Abstract
Aminopeptidases can selectively catalyze the cleavage of the N-terminal amino acid residues from peptides and proteins. Bacillus subtilis aminopeptidase (BSAP) is most active toward p-nitroanilides (pNAs) derivatives of Leu, Arg, and Lys. The BSAP with broad substrate specificity is expected to improve its application. Based on an analysis of the predicted structure of BSAP, four residues (Leu 370, Asn 385, Ile 387, and Val 396) located in the substrate binding region were selected for saturation mutagenesis. The hydrolytic activity toward different aminoacyl-pNAs of each mutant BSAP in the culture supernatant was measured. Although the mutations resulted in a decrease of hydrolytic activity toward Leu-pNA, N385L BSAP exhibited higher hydrolytic activities toward Lys-pNA (2.2-fold) and Ile-pNA (9.1-fold) than wild-type BSAP. Three mutant enzymes (I387A, I387C and I387S BSAPs) specially hydrolyzed Phe-pNA, which was undetectable in wild-type BSAP. Among these mutant BSAPs, N385L and I387A BSAPs were selected for further characterized and used for protein hydrolysis application. Both of N385L and I387A BSAPs showed higher hydrolysis efficiency than the wild-type BASP and a combination of the wild-type and N385L and I387A BSAPs exhibited the highest hydrolysis efficiency for protein hydrolysis. This study will greatly facilitate studies aimed on change the substrate specificity and our results obtained here should be useful for BSAP application in food industry.
Keywords: Bacillus subtilis, aminopeptidase, substrate specificity, saturation mutagenesis, protein hydrolysis
Introduction
Aminopeptidases (APs; EC 3.4.11) can selectively catalyze the cleavage of the N-terminal amino acid residues from peptides and proteins. These enzymes widely exist in all organisms from bacteria to mammals with different substrate specificities. Based on the substrate specificity, APs can be classified into two categories: broad or narrow. The former type is usually named according to their optimal substrates, such as leucine aminopeptidase and lysine aminopeptidase, which preferentially releases leucine and lysine from peptides, respectively. The latter type, such as proline-specific aminopeptidase, removes certain N-terminal amino acid residues from peptides specifically.1,2
APs are associated with many human diseases and play an important role in a wide range of biological processes.3-6 The research to elucidate the catalytic mechanisms of APs is significant for medicine and pharmacology. In the food industry, APs have been widely used to debitter protein hydrolysate and applied to hydrolyze proteins extensively.7-10 APs can increase the content of free amino acids and small peptides in protein hydrolysate. Furthermore, APs with different and broad substrate specificities are required for improving the process efficiency.
In our previous study, we cloned and overexpressed the gene of Bacillus subtilis aminopeptidase (BSAP) in recombinant B. subtili.s 11 BSAP (MEROPS ID: M28.UPA) belongs to the M28 family, in which APs contain two zinc ions in their active sites and share a common catalytic mechanism. Based on the researches of crystallographic structure and site-directed mutagenesis, the roles of two zinc ions and residues that compose the active center have been elucidated in detail.12-17 However, little information is currently available concerning the interaction between APs and the substrate, and an AP with broad substrate specificity is expected to improve its application.
In this study, we aim to change the substrate specificity of BSAP and evaluate the effect of BSAPs with different substrate specificities on protein hydrolysis. Based on the predicted structure of BSAP, four residues were selected as the saturation mutation sites due to their direct contact with the side chain of the bound substrate. Five positive mutant BSAPs with different substrate specificities were obtained from these 76 mutant BSAPs. Two positive mutants were selected for application in soybean protein hydrolysis. Both of the two exhibited higher hydrolysis efficiency than wild-type BSAP, and the combination of the wild-type and two mutant BSAPs showed the highest hydrolysis efficiency for protein hydrolysis.
Results
Selection of mutation sites based on homology modeling of the BSAP structure
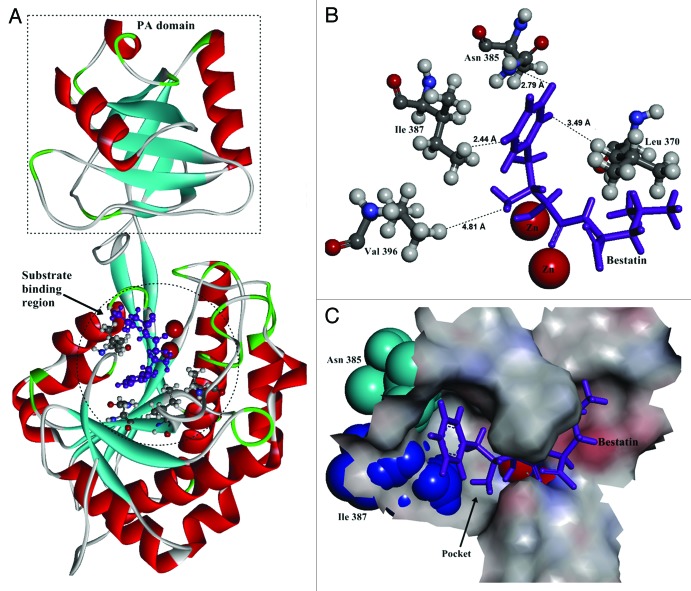
The crystal structure of AP from Aneurinibacillus sp AM-1 (PDB ID: 2EK8) was used as the template for homology modeling of BSAP. The predicted structure of BSAP including the protease-associated domain18,19 in the non-catalytic region is shown in Figure 1A. In the catalytic region, the two zinc ions and substrate binding site were indicated.
Figure 1. The structure of BSAP obtained by homology modeling. (A) Overall structure of BSAP. The α helices and β sheets are shown in red and cyan, respectively. Bestatin is shown in purple as the substrate. The PA-domain and substrate binding region are shown by dashed pane and dashed circle. (B) Local model of mutation sites in BSAP. The residues (Leu370, Asn385, Ile387, and Val396) in direct contact with the side chain of bound substrate are shown in balls and sticks, in which the oxygen atoms, nitrogen atoms, carbon atoms, and hydrogen atoms are in red, blue, deep gray, and light gray, respectively. The purple sticks indicate the bound substrate (bestatin). The two zinc atoms are shown in the form of magnified red balls. The distances (Å) from the bound substrate to the mutation sites in BSAP are shown. (C) The surface of the substrate binding region around the substrate in BSAP. The bound substrate is indicated by purple sticks. The residues Asn385 and Ile387 are displayed in the form of cyan and blue “CPK (Corey-Pauling-Koltun),” respectively. The pocket formed by the substrate binding sites is indicated by arrow.
In docking simulation, bestatin was docked into the predicted structure of BSAP as the substrate analog. The result shows that there are several residues in direct contact with the side chain of the substrate. The four closest residues (Leu 370, Asn 385, Ile 387 and Val 396) were chosen for saturation mutagenesis to investigate their effects on the substrate preference (Fig. 1B).
Saturation mutagenesis and screening the positive mutants
The residues at the four positions of BSAP were replaced with 19 other amino acids, respectively. The mutant enzymes were expressed in recombinant B. subtilis and secreted into the culture supernatant. The hydrolytic activity toward different aminoacyl-pNAs of the culture supernatant containing each mutant BSAP was tested and compared with that of wild-type BSAP. Saturation mutagenesis of Leu 370 and Val 396 did not generate any positive mutants, and the activities of these mutants were lower than that of wild-type BSAP. Five positive mutants (N385L, N385W, I387A, I387C, and I387S) were obtained via saturation mutagenesis at the position N385 and I387.
Using purified enzymes of these mutants (Fig. 2A), the specific activities toward different aminoacyl-pNAs were assayed precisely and compared with the wild-type enzyme. As shown in Figure 2B, although the hydrolytic activity of all mutations toward Leu-pNA decreased, N385L BSAP showed higher activity toward Lys-pNA than wild-type BSAP (2.2-fold). Three mutants (I387A, I387C, and I387S BSAPs) exhibited higher activity toward Met-pNA than wild-type BSAP (2‒3 folds), and similar hydrolytic activities of these three mutants toward Phe-pNA were also observed, which was not detected in wild-type BSAP. In addition, an increase in hydrolytic activities toward other four aminoacyl-pNAs (Ala-pNA, Ile-pNA, Val-pNA and Pro-pNA) was also observed in N385L BSAP (data not shown). According to these results, we chose N385L and I387A BSAPs for further study due to their high hydrolytic activities toward Lys-pNA and Phe-pNA, respectively.
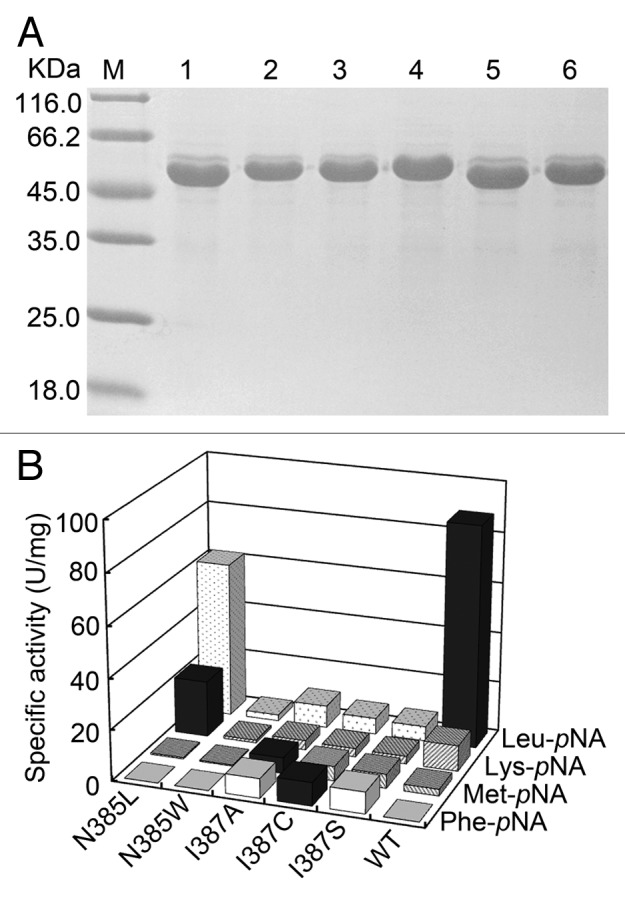
Figure 2. SDS-PAGE analysis and specific activity of the purified mutant and wild-type BSAPs toward several aminoacyl-pNAs. (A) SDS-PAGE analysis of the purified mutant and wild-type BSAPs. Samples (15 μg of protein) were loaded on a 12% gel. Lanes: M, molecular mass marker; 1, N385L BSAP; 2, N385W BSAP; 3, I387A BSAP; 4, I387C BSAP; 5, I387S BSAP; 6, wild-type BSAP. (B) Substrate specificities of the positive mutant (N385L, N385W, I387A, I387C and I387S) and wild-type BSAPs toward four aminoacyl-pNAs (Leu-pNA, Lys-pNA, Met-pNA and Phe-pNA). The highest hydrolytic activity toward each aminoacyl-pNA is indicated by black bar. The values are the representative of three independent experiments. In all cases, the standard deviation was less than 5% of the mean. WT, wild-type.
Thermostability and secondary structural analysis of the positive mutants
To compare the thermostability of wild-type, N385L and I387A BSAPs, the purified enzymes were incubated at temperatures ranging from 30 to 80 °C for 30 min. As shown in Figure 3A, the activities of the wild-type and mutant BSAPs decreased during the temperature increasing. The decrease of the activity of wild-type BSAP was slower than that of I387A BSAP, but faster than that of N385L BSAP. After 60 min of incubation at 60 °C, wild-type, N385L, and I387A BSAPs retained 43%, 59%, and 29% of their original activity (Fig. 3B), respectively. These findings indicated that N385L BSAP exhibited higher thermostability than the wild-type BSAP.
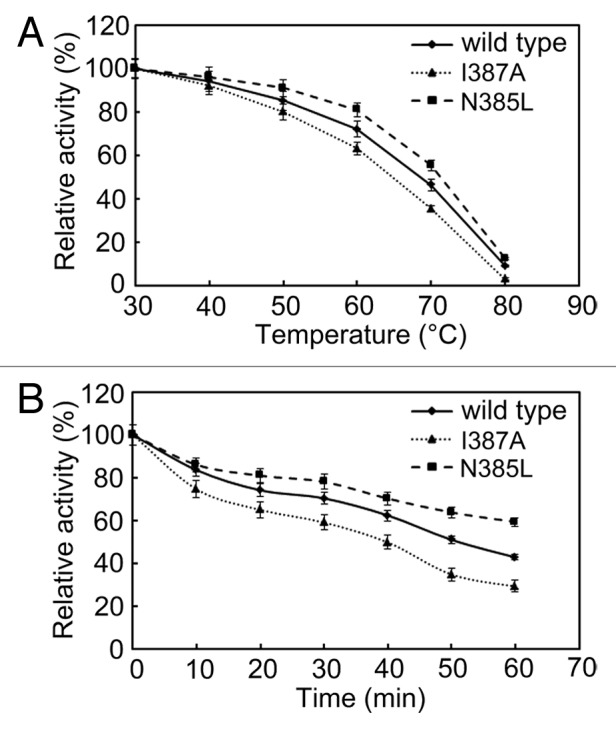
Figure 3. Effect of temperature on the stability of N385L (), I387A () and wild-type () BSAPs. (A) Thermostability of those three BSAPs. (B) Thermal inactivation of those three BSAPs. The results are the mean of three independent experiments.
The far UV circular dichroism (CD) spectra were analyzed to evaluate the effect of mutation on secondary structure. As shown in Figure 4, the spectra for wild-type, N385L, and I387A BSAPs were similar, suggesting that the secondary structure changed negligibly due to these single-point mutations.
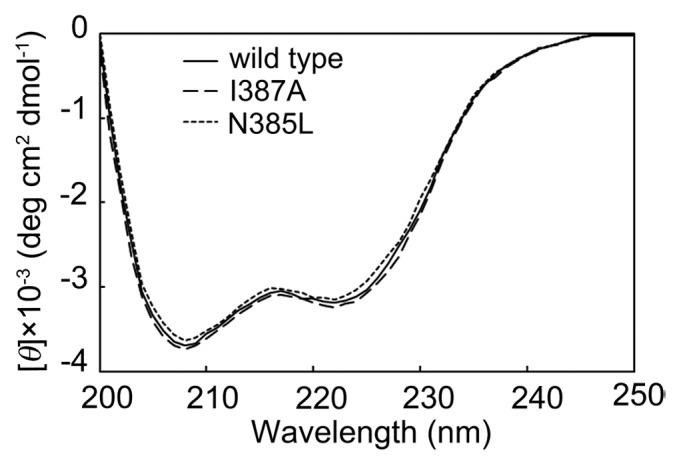
Figure 4. CD spectra of N385L, I387A and wild-type BSAPs. The secondary structural change of the mutants was checked by CD spectra in the far UV region (200‒250 nm).
Kinetic analysis
The kinetic parameters (Km, kcat, and kcat/Km) of wild-type, N385L, and I387A BSAPs toward three aminoacyl-pNAs (Leu-pNA, Lys-pNA, and Phe-pNA) were shown in Table 1. Although N385L BSAP displayed the reduction in catalytic efficiency (kcat/Km) toward Leu-pNA, its catalytic efficiency toward Lys-pNA was approximately 2.6-fold higher than that of the wild-type BSAP, and this efficiency was mainly caused by kcat (2.45-fold) but not Km (0.94-fold). In the case of I387A BSAP, this mutation resulted in the reduction in catalytic efficiency toward Leu-pNA and Lys-pNA in varying degrees. In contrast, I387A BSAP showed catalytic efficiency toward Phe-pNA with Km of 4.08 mM and kcat of 7.54 S−1, which was undetectable in the wild-type BSAP.
Table 1. Kinetic parameters of wild-type, N385L, and I387A BSAPsa.
| Substrate | BSAP variant | kcat (s−1) | kcat relative to WT | Km (mM) | Km relative to WT | kcat/ Km (mM−1 s−1) | kcat/ Km relative to WT |
|---|---|---|---|---|---|---|---|
| Leu-pNA | WTb | 70.66 ± 3.58 | 1 | 2.52 ± 0.11 | 1 | 28.1 | 1 |
| N385L | 52.58 ± 2.49 | 0.74 | 1.78 ± 0.09 | 0.73 | 29.5 | 1.01 | |
| I387A | 11.65 ± 0.93 | 0.17 | 12.27 ± 0.98 | 4.96 | 0.95 | 0.03 | |
| Lys-pNA | WT | 10.71 ± 0.54 | 1 | 4.12 ± 0.24 | 1 | 2.60 | 1 |
| N385L | 26.27 ± 1.46 | 2.45 | 3.86 ± 0.19 | 0.94 | 6.81 | 2.61 | |
| I387A | 6.82 ± 0.51 | 0.62 | 7.56 ± 0.41 | 1.82 | 0.91 | 0.34 | |
| Phe-pNA | WT | NDc | ND | ND | ND | ND | ND |
| N385L | ND | ND | ND | ND | ND | ND | |
| I387A | 7.54 ± 0.35 | ND | 4.08 ± 0.23 | ND | 1.85 | ND |
aKm and kcat values were calculated by nonlinear regression analysis. The values are means ± standard deviations of three independent experiments; bWT, wild-type; cND, not detected.
Comparison of the hydrolytic ability of the wild-type and the mutant BSAPs
Using aminoacyl-pNAs as the substrate, some mutant BSAPs with different substrate preferences were obtained. To investigate the effect of these positive mutants on protein hydrolysis, N385L, I387A and wild-type BSAPs were selected to hydrolyze soybean protein. The free amino concentration in each reaction was determined and the DH was calculated.
As shown in Table 2, the total amino acid concentration of the wild-type group (9.31 mg/mL) was lower than those of other three groups (11.02 mg/mL of N385L, 10.54 mg/mL of I387A, and 11.14 mg/mL of combination). The DH of combination group was highest (27.5%), which was 1.26-fold higher than that of wild-type group (21.9%), slightly higher than that of N385L group (26.3%), and that of I387A group (24.7%). Almost each amino acid concentration in the reactions of N385L and I387A, and the combination groups, was much higher than that of the wild-type group, especially the Thr (1.61 folds), Gly (1.59 folds), Val (1.56 folds), and Ile (1.72 folds) in the reaction of N385L group, the Ser (1.38 folds), Gly (1.49 folds), Ala (1.41 folds), and Phe (1.24 folds) in the reaction of I387A group and the Thr (1.41 folds), Gly (1.41 folds), Val (1.34 folds), and Ile (1.44 folds) in the reaction of the combination group. These findings demonstrated that the two mutant BASPs exhibited higher hydrolysis efficiency than wild-type BASP, and a combination BSAPs was much more conducive for protein hydrolysis.
Table 2. Free amino acid compositions of soybean protein hydrolysates and DH of different groups (mg/100 mL)a.
| Amino acid | None | WTb | N385L | I387A | Combination | N385L relative to WT | I387A relative to WT | Combination relative to WT |
|---|---|---|---|---|---|---|---|---|
| Asp | NDc | 16.92 ± 0.84 | 20.48 ± 0.62 | 17.26 ± 0.57 | 19.88 ± 0.68 | 1.21 | 1.02 | 1.17 |
| Thr | ND | 27.50 ± 1.17 | 44.38 ± 1.99 | 30.28 ± 1.43 | 38.79 ± 1.57 | 1.61 | 1.10 | 1.41 |
| Ser | ND | 50.54 ± 1.95 | 64.09 ± 2.87 | 69.52 ± 3.02 | 64.41 ± 3.15 | 1.26 | 1.38 | 1.27 |
| Glu | ND | 62.42 ± 2.74 | 75.52 ± 3.17 | 65.98 ± 2.53 | 73.02 ± 3.08 | 1.21 | 1.06 | 1.17 |
| Gly | ND | 19.68 ± 0.58 | 26.22 ± 0.84 | 31.33 ± 1.08 | 27.72 ± 0.95 | 1.59 | 1.49 | 1.41 |
| Ala | ND | 42.04 ± 1.05 | 55.18 ± 2.12 | 59.36 ± 2.24 | 55.89 ± 1.98 | 1.31 | 1.41 | 1.33 |
| Cys | ND | 11.76 ± 0.35 | 14.14 ± 0.62 | 12.76 ± 0.38 | 13.68 ± 0.42 | 1.20 | 1.09 | 1.16 |
| Val | 5.86 ± 0.22 | 43.26 ± 2.03 | 67.38 ± 2.86 | 43.79 ± 1.73 | 57.70 ± 2.26 | 1.56 | 1.02 | 1.34 |
| Met | ND | 20.02 ± 0.92 | 24.78 ± 0.93 | 22.31 ± 0.84 | 23.64 ± 0.72 | 1.24 | 1.11 | 1.18 |
| Ile | ND | 34.48 ± 1.58 | 59.26 ± 1.96 | 36.02 ± 1.76 | 49.81 ± 1.83 | 1.72 | 1.05 | 1.44 |
| Leu | ND | 113.22 ± 3.76 | 131.44 ± 5.57 | 114.34 ± 4.68 | 130.05 ± 5.03 | 1.16 | 1.01 | 1.15 |
| Tyr | 19.88 ± 0.74 | 94.72 ± 3.13 | 91.38 ± 2.94 | 115.96 ± 4.43 | 110.62 ± 4.27 | 0.96 | 1.22 | 1.17 |
| Phe | 7.36 ± 0.32 | 140.32 ± 4.83 | 141.48 ± 4.87 | 173.94 ± 5.71 | 165.64 ± 5.66 | 1.01 | 1.24 | 1.18 |
| Lys | 6.48 ± 0.24 | 87.88 ± 3.64 | 108.92 ± 2.54 | 90.10 ± 3.19 | 102.16 ± 2.97 | 1.24 | 1.02 | 1.16 |
| His | 0.76 ± 0.03 | 24.86 ± 1.06 | 26.86 ± 1.25 | 30.05 ± 1.36 | 29.04 ± 1.28 | 1.08 | 1.21 | 1.17 |
| Arg | 41.84 ± 1.97 | 141.49 ± 5.61 | 152.02 ± 5.33 | 141.12 ± 5.04 | 151.84 ± 5.18 | 1.07 | 1.01 | 1.07 |
| Total | 82.18 | 931.10 | 1101.54 | 1054.12 | 1113.86 | 1.18 | 1.13 | 1.20 |
| DH (%) | ND | 21.9 ± 1.04 | 26.3 ± 1.16 | 24.7 ± 1.03 | 27.5 ± 1.21 | 1.20 | 1.13 | 1.26 |
a, Relative to the amino acid composition of WT group, the amino acids of other three groups exhibiting high ratio were underlined and in bold. The values are means ± standard deviations of three independent experiments; b, WT, wild-type; c, ND, not detected.
Discussion
The APs with broad substrate specificity are expected in order to improve its application. However, the interaction between the substrate and the enzyme is so complex that only few researches on modifying the substrate specificity of enzymes have been reported, such as methyltransferase, Streptomyces aminopeptidase and mitochondrial ornithine carriers.20-24 Based on those researches we changed the substrate specificity of BSAP by replacing the residues in substrate binding region in this study. Soybean protein hydrolysis was performed to evaluate the effect of mutant BSAPs with different substrate preference for practical application. We provide evidence that combination of BSAPs with different substrate specificities can increase the content of free amino acids, which benefits the protein hydrolysis. This study revealed enzymology properties of BSAP, which should be significant for the theoretical investigation and its practical application.
By saturation mutagenesis of BSAP at position 370, 385, 387, and 396, several mutants exhibiting different substrate preference were obtained. Among five positive mutants, N385L and I387A BSAPs exhibited better catalytic efficiency toward Lys-pNA and Phe-pNA than other three mutants, respectively. Kinetic analysis revealed that the effect of N385L mutation on Lys-pNA hydrolysis is mostly on kcat and not on Km. Thereby we speculate that the change in the environment of the substrate binding region by this mutation leads to a subtle orientation shift of the bound substrate. This shift changes the distance between the substrate and the catalytic residues. In the case of I387A BSAP, the mutation led to the catalytic activity toward Phe-pNA which was undetectable in wild-type BSAP. As shown in Figure 1C, the substrate was located in a pocket formed by the residues of substrate binding region. The substrate specificity of wild-type BSAP showed that BSAP are most active toward hydrophobic amino acids with large aliphatic chain (Leu) and basic amino acids (Arg and Lys). The Phe residue does not suit the substrate binding pocket due to its large side chain. I387A mutation can expand the pocket by reducing the side chain of the residue, which makes the pocket suitable for the interaction with Phe-pNA. The Cys and Ser mutations led to the similar catalytic activity toward Phe-pNA, suggesting that replacement of Ile387 by a small residue leads to the catalytic activity toward Phe-pNA.
Although CD spectra showed that the secondary structures have no significant difference among wild-type, N385L, and I387A BSAPs, the apparent differential thermostability of these BSAPs was observed. It is well known that the hydrophobic interaction plays an important role in the stability of protein structure. N385L BSAP exhibited better thermostability than that of wild-type BSAP due to the enhanced hydrophobicity of substrate binding region caused by N385L mutation. In contrast, I387A mutation weakened the hydrophobic interaction in substrate binding region, which led to the poorer thermostability.
Here we obtained the BSAPs with different substrate preferences via site-directed mutagenesis. However, the substrate specificity did not exhibit expected variation by single-site mutation. The combination of several positive single-site mutations at position 385 and 387 was also performed. Unfortunately, it did not generate any mutant BSAPs with higher performance. The electrostatic environment, hydrophobicity and steric hindrance determined the interaction between the substrate and the enzyme simultaneously, which indicated that the positive mutations were non-cumulative. Therefore, further studies using the combination of site-directed mutagenesis and other directed evolution means to remodel BSAP are required to improve their performance and fulfill industrial requirements.
Materials and Methods
Materials, bacterial strains and plasmids
All aminoacyl-pNAs were purchased from Bachem AG. Plasmid PMA5-BSAP (with the BSAP gene inserted into the NdeI-BamHI gap of PMA525) was used for expression of wild-type BSAP and as a template of mutagenesis. Escherichia coli JM109 was the host for cloning work. B. subtilis WB60026 was used as the host strain for gene expression.
Homology modeling and docking study
Based on the structure of AP from Aneurinibacillus sp strain AM-1 (Protein Data Bank accession number 2EK8.pdb), the predicted structure of BSAP was obtained by homology modeling using Discovery Studio software 2.5 (Accelrys Software Inc.). Stereo chemical analysis of the structures was performed using PROCHECK (http://nihserver.mbi.ucla.edu/SAVS/). The final model displays good geometry with less than 2% of residues in the disallowed region and was used in this study. The docking study was performed using Discovery Studio 2.5 (Accelrys Software Inc.). The structure of bestatin was built into the software as the substrate, and simulation of interactive docking was performed using the rigid form of BSAP and flexible form of bestatin.
Saturation mutagenesis
Saturation mutagenesis of each residue was conducted by inverse PCR using 19 pairs of oligonucleotide primers containing a point mutation. The PCR program was as follows: 18 cycles of 20 sec at 98 °C and 9 min at 68 °C. The PCR product was treated with DpnI at 37 °C for 1 h. Then the PCR product was transformed into competent cell of E. coli JM109 according to the manufacturer’s protocol. After the confirmation of the sequence, the extracted plasmid was transformed into B. subtilis WB600 following the method of Spizizen et al.27
Activity assay and determination of kinetic parameters
The activity was determined following our previous study.11 Kinetic parameters for aminoacyl-pNA hydrolysis by the enzymes were determined with different concentrations from 0.8 to 4 mM (Leu-pNA and Lys-pNA) or from 0.2 to 2 mM (Phe-pNA). The protein content of the samples was measured by the Bradford method28 with BSA as a standard.
Screening and preparation of mutant BSAPs
B.subtilis WB600 harboring the constructed plasmid for mutant BSAP expression was cultivated at 37 °C for 24 h in 20 mL TB medium containing 50 μg/mL kanamycin. The activity of the culture supernatant toward several aminoacyl-pNAs was measured and compared with that of wild-type BSAP to screen the positive mutants with different substrate specificities. The purified BSAPs were prepared following the method of our previous study.11
Thermostability assay and CD spectrometry
To detect thermal inactivation, the purified enzymes (2 μg/mL) were incubated in the range of 30‒80 °C for 30 min. After heating, the enzyme was placed on ice for 10 min immediately, and the remaining activity was measured. The remaining activity was recorded as percentage of the original activity. The time courses of activity of purified BSAPs were measured by incubating the enzyme in buffer A at 60 °C. At regular intervals (0‒60 min), samples were removed and cooled on ice for 10 min, and the remaining activity was determined. The activity of the enzymes kept on ice was considered as the control (100%).
CD spectroscopy was performed on MOS-450/AF-CD-STP-A (Bio-Logic). Spectra were recorded from 200 to 260 nm using a 1 mm path-length quartz cuvette with a protein concentration of 0.1 mg/mL in buffer A. In order to minimize signal baseline drift, the spectropolarimeter and xenon lamp were warmed up for at least 30 min prior to each experiment. The results are expressed as mean residue ellipticity [θ], which is defined as [θ] = 100 θobs/lc, where θobs is observed ellipticity in degrees, c is the concentration in residue moles per liter, and l is the length of the light path in centimeters.
Hydrolysis of soybean protein
Soybean protein was prepared with deionized water at the final concentration of 5% (w/v). After heated at 90 °C for 10 min, the pH of the solution was adjusted to 8.5 by NaOH. The purified BSAPs (wild-type, I387A and N385L BSAPs) (0.1 g/L, finial) were used to hydrolyze soybean protein, respectively. In addition, a combination of wild-type, I387A and N385L BSAPs (ratio 2:1:1, total 0.1 g/L) was also used to hydrolyze soybean protein. In order to improve hydrolysis efficiency, alkaline protease (1 g/L) was added together with the BSAPs. The reaction was treated under 50 °C for 5 h.
The degree of hydrolysis (DH) was defined as the percentage of free amino groups cleaved from protein, which was calculated from the ratio of α-amino nitrogen to total nitrogen. The α-amino nitrogen content was determined by formaldehyde titration method.29 The total nitrogen content was determined by Kjeldahl method.30 Amino acid composition was determined using HITACHI L-8900 amino acid analyzer (Hitachi).
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
This work is financially supported by the National High Technology Research and Development Program of China (863 Program, 2011AA100905), the Program for New Century Excellent Talents in University (NCET-10-0461), the Key project of Chinese Ministry of Education (311023), the Priority Academic Program Development of Jiangsu Higher Education Institution and the 111 Project (No. 111-2-06).
Glossary
Abbreviations:
- aminopeptidases
APs
- Bacillus subtilis aminopeptidase
BSAP
- p-nitroanilides
pNAs
- degree of hydrolysis
DH
- circular dichroism
CD
- sodium dodecyl sulfate-polyacrylamide gel electrophoresis
SDS-PAGE
Submitted
03/18/2013
Revised
05/07/2013
Accepted
05/22/2013
Footnotes
These authors contributed equally to this work.
Previously published online: www.landesbioscience.com/journals/prion/article/25147
References
- 1.Gonzales T, Robert-Baudouy J. Bacterial aminopeptidases: properties and functions. FEMS Microbiol Rev. 1996;18:319–44. doi: 10.1111/j.1574-6976.1996.tb00247.x. [DOI] [PubMed] [Google Scholar]
- 2.Kilcawley KN, Wilkinson MG, Fox PF. Determination of key enzyme activities in commercial peptidase and lipase preparations from microbial or animal sources. Enzyme Microb Technol. 2002;31:310–20. doi: 10.1016/S0141-0229(02)00136-9. [DOI] [Google Scholar]
- 3.Taylor A. Aminopeptidases: towards a mechanism of action. Trends Biochem Sci. 1993;18:167–71. [PubMed] [Google Scholar]
- 4.Pulido-Cejudo G, Conway B, Proulx P, Brown R, Izaguirre CA. Bestatin-mediated inhibition of leucine aminopeptidase may hinder HIV infection. Antiviral Res. 1997;36:167–77. doi: 10.1016/S0166-3542(97)00052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holz RC, Bzymek KP, Swierczek SI. Co-catalytic metallopeptidases as pharmaceutical targets. Curr Opin Chem Biol. 2003;7:197–206. doi: 10.1016/S1367-5931(03)00033-4. [DOI] [PubMed] [Google Scholar]
- 6.Bauvois B. Transmembrane proteases in cell growth and invasion: new contributors to angiogenesis? Oncogene. 2004;23:317–29. doi: 10.1038/sj.onc.1207124. [DOI] [PubMed] [Google Scholar]
- 7.Guo Y, Pan D, Tanokura M. Optimisation of hydrolysis conditions for the production of the angiotensin-I converting enzyme (ACE) inhibitory peptides from whey protein using response surface methodology. Food Chem. 2009;114:328–33. doi: 10.1016/j.foodchem.2008.09.041. [DOI] [Google Scholar]
- 8.Su G, Ren J, Yang B, Cui C, Zhao M. Comparison of hydrolysis characteristics on defatted peanut meal proteins between a protease extract from Aspergillus oryzae and commercial proteases. Food Chem. 2011;126:1306–11. doi: 10.1016/j.foodchem.2010.11.083. [DOI] [Google Scholar]
- 9.Wang F, Ning Z, Lan D, Liu Y, Yang B, Wang Y. Biochemical properties of recombinant leucine aminopeptidase II from Bacillus stearothermophilus and potential applications in the hydrolysis of Chinese anchovy (Engraulis japonicus) proteins. J Agric Food Chem. 2012;60:165–72. doi: 10.1021/jf204002e. [DOI] [PubMed] [Google Scholar]
- 10.Zhu X, Barman A, Ozbil M, Zhang T, Li S, Prabhakar R. Mechanism of peptide hydrolysis by co-catalytic metal centers containing leucine aminopeptidase enzyme: a DFT approach. J Biol Inorg Chem. 2012;17:209–22. doi: 10.1007/s00775-011-0843-2. [DOI] [PubMed] [Google Scholar]
- 11.Gao X, Cui W, Tian Y, Zhou Z. Over-expression, secretion, biochemical characterisation, and structure analysis of Bacillus subtilis aminopeptidase. J Sci Food Agric. 2013 doi: 10.1002/jsfa.6105. accepted. [DOI] [PubMed] [Google Scholar]
- 12.Chen G, Edwards T, D’souza VM, Holz RC. Mechanistic studies on the aminopeptidase from Aeromonas proteolytica: a two-metal ion mechanism for peptide hydrolysis. Biochemistry. 1997;36:4278–86. doi: 10.1021/bi9618676. [DOI] [PubMed] [Google Scholar]
- 13.Fundoiano-Hershcovitz Y, Rabinovitch L, Langut Y, Reiland V, Shoham G, Shoham Y. Identification of the catalytic residues in the double-zinc aminopeptidase from Streptomyces griseus. FEBS Lett. 2004;571:192–6. doi: 10.1016/j.febslet.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Gilboa R, Spungin-Bialik A, Wohlfahrt G, Schomburg D, Blumberg S, Shoham G. Interactions of Streptomyces griseus aminopeptidase with amino acid reaction products and their implications toward a catalytic mechanism. Proteins. 2001;44:490–504. doi: 10.1002/prot.1115. [DOI] [PubMed] [Google Scholar]
- 15.Hershcovitz YF, Gilboa R, Reiland V, Shoham G, Shoham Y. Catalytic mechanism of SGAP, a double-zinc aminopeptidase from Streptomyces griseus. FEBS J. 2007;274:3864–76. doi: 10.1111/j.1742-4658.2007.05912.x. [DOI] [PubMed] [Google Scholar]
- 16.Munih P, Moulin A, Stamper CC, Bennett B, Ringe D, Petsko GA, et al. X-ray crystallographic characterization of the Co(II)-substituted Tris-bound form of the aminopeptidase from Aeromonas proteolytica. J Inorg Biochem. 2007;101:1099–107. doi: 10.1016/j.jinorgbio.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Chen S-L, Marino T, Fang W-H, Russo N, Himo F. Peptide hydrolysis by the binuclear zinc enzyme aminopeptidase from Aeromonas proteolytica: a density functional theory study. J Phys Chem B. 2008;112:2494–500. doi: 10.1021/jp710035j. [DOI] [PubMed] [Google Scholar]
- 18.Luo X, Hofmann K. The protease-associated domain: a homology domain associated with multiple classes of proteases. Trends Biochem Sci. 2001;26:147–8. doi: 10.1016/S0968-0004(00)01768-0. [DOI] [PubMed] [Google Scholar]
- 19.Mahon P, Bateman A. The PA domain: a protease-associated domain. Protein Sci. 2000;9:1930–4. doi: 10.1110/ps.9.10.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Farrell HC, Musayev FN, Scarsdale JN, Rife JP. Control of substrate specificity by a single active site residue of the KsgA methyltransferase. Biochemistry. 2012;51:466–74. doi: 10.1021/bi201539j. [DOI] [PubMed] [Google Scholar]
- 21.Arima J, Uesugi Y, Iwabuchi M, Hatanaka T. Alteration of leucine aminopeptidase from Streptomyces septatus TH-2 to phenylalanine aminopeptidase by site-directed mutagenesis. Appl Environ Microbiol. 2005;71:7229–35. doi: 10.1128/AEM.71.11.7229-7235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arima J, Uesugi Y, Iwabuchi M, Hatanaka T. Change in substrate preference of Streptomyces aminopeptidase through modification of the environment around the substrate binding site. Appl Environ Microbiol. 2006;72:7962–7. doi: 10.1128/AEM.01460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arima J, Uesugi Y, Uraji M, Iwabuchi M, Hatanaka T. The role of Glu196 in the environment around the substrate binding site of leucine aminopeptidase from Streptomyces griseus. FEBS Lett. 2006;580:912–7. doi: 10.1016/j.febslet.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 24.Monné M, Miniero DV, Daddabbo L, Robinson AJ, Kunji ERS, Palmieri F. Substrate specificity of the two mitochondrial ornithine carriers can be swapped by single mutation in substrate binding site. J Biol Chem. 2012;287:7925–34. doi: 10.1074/jbc.M111.324855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zyprian E, Matzura H. Characterization of signals promoting gene expression on the Staphylococcus aureus plasmid pUB110 and development of a gram-positive expression vector system. DNA. 1986;5:219–25. doi: 10.1089/dna.1986.5.219. [DOI] [PubMed] [Google Scholar]
- 26.Wu XC, Lee W, Tran L, Wong SL. Engineering a Bacillus subtilis expression-secretion system with a strain deficient in six extracellular proteases. J Bacteriol. 1991;173:4952–8. doi: 10.1128/jb.173.16.4952-4958.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spizizen J. Transformation of Biochemically Deficient Strains of Bacillus Subtilis by Deoxyribonucleate. Proc Natl Acad Sci U S A. 1958;44:1072–8. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 29.Nilsang S, Lertsiri S, Suphantharika M, Assavanig A. Optimization of enzymatic hydrolysis of fish soluble concentrate by commercial proteases. J Food Eng. 2005;70:571–8. doi: 10.1016/j.jfoodeng.2004.10.011. [DOI] [Google Scholar]
- 30.Miller J, Young CT. Protein nutritional quality of Florunner peanut meal as measured by rat bioassay. J Agric Food Chem. 1977;25:653–7. doi: 10.1021/jf60211a051. [DOI] [PubMed] [Google Scholar]