Abstract
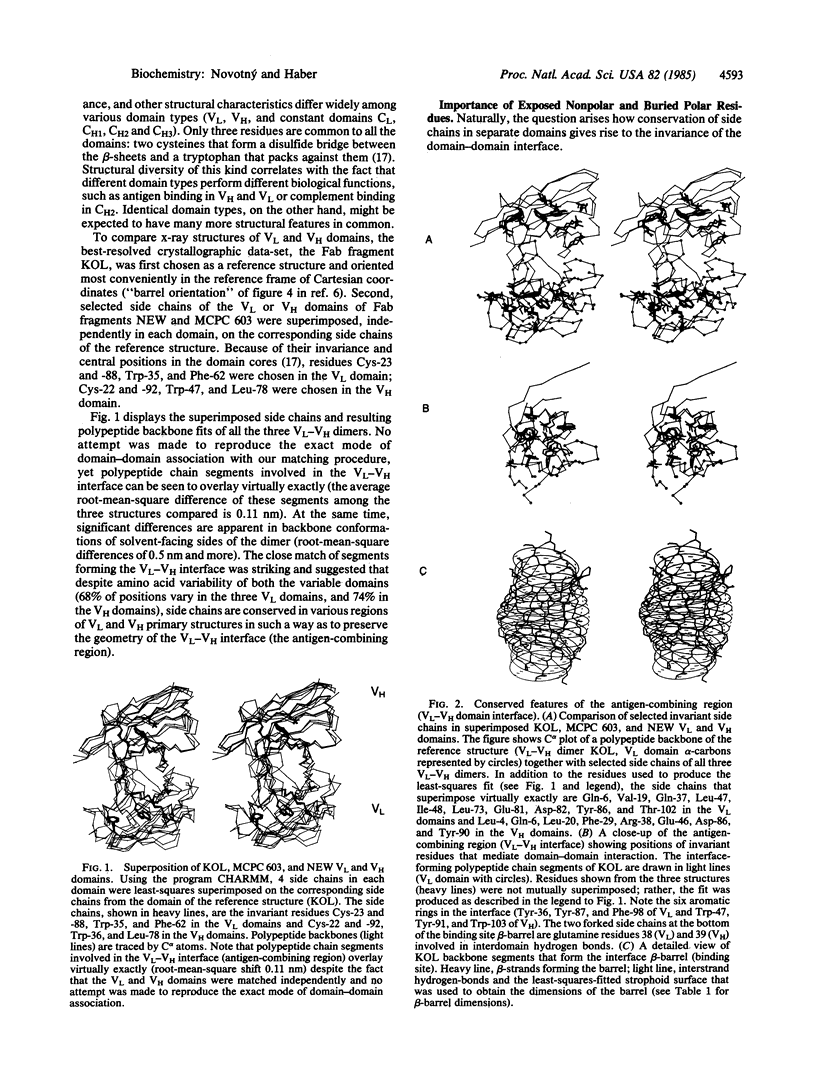
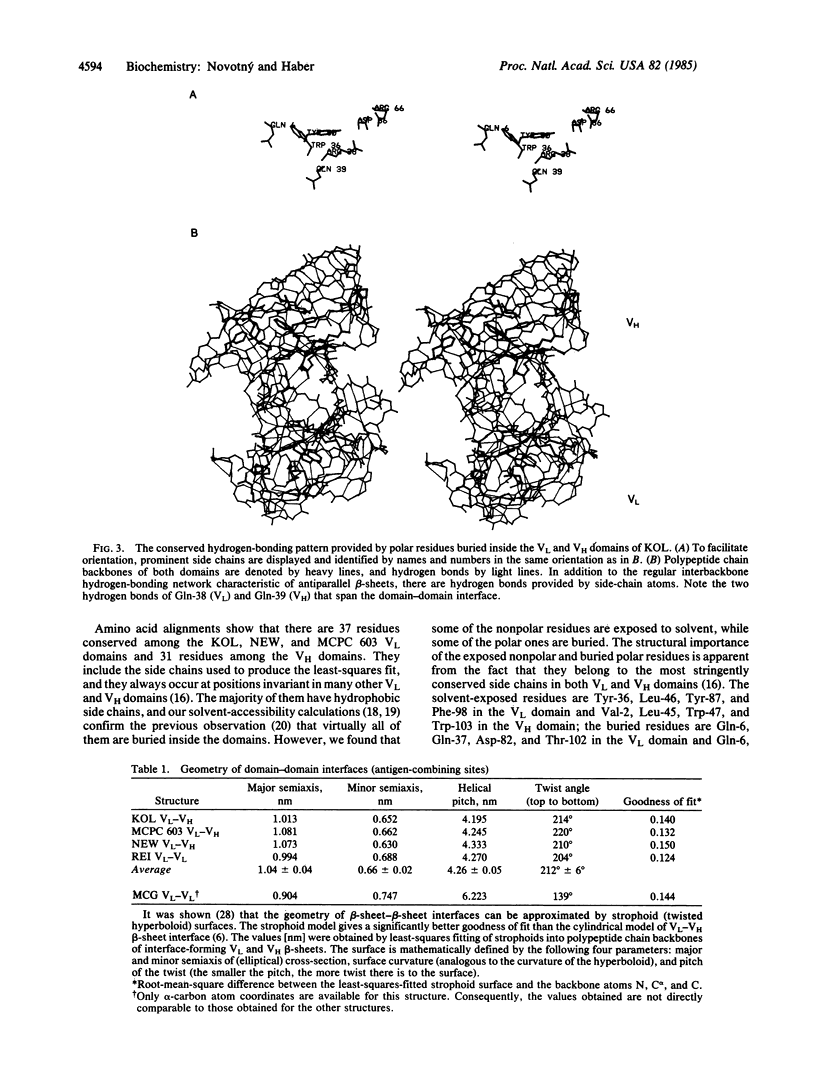
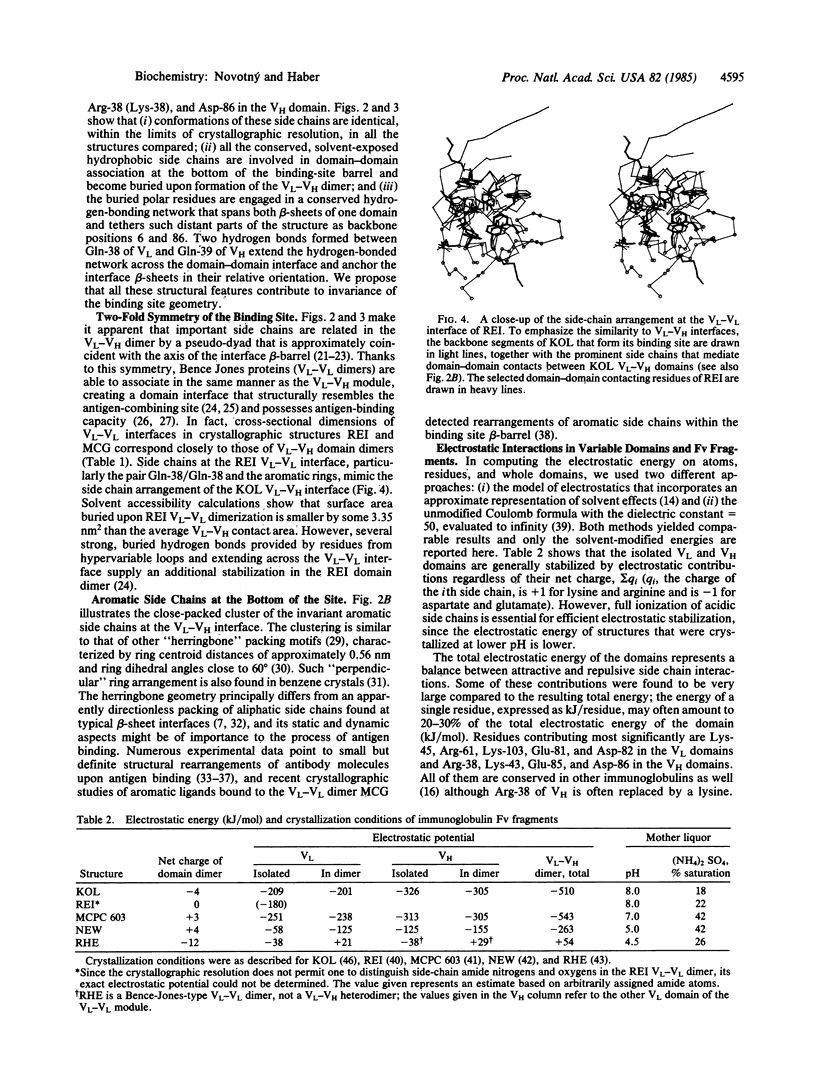
Antigen-combining site arises by noncovalent association of the variable domain of the immunoglobulin heavy chain (VH) with that of the light chain (VL). To analyze the invariant features of the binding region (VL-VH domain interface), we compared the known immunoglobulin three-dimensional structures by a variety of methods. The interface forms a close-packed, twisted, prism-shaped "beta-barrel" characterized by cross-sectional dimensions 1.04 X 0.66 nm and a top-to-bottom twist angle of 212 degrees. The geometry of the interface is preserved via invariance of some 15 side chains, both inside the domains and on their surface. Buried polar residues form a conserved hydrogen-bonding network that has a similar topological connectivity in the two domain types; two hydrogen bonds contributed by invariant side chains extend across the interface and anchor the beta-sheets in their relative orientation. Invariant aromatic residues close-pack at the bottom of the binding-site beta-barrel with their ring planes oriented perpendicularly in the characteristic "herringbone" packing mode. Electrostatic computations that implicitly include solvent effects show the domains to be stabilized by large electrostatic forces. However, structures that were crystallized at lower pH have their electrostatic energies appropriately lowered, implying that full ionization of carboxyl side chains is essential for efficient electrostatic stabilization. The unusual mode of domain-domain association in the VL-VL dimer RHE correlates with its overall repulsive electrostatic energy (+54 kJ/mol), as opposed to negative (i.e., stabilizing) energy values (-263 to -543 kJ/mol) found in the domains of the other structures. The VL-VL dimer REI mimics closely the interface geometry of VL-VH dimers although its domain-domain contact area is lower by 18%.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amzel L. M., Poljak R. J. Three-dimensional structure of immunoglobulins. Annu Rev Biochem. 1979;48:961–997. doi: 10.1146/annurev.bi.48.070179.004525. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Chothia C., Janin J. Relative orientation of close-packed beta-pleated sheets in proteins. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4146–4150. doi: 10.1073/pnas.78.7.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C., Lesk A. M. Evolution of proteins formed by beta-sheets. I. Plastocyanin and azurin. J Mol Biol. 1982 Sep 15;160(2):309–323. doi: 10.1016/0022-2836(82)90178-4. [DOI] [PubMed] [Google Scholar]
- Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- Cohen F. E., Sternberg M. J., Taylor W. R. Analysis of the tertiary structure of protein beta-sheet sandwiches. J Mol Biol. 1981 May 25;148(3):253–272. doi: 10.1016/0022-2836(81)90538-6. [DOI] [PubMed] [Google Scholar]
- Davies D. R., Metzger H. Structural basis of antibody function. Annu Rev Immunol. 1983;1:87–117. doi: 10.1146/annurev.iy.01.040183.000511. [DOI] [PubMed] [Google Scholar]
- Davies D. R., Padlan E. A., Segal D. M. Three-dimensional structure of immunoglobulins. Annu Rev Biochem. 1975;44:639–667. doi: 10.1146/annurev.bi.44.070175.003231. [DOI] [PubMed] [Google Scholar]
- Edmundson A. B., Ely K. R., Girling R. L., Abola E. E., Schiffer M., Westholm F. A., Fausch M. D., Deutsch H. F. Binding of 2,4-dinitrophenyl compounds and other small molecules to a crystalline lambda-type Bence-Jones dimer. Biochemistry. 1974 Aug 27;13(18):3816–3827. doi: 10.1021/bi00715a031. [DOI] [PubMed] [Google Scholar]
- Edmundson A. B., Ely K. R., Herron J. N. A search for site-filling ligands in the Mcg Bence-Jones dimer: crystal binding studies of fluorescent compounds. Mol Immunol. 1984 Jul;21(7):561–576. doi: 10.1016/0161-5890(84)90041-5. [DOI] [PubMed] [Google Scholar]
- Epp O., Colman P., Fehlhammer H., Bode W., Schiffer M., Huber R., Palm W. Crystal and molecular structure of a dimer composed of the variable portions of the Bence-Jones protein REI. Eur J Biochem. 1974 Jun 15;45(2):513–524. doi: 10.1111/j.1432-1033.1974.tb03576.x. [DOI] [PubMed] [Google Scholar]
- Furey W., Jr, Wang B. C., Yoo C. S., Sax M. Structure of a novel Bence-Jones protein (Rhe) fragment at 1.6 A resolution. J Mol Biol. 1983 Jul 5;167(3):661–692. doi: 10.1016/s0022-2836(83)80104-1. [DOI] [PubMed] [Google Scholar]
- Holowka D. A., Strosberg A. D., Kimball J. W., Haber E., Cathou R. E. Changes in intrinsic circular dichroism of several homogeneous anti-type 3 pneumococcal antibodies on binding of a small hapten. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3399–3403. doi: 10.1073/pnas.69.11.3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancet D., Pecht I. Kinetic evidence for hapten-induced conformational transition in immunoglobin MOPC 460. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3549–3553. doi: 10.1073/pnas.73.10.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Lesk A. M., Chothia C. Evolution of proteins formed by beta-sheets. II. The core of the immunoglobulin domains. J Mol Biol. 1982 Sep 15;160(2):325–342. doi: 10.1016/0022-2836(82)90179-6. [DOI] [PubMed] [Google Scholar]
- Levison S. A., Hicks A. N., Portmann A. J., Dandliker W. B. Fluorescence polarization and intensity kinetic studies of antifluorescein antibody obtained at different stages of the immune response. Biochemistry. 1975 Aug 26;14(17):3778–3786. doi: 10.1021/bi00688a009. [DOI] [PubMed] [Google Scholar]
- Marquart M., Deisenhofer J., Huber R., Palm W. Crystallographic refinement and atomic models of the intact immunoglobulin molecule Kol and its antigen-binding fragment at 3.0 A and 1.0 A resolution. J Mol Biol. 1980 Aug 25;141(4):369–391. doi: 10.1016/0022-2836(80)90252-1. [DOI] [PubMed] [Google Scholar]
- Nockolds C. E., Kretsinger R. H., Coffee C. J., Bradshaw R. A. Structure of a calcium-binding carp myogen. Proc Natl Acad Sci U S A. 1972 Mar;69(3):581–584. doi: 10.1073/pnas.69.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrup S. H., Pear M. R., Morgan J. D., McCammon J. A., Karplus M. Molecular dynamics of ferrocytochrome c. Magnitude and anisotropy of atomic displacements. J Mol Biol. 1981 Dec 25;153(4):1087–1109. doi: 10.1016/0022-2836(81)90469-1. [DOI] [PubMed] [Google Scholar]
- Novotný J., Bruccoleri R. E., Newell J. Twisted hyperboloid (Strophoid) as a model of beta-barrels in proteins. J Mol Biol. 1984 Aug 15;177(3):567–573. doi: 10.1016/0022-2836(84)90301-2. [DOI] [PubMed] [Google Scholar]
- Novotný J., Bruccoleri R., Karplus M. An analysis of incorrectly folded protein models. Implications for structure predictions. J Mol Biol. 1984 Aug 25;177(4):787–818. doi: 10.1016/0022-2836(84)90049-4. [DOI] [PubMed] [Google Scholar]
- Novotný J., Bruccoleri R., Newell J., Murphy D., Haber E., Karplus M. Molecular anatomy of the antibody binding site. J Biol Chem. 1983 Dec 10;258(23):14433–14437. [PubMed] [Google Scholar]
- Padlan E. A., Davies D. R. Variability of three-dimensional structure in immunoglobulins. Proc Natl Acad Sci U S A. 1975 Mar;72(3):819–823. doi: 10.1073/pnas.72.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padlan E. A. Evaluation of the structural variation among light chain variable domains. Mol Immunol. 1979 May;16(5):287–296. doi: 10.1016/0161-5890(79)90128-7. [DOI] [PubMed] [Google Scholar]
- Padlan E. A. Structural basis for the specificity of antibody-antigen reactions and structural mechanisms for the diversification of antigen-binding specificities. Q Rev Biophys. 1977 Feb;10(1):35–65. doi: 10.1017/s0033583500000135. [DOI] [PubMed] [Google Scholar]
- Palm W. Uber die physikalisch-chemische Charakterisierung und nachfolgende Kristallisation von Immunglobulinen und deren Fragmenten: Ein Beitrag zur dreidimensionalen Strukturaufklärung der Antikörper, IV. Das Fab-Fragment und das Fc-Fragment des humanen Immunglobulins Kol; Isolierung und Kristallisation. Hoppe Seylers Z Physiol Chem. 1976 Jun;357(6):799–802. [PubMed] [Google Scholar]
- Palm Walter H. On the isolation, characterisation, and crystallisation of a human Bence-Jones protein of kappa type. FEBS Lett. 1970 Sep 18;10(1):46–48. doi: 10.1016/0014-5793(70)80412-4. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Richards F. M. Packing of alpha-helices: geometrical constraints and contact areas. J Mol Biol. 1978 Mar 15;119(4):537–555. doi: 10.1016/0022-2836(78)90201-2. [DOI] [PubMed] [Google Scholar]
- Rossi G., Choi T. K., Nisonoff A. Crystals of fragment Fab': preparation from pepsin digests of human IgG myeloma proteins. Nature. 1969 Aug 23;223(5208):837–838. doi: 10.1038/223837a0. [DOI] [PubMed] [Google Scholar]
- Rudikoff S., Potter M., Segal D. M., Padlan E. A., Davies D. R. Crystals of phosphorylcholine-binding Fab-fragments from mouse myeloma proteins: preparation and x-ray analysis. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3689–3692. doi: 10.1073/pnas.69.12.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul F. A., Amzel L. M., Poljak R. J. Preliminary refinement and structural analysis of the Fab fragment from human immunoglobulin new at 2.0 A resolution. J Biol Chem. 1978 Jan 25;253(2):585–597. [PubMed] [Google Scholar]
- Schechter I., Ziv E. Binding of 2,4-dinitrophenyl derivatives by the light chain dimer obtained from immunoglobulin A produced by MOPC-315 mouse myeloma. Biochemistry. 1976 Jun 29;15(13):2785–2790. doi: 10.1021/bi00658a013. [DOI] [PubMed] [Google Scholar]
- Schlessinger J., Steinberg I. Z., Givol D., Hochman J., Pecht I. Antigen-induced conformational changes in antibodies and their Fab fragments studied by circular polarization of fluorescence. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2775–2779. doi: 10.1073/pnas.72.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal D. M., Padlan E. A., Cohen G. H., Rudikoff S., Potter M., Davies D. R. The three-dimensional structure of a phosphorylcholine-binding mouse immunoglobulin Fab and the nature of the antigen binding site. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4298–4302. doi: 10.1073/pnas.71.11.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. C., Sax M. Structure of a dimeric fragment related to the lambda-type Bence-Jones protein: a preliminary study. J Mol Biol. 1974 Aug 15;87(3):505–508. doi: 10.1016/0022-2836(74)90100-4. [DOI] [PubMed] [Google Scholar]
- Wang B. C., Yoo C. S., Sax M. Crystal structure of Bence Jones protein rhe (3 A) and its unique domain-domain association. J Mol Biol. 1979 Apr 25;129(4):657–674. doi: 10.1016/0022-2836(79)90475-3. [DOI] [PubMed] [Google Scholar]
- Warshel A., Russell S. T., Churg A. K. Macroscopic models for studies of electrostatic interactions in proteins: limitations and applicability. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4785–4789. doi: 10.1073/pnas.81.15.4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidovetzki R., Blatt Y., Pecht I. A heterologous immunoglobulin chain recombinant carries a distinct site for dinitrophenyl and obeys the common hapten binding mechanism. Biochemistry. 1981 Aug 18;20(17):5011–5018. doi: 10.1021/bi00520a030. [DOI] [PubMed] [Google Scholar]


