and the filter paper at -80 °C for 15 min.
Make X-gal solution. Add 50 μl 20 mg/ml X-gal and 15 μl β-mercaptoethanol to 5 ml of Z-buffer (1 L buffer contains 16.1 g Na2HPO47H2O, 5.5 g NaH2PO4 H2O, 0.75 g KCl , 0.25 g MgSO4•7H2O) . If Z Buffer was kept at 4 °C, bring it to room temperature before adding these reagents.
Place two circles of 3 mm filter paper in a Petri dish and soak them with 5 ml X-gal solution. Drain excess buffer from the Petri dish and place the nitrocellulose membrane with the colony side up.
Close the lid, ensure that the membrane is making complete contact with the filter paper and it is completely wet. Wrap the Petri dish in foil and incubate at 37 °C for 30 min to overnight.
4. Liquid β-gal Assay
Grow Yeast in -His/-Leu Dropout liquid medium to OD600 0.7.
Transfer 1 ml medium into a 1.5 ml microcentrifuge tube and centrifuge at 3,000 rpm for 2 min. Discard the supernatant.
Add 1 ml of Z Buffer to resuspend the pellet, centrifuge at 3,000 rpm again for 2 min. Discard the supernatant.
Add 150 μl of Z Buffer with 2ME to resuspend the cell pellet, mix then add 50 μl chloroform and 20 μl 0.1% SDS. Vigorously vortex the sample for 15 sec.
Add 700 μl of ONPG Solution (1 mg/ml in Z buffer). Set the timer, incubate at 30 °C long enough to allow the yellow colour to develop and note total reaction time.
Add 500 μl of 1 M Na2CO3 to stop the reaction and determine the absorbance at 420 nm. The blank is 700 μl ONPG solution plus 500 μl 1 M Na2CO3.
Calculate Miller Units as follows: Miller Units = (Å420*1,000)/(Å600*min*ml)
Å420: absorbance units at 420 nanometers; Å600: absorbance units at 600 nanometers; min: the reaction time in minutes; ml: the reaction volume in ml
Representative Results
We performed an assay to see if we could detect a colorimetric readout of the association of PXR and steroid receptor coactivator-1 (SRC-1). Since yeast has significant sterol production, it has previously been shown that lacZ expression in yeast can be induced without the need for additional exogenous ligand. We found that lacZ expression (blue colonies) is also induced in the yeast strain transformed with PXR and SRC-1; however, there is no induction of LacZ expression (white colonies) in yeast transformed with empty vectors, PXR or SRC-1 individually (Figure 3). To determine whether ketoconazole (25 μM) disrupted PXR and SRC-1 interaction in yeast, replica plates containing ketoconazole were soaked with nitrocellulose and X-gal filter, lift, and β-galactosidase liquid assays were performed. We show that ketoconazole disrupts PXR and SRC-1 interactions in yeast since all the colonies from the replica filter were now white (which was also shown by the significantly reduced β-galactosidase activity by liquid enzymatic assays) (Figure 4A). We had previously shown in mammalian assays that the PXR mutant (T248E/K277Q) can be activated by a strong ligand (e.g. rifampicin) but is immune to the antagonistic effects of ketoconazole17. Similarly, when we engineered the PXR double mutant (T248E/K277Q) in the yeast plasmid and then performed yeast transformations with SRC-1, we were able to show that the colonies exposed to ketoconazole still retain LacZ expression (Figure 4B).
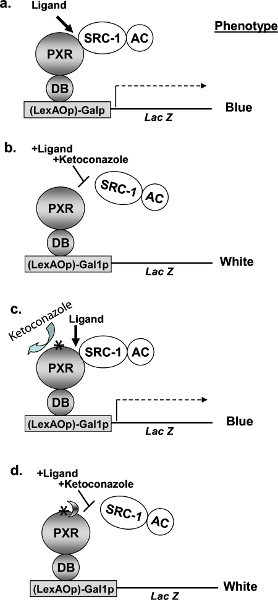 Figure 1. Schematic illustrating the principles of the yeast two-hybrid (Y2H) assay. The yeast assay has been established as a functional assay that allows for isolation of PXR mutants that are resistant to ketoconazole mediated inhibition of SRC-1 interaction. A, rifampicin-induced two-hybrid interaction of LexA-DB-hPXR fusion protein with SRC-1 fused to GAL4-activation domain (GAL4AC-SRC-1). The interaction results in blue colonies in an X-gal assay because of the LacZ reporter. B, presence of ketoconazole disrupts the rifampicin-induced interaction of hPXR with SRC-1. X-gal assay results in white colonies. C, presence of a mutation (indicated with an asterisk) in hPXR that renders it immune to the action of ketoconazole will yield a blue colony. D, a further modification of our assay in the future could incorporate additional second site suppressor mutations (illustrated with half moon) nullifying the effect of first mutation that could result in the disruption of hPXR-SRC-1 interaction and hence a white colony in the X-gal assay. LexAOp, LexA operon; Galp, Gal4 promoter, Lac Z, beta-glalactosidase; DB, LexAOp DNA-binding domain of Galp; AC, activation domain; *, mutation(s). (Reproduced from reference6). Click here to view larger figure.
Figure 1. Schematic illustrating the principles of the yeast two-hybrid (Y2H) assay. The yeast assay has been established as a functional assay that allows for isolation of PXR mutants that are resistant to ketoconazole mediated inhibition of SRC-1 interaction. A, rifampicin-induced two-hybrid interaction of LexA-DB-hPXR fusion protein with SRC-1 fused to GAL4-activation domain (GAL4AC-SRC-1). The interaction results in blue colonies in an X-gal assay because of the LacZ reporter. B, presence of ketoconazole disrupts the rifampicin-induced interaction of hPXR with SRC-1. X-gal assay results in white colonies. C, presence of a mutation (indicated with an asterisk) in hPXR that renders it immune to the action of ketoconazole will yield a blue colony. D, a further modification of our assay in the future could incorporate additional second site suppressor mutations (illustrated with half moon) nullifying the effect of first mutation that could result in the disruption of hPXR-SRC-1 interaction and hence a white colony in the X-gal assay. LexAOp, LexA operon; Galp, Gal4 promoter, Lac Z, beta-glalactosidase; DB, LexAOp DNA-binding domain of Galp; AC, activation domain; *, mutation(s). (Reproduced from reference6). Click here to view larger figure.
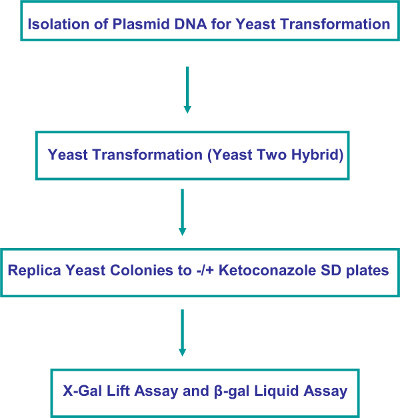 Figure 2. Experimental Flow Chart.
Figure 2. Experimental Flow Chart.
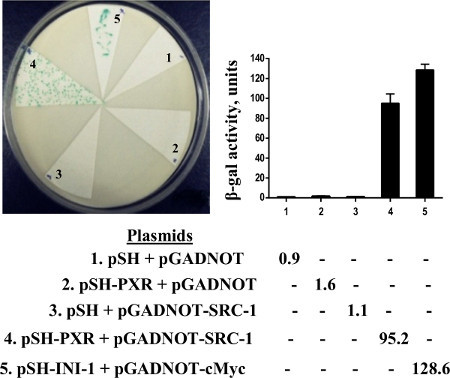 Figure 3. X-gal, lift assay, and β-gal liquid assay. Yeast was transformed with indicated plasmids and plated colonies subjected to X-gal lift assay (left panel) and β-gal liquid assay (right panel). pSH empty vector + pGADNOT empty vector (lane 1); pSH-PXR + pGADNOT empty vector (lane 2); pSH empty vector + pGADNOT-SRC-1 (lane 3); pSH-PXR + pGADNOT-SRC-1 (lane 4); 5. pSH-INI-1 + pGADNOT-c-Myc (positive control; lane 5). (reproduced from reference6).
Figure 3. X-gal, lift assay, and β-gal liquid assay. Yeast was transformed with indicated plasmids and plated colonies subjected to X-gal lift assay (left panel) and β-gal liquid assay (right panel). pSH empty vector + pGADNOT empty vector (lane 1); pSH-PXR + pGADNOT empty vector (lane 2); pSH empty vector + pGADNOT-SRC-1 (lane 3); pSH-PXR + pGADNOT-SRC-1 (lane 4); 5. pSH-INI-1 + pGADNOT-c-Myc (positive control; lane 5). (reproduced from reference6).
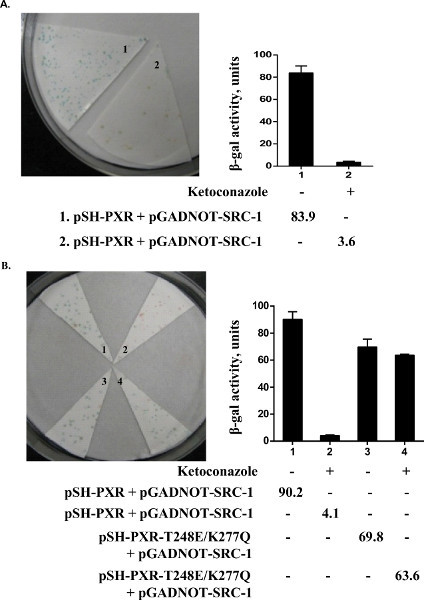 Figure 4. Ketoconazole disrupts wild-type but not mutant PXR association with coactivator, SRC-1. Yeast colonies were replica in plates containing vehicle (0.2% DMSO; lane 1) or ketoconazole (25 μM; lane 2). X-gal lift assay (left panel) and β-gal liquid assay (right panel) were then performed. Wild type PXR (A, B line 1 and 2) and ketoconazole mute mutant T248E/K277Q (B line 3 and 4) were shown in this figure. (reproduced from reference6).
Figure 4. Ketoconazole disrupts wild-type but not mutant PXR association with coactivator, SRC-1. Yeast colonies were replica in plates containing vehicle (0.2% DMSO; lane 1) or ketoconazole (25 μM; lane 2). X-gal lift assay (left panel) and β-gal liquid assay (right panel) were then performed. Wild type PXR (A, B line 1 and 2) and ketoconazole mute mutant T248E/K277Q (B line 3 and 4) were shown in this figure. (reproduced from reference6).
Discussion
In our modified Y2H assay, we have identified important residues for ketoconazole interactions on PXR6. Since SRC-1 is a coactivator (and was cloned into the pGADNot vector), we also tested whether SRC-1 could activate lacZ expression when cloned into the pSH vector system and whether this would change the activation profile and/or affect the leakiness of the yeast two-hybrid assay. Using our redesigned plasmids we performed two-hybrid assays in erg3Δ/erg11Δ yeast. As before, we show that lacZ expression (blue colonies) is also induced in erg3Δ/erg11Δ yeast strain transformed with PXR and SRC-1; however, there is no induction of LacZ expression (white colonies) in yeast transformed with empty vectors, PXR or SRC-1 individually (data not shown). Our approach provides a powerful new method for isolation of genetic interaction allosteric ligand-protein residues for proteins not amenable to conventional structural biology and/or proteomic approaches5,21.
The protocol as described has several pitfalls that must be acknowledged. First, sensitivity of detection, especially those partial antagonist effects on protein-protein interaction residues can limit the resolution of the assay. Colorimetric scanning might help resolve some of the detection issues21. Second, the presence of blue colonies may not always indicate ketoconazole resistant PXR mutations (false positive). The lacZ expression could be rescued in the presence of ketoconazole due to mutation(s) in PXR that makes the protein constitutively active or active independent of SRC-1 interaction. Such mutants of PXR can be distinguished from the true ketoconazole resistant mutants by testing the ability of the LexA-DB-PXR to self activate lacZ expression, i.e. these mutants will yield blue colonies in the absence of GAL4AC-SRC-1. As a corollary, the mutants can also introduced into yeast using the GAL4AC-plasmid in the absence of LexA-DB-SRC-1 to further describe evidence for self-activation of LacZ. Third, it remains unknown if the same result would be obtained if other Y2H protocols/vectors were used13. Fourth, the study of intramolecular revertant mutants (second site suppressor mutants of ketoconazole-resistant mutants) can add to the understanding of antagonist binding by defining neighboring residues that impact the binding pharmacophore. With a modification of the library of mutants, one can define this more precisely using a ketoconazole-resistant mutant as the mutagenesis template.
This method can be modified to incorporate any drug whereby the cytotoxic target in yeast is well established or to a drug(s) that has no potential toxicity towards yeast. The yeast can be modified, as we have shown6, and still be viable for Y2H assays. The power of this method also relies on ancillary information from other assays (e.g. molecular docking) that inform site-specific interactions. Furthermore, specific nuclear receptor/coregulator interactions (e.g. nurr77/LKB1; AR/PELP1) may be studied22,23. The tractability of this system is that it is portable, can be performed within days in a high throughput manner and does not require expensive reagents or methods.
Disclosures
No conflicts of interest declared.
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants CA127231 and The Damon Runyon Foundation Clinical Investigator Award (CI 1502) (to S.M). We would like to thank Professor Zdenek Dvorak from Palacky University Olomouc, Czech Republic for his helpful insights into discussing portability of this technique to their institution and standardization of protocol.
References
- Kliewer SA, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92(1):73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Blumberg B, et al. a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998;12(20):3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas A, Mani S, Redinbo MR, Krasowski MD, Li H, Ekins S. Elucidating the 'Jekyll and Hyde' Nature of PXR: The Case for Discovering Antagonists. Pharm. Res. 2009;26(8):1807–1815. doi: 10.1007/s11095-009-9901-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pondugula SR, Mani S. Pregnane xenobiotic receptor in cancer pathogenesis and therapeutic response. Cancer Lett. 2013;328(1):1–9. doi: 10.1016/j.canlet.2012.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S, Dou V, Redinbo MR. PXR antagonists and implications for drug metabolism. Drug Metab. Rev. 2013;45(1):60–72. doi: 10.3109/03602532.2012.746363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. Novel Yeast-Based Strategy Unveils Antagonist Binding Regions on the Nuclear Xenobiotic Receptor PXR. J. Biol. Chem. 2013;288(19):13655–13668. doi: 10.1074/jbc.M113.455485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg SM, et al. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 1995;15(7):3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74(1):205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Wang H, et al. The phytoestrogen coumestrol is a naturally occurring antagonist of the human pregnane X receptor. Mol. Endocrinol. 2008;22(4):838–857. doi: 10.1210/me.2007-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalpana GV, Goff SP. Genetic analysis of homomeric interactions of human immunodeficiency virus type 1 integrase using the yeast two-hybrid system. Proc. Natl. Acad. Sci. U.S.A. 1993;90(22):10593–10597. doi: 10.1073/pnas.90.22.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdi A, Colas P. Yeast two-hybrid methods and their applications in drug discovery. TiPS. 2012;33(2):109–118. doi: 10.1016/j.tips.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Battesti A, Bouveret E. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods. 2012;58(4):325–334. doi: 10.1016/j.ymeth.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Caufield JH, Sakhawalkar N, Uetz P. A comparison and optimization of yeast two-hybrid systems. Methods. 2012;58(4):317–324. doi: 10.1016/j.ymeth.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers M, et al. Automated yeast two-hybrid screening for nuclear receptor-interacting proteins. Mol. Cell Proteomics. 2005;4(2):205–213. doi: 10.1074/mcp.M400169-MCP200. [DOI] [PubMed] [Google Scholar]
- Takeshita A, Taguchi M, Koibuchi N, Ozawa Y. Putative role of the orphan nuclear receptor SXR (steroid and xenobiotic receptor) in the mechanism of CYP3A4 inhibition by xenobiotics. J. Biol. Chem. 2002;277(36):32453–32458. doi: 10.1074/jbc.M111245200. [DOI] [PubMed] [Google Scholar]
- Huang H, et al. Inhibition of drug metabolism by blocking the activation of nuclear receptors by ketoconazole. Oncogene. 2007;26(2):258–268. doi: 10.1038/sj.onc.1209788. [DOI] [PubMed] [Google Scholar]
- Wang H, et al. Activated pregnenolone X- receptor is a target for ketoconazole and Its analogs. Clin. Cancer Res. 2007;13(8):2488–2495. doi: 10.1158/1078-0432.CCR-06-1592. [DOI] [PubMed] [Google Scholar]
- Ghannoum MA, Rice LB. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 1999;12(4):501–517. doi: 10.1128/cmr.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R, Bachhawat AK. The yeast multidrug resistance pump, Pdr5p, confers reduced drug resistance in erg mutants of Saccharomyces cerevisiae. Microbiology. 1999;145:809–818. doi: 10.1099/13500872-145-4-809. [DOI] [PubMed] [Google Scholar]
- White TC, Marr KA, Bowden RA, cellular Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 1998;11(2):382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebriiskii IG, et al. Detection of peptides, proteins, and drugs that selectively interact with protein targets. Genome Res. 2002;12:1785–1791. doi: 10.1101/gr.450702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, et al. Central role for PELP1 in nonandrogenic activation of the androgen receptor in prostate cancer. Mol Endocrinol. 2012;26(4):550–561. doi: 10.1210/me.2011-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan YY, et al. The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK. Nat Chem Biol. 2012;8(11):897–904. doi: 10.1038/nchembio.1069. [DOI] [PubMed] [Google Scholar]


