Abstract
DNA adducts represent internal dosimeters to measure exposure to environmental and endogenous genotoxicants. Unfortunately, in molecular epidemiologic studies, measurements of DNA adducts often are precluded by the unavailability of fresh tissue. In contrast, formalin-fixed paraffin embedded (FFPE) tissues frequently are accessible for biomarker discovery. We report here that DNA adducts of aristolochic acids (AAs) can be measured in FFPE tissues at a level of sensitivity comparable to freshly frozen tissue. AAs are nephrotoxic and carcinogenic compounds found in Aristolochia herbaceous plants, many of which have been used worldwide for medicinal purposes. AAs are implicated in the etiology of aristolochic acid nephropathy and upper urinary tract carcinoma. 8-Methoxy-6-nitrophenanthro-[3,4-d]-1,3-dioxole-5-carboxylic acid (AA-I) is a component of Aristolochia herbs and a potent human urothelial carcinogen. AA-I reacts with DNA to form the aristolactam (AL-I)-DNA adduct 7-(deoxyadenosin-N6-yl) aristolactam I (dA-AL-I). We established a method to quantitatively retrieve dA-AL-I from FFPE tissue. Adducts were measured, using ultraperformance liquid chromatography/mass spectrometry, in liver and kidney tissues of mice exposed to AA-I, at doses ranging from 0.001 to 1 mg/kg body weight. dA-AL-I was then measured in 10-µm thick tissue-sections of FFPE kidney from patients with upper urinary tract cancers; the values were comparable to those observed in fresh frozen samples. The limit of quantification of dA-AL-I was 3 adducts per 109 DNA bases per 2.5 µg of DNA. The ability to retrospectively analyze FFPE tissues for DNA adducts may provide clues to the origin of human cancers for which an environmental cause is suspected.
Introduction
The covalent modification of DNA by chemical mutagens is an initiating step in chemical carcinogenesis.1,2 DNA adduct formation is an important end point for cross-species extrapolation of the biologically effective dose and for human risk assessment of exposure to chemical carcinogens.3,4 Identification and quantification of DNA adducts often is the first step in elucidating the potential role of a genotoxic chemical in the etiology of human cancer.5 The paucity of accessible frozen tissue restricts the implementation of DNA adduct biomarkers to assess carcinogen exposure and DNA damage in molecular epidemiology studies. In contrast, formalin-fixed samples from patients with cancer are frequently available.
Formalin fixation, followed by paraffin embedding (FFPE) has been used as the standard storage technique for more than a century in laboratories world-wide. Chemical fixation preserves cell morphology; thus, cellular components remain intact. Details of the preserved cells can be visualized, following various staining techniques, by light and electron microscopy, or by immunohistochemistry.6 Formaldehyde is the most commonly used agent for fixation: its reaction with macromolecules introduces intramolecular and intermolecular crosslinks between proteins and DNA.7–9
Considerable efforts have been made to retrieve proteins, DNA and RNA from FFPE tissue for biomarker discovery. Various conditions, including elevated temperature and alkaline pH,10,11 have been employed to reverse the crosslinks of these macromolecules in FFPE tissue. A major limitation to the use of FFPE tissues in genomic studies is the highly variable and often poor quality of the DNA used for polymerase chain reaction (PCR) amplification.6,11 However, recent improvements in retrieval techniques, both for DNA12,13 and proteins7,14 isolated from archived FFPE samples have been used successfully for biomarker discovery in genomics and proteomics.
The screening of putative carcinogen DNA adducts in human FFPE tissues has been reported. These studies primarily employed immunohistochemical (IHC) techniques for the detection of DNA damage in fixed cells or formalin fixed tissue-sections.15–18 DNA adducts can be detected, by IHC, in specific cell types within a tissue. Moreover, the applicability of IHC to screen paraffin-embedded tissue-sections makes IHC an attractive method for the analysis of DNA adducts.15,16 However, an important drawback of IHC is that the specificity of many antibodies, even monoclonal antibodies, for DNA adducts is uncertain as they may cross-react with other DNA lesions, leading to errors in identification and quantification.
The quantitative measurement of DNA adducts in DNA obtained from FFPE tissue requires the retrieval of high quality, fully digestible DNA without cross links. However, many carcinogen DNA adducts are unstable at the elevated temperature or alkaline pH commonly used to retrieve DNA from FFPE tissue.19,20 As a consequence, the development of robust analytical methods to quantitatively measure carcinogen DNA adducts in FFPE tissue has been challenging. To our knowledge, there are only two reports in the literature on the measurement of DNA adducts in archived FFPE biospecimens using semi-quantitative 32P-postlabeling methods.21,22 Moreover, the measurement of DNA adducts by mass spectrometric methods in human FFPE tissue has not been reported.
Recently vendors of several commercial kits have developed mild retrieval conditions to recover DNA free of cross links, which can serve as high fidelity templates for amplification with PCR. These improvements in retrieval of DNA from archival FFPE tissues prompted us to examine the suitability of such tissues for analysis of carcinogen-DNA adducts by mass spectrometric analysis, following enzymatic digestion of DNA. We selected aristolochic acids (AAs) as the model human carcinogen for development of a DNA adduct biomarker in view of the unique opportunity to obtain tissues from human subjects with upper urothelial cancer and known exposure to AA.22–24
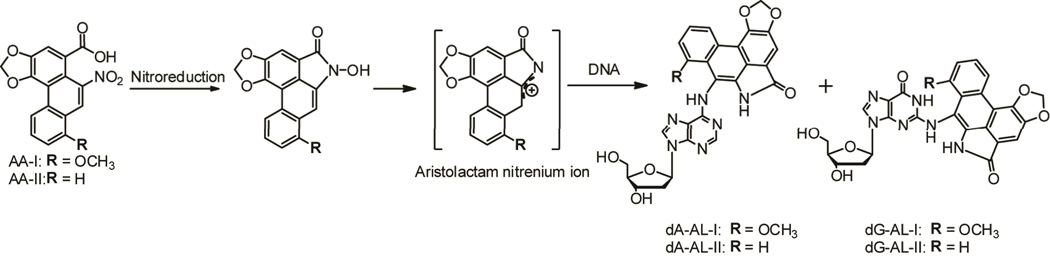
Aristolochic acids (AAs) are a family of structurally-related nephrotoxic and carcinogenic nitrophenanthrene compounds found in Aristolochia herbaceous plants, some of which have been used worldwide for medicinal purposes for over 2000 years. AAs have been implicated in the etiology of aristolochic acid nephropathy and of Endemic (Balkan) Nephropathy (BEN), a devastating environmental disease.22,24,25 8-Methoxy-6-nitrophenanthro-[3,4-d]-1,3-dioxole-5-carboxylic acid (AA-I) and 6-nitrophenanthrene-[3,4-d]-1,3-dioxole-5-carboxylic acid (AA-II) are enzymatically metabolized via reduction of the nitro moieties of the phenanthrene ring to produce reactive N-hydroxyaristolactams (AL) and the postulated nitrenium intermediates, which covalently bind to the exocyclic amino groups of the purine nucleobases (Figure 1).26,27 In rodents, the major AL-DNA adducts have been identified as 7-(deoxyadenosin-N6-yl) aristolactam I (dA-AL-I), 7-(deoxyguanosin-N2-yl) aristolactam I (dG-AL-I), 7-(deoxyadenosin-N6-yl) aristolactam II (dA-AL-II), and 7-(deoxyguanosin-N2-yl) aristolactam II (dG-AL-II) (Figure 1).26–28 dA-AL-I is by far the most abundant adduct found in human kidney.22–24,29–31 The dA-AL lesions are responsible, via translesion synthesis, for generating otherwise rare A:T → T:A transversions in the TP53 tumor suppressor gene in urothelial carcinomas of patients with AA-induced urothelial carcinomas of the upper urinary tract (UUC).22–24 This mutation also predominates in human TP53 knock-in mouse cells,32 and in the H-ras gene of rats treated with AA,33 and in site-specific mutagenesis experiments in which a plasmid DNA contained a single dA-AL adduct.34
Figure 1.
Metabolic activation of AAs. AL-DNA adduct formation occurs after reduction of the nitro moiety of the phenanthrene rings of AA-I and AA-II to form the N-hydroxyaristolactams and proposed nitrenium ions, which react with the exocyclic amino groups of dG and dA to form dA-AL and dG-AL adducts.
We previously established a robust method to quantify AL-DNA adducts in freshly frozen human kidney tissue from patients with AAN and UUC, using ultraperformance liquid chromatography with electrospray ionization and multistage scan mass spectrometry (UPLC-ESI-MS/MSn).31 In this study, we describe a method to measure AL-DNA adducts in FFPE tissues of mice and humans. The results show that AL-DNA adducts can be retrieved in high yield from FFPE tissue.
Experimental Section
Caution: Aristolochic acids are human carcinogens and should be handled with caution in a well-ventilated fume hood with the appropriate protective clothing.
Materials
8-Methoxy-6-nitrophenanthro-(3,4-d)-1,3-dioxole-5-carboxylic (AA-I) was a kind gift from Dr. Horacio Priestap, Department of Biological Sciences, Florida International University. 15N-labeled 2′-deoxyadenosine, [15N5]-dA (isotopic purity = 99%) was purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA). Calf thymus DNA (CT DNA), DNase I (Type IV, from bovine pancreas), alkaline phosphatase (from Escherichia coli), nuclease P1 (from Penicillium citrinum), RNase A, and RNase T1, were purchased from Sigma (St. Louis, MO). Phosphodiesterase I (from Crotalus adamanteus venom) was purchased from Worthington Biochemicals Corp. (Newark, NJ, USA). Dimethyl sulfoxide (DMSO) (>99.9%) and LC/MS grade formic acid (99.5%) were purchased from Fischer Scientific (Harnover Park, IL). Formalin cups containing 10% neutral-buffered formalin (20 mL) were purchased from Richard Allen Scientific through Thermo Fisher Scientific (Kalamazoo, MI). All solvents were high-purity B & J Brand from Honeywell Burdick and Jackson (Muskegon, MI) or Optima LC/MS brand from Fisher Scientific (Fair Lawn, NJ). ZR FFPE DNA MiniPrep™ kit was from Zymo Research (Irvine, CA). Amicon Ultrafree-MC centrifugal filters were from Millipore (Bedford, MA). Microliter CapLC vials with silylated inserts were from Microliter Analytical Supplies (Suwanee, GA)
Syntheses of AL-DNA Adducts
The syntheses of 7-(deoxyadenosin-N6-yl)aristolactam-I (dA-AL-I) and ([15N5]-dA-AL-I) were performed by reacting AA-I with dA or [15N5]-dA, employing previously described protocols.31,35
Animal Studies
The guidelines established by the National Institutes of Health Office of Laboratory Animal Welfare, were adhered to for the use of animals. Mice were acclimatized to temperature (22 ± 2 °C) and humidity (55 ± 5%) in rooms with a 12 h light/dark cycle for at least 1 week before the experiment. Laboratory chow and tap water were given ad libitum. Male C57BL/6J mice (Jackson Laboratories, Bar Harbor, Maine), 8–10 weeks old (N = 4 per group), were dosed (i.p.) with AA-I (0, 0.001, 0.01, 0.1, or 1 mg/kg, in phosphate buffered saline). After 20 h, mice were euthanized by asphyxiation with CO2, followed by cervical dislocation. The whole livers and kidneys were immediately excised, and stored at −80 °C. Organs from a second set of animals were immersed in 10% neutral-buffered formalin (20 mL) for 24 h at room temperature. A sagittal section of the kidney was cut prior to fixation, whereas the entire liver was fixed in formalin. Tissues were embedded in paraffin in a tissue processor at the Histology Core Facility at Stony Brook University. The processing conditions are provided in Supporting Information (Table S-1). The FFPE tissue blocks were stored at ambient temperature prior to chemical analysis.
Human Tissue Samples
The research protocol was reviewed and approved by the Institutional Review Boards at Stony Brook University, the Wadsworth Center, and the School of Medicine, University of Zagreb. Kidney samples, containing both renal cortex and medulla, were obtained from Croatian and Serbian patients from the Balkan endemic regions undergoing nephroureterectomy. Portions of the excised kidney were immediately stored frozen at −80 °C or fixed in neutral-buffered formalin. The formalin fixation time (24 h) and ensuing paraffin embedding conditions were slightly different for Croatian and Serbian patients, but the procedures largely followed the processing conditions described in Supporting Information (Table S-1). The human FFPE tissue blocks were prepared between 2007 – 2009 and stored at ambient temperature. Ten-µm thick sections were cut from the FFPE tissue blocks with a Rotary Microtome and used for DNA extraction. These section cuts were stored at room temperature for up to 2 months prior to analysis of DNA adducts.
Isolation of DNA from Freshly Frozen Mouse Tissue
The tissues were thawed in a water bath at 4 °C, and then immediately homogenized in 10 mM Tris-HCl buffer (pH 8.3) containing 10 mM EDTA (TE buffer, 3 mL per g tissue) with a Potter-Elvehjem homogenizer. The homogenate was centrifuged at 3000 g for 10 min. Genomic DNA in the nuclear pellet (equivalent of 50 mg of wet tissue weight) was isolated by CHCl3/phenol extraction36 with modifications. The entire protocol is described in Supporting Information (Protocol S-1).
Deparaffinization, Rehydration, and Isolation of DNA from Mouse FFPE Tissues
The organ samples were cut out of the block intact, and the bulk of the paraffin was removed with a scalpel. The tissues were transferred to a glass vial and submerged with p-xylene (~3.5 mL), washed with p-xylene, and then washed with ethanol/water, followed by rehydration with water, and then TE buffer. The full protocol is provided in Supporting Information (Protocol S-2). The rehydrated tissues (20 – 25 mg of dry weight deparaffinized tissue) were homogenized with a Potter-Elvehjem homogenizer, followed by centrifugation at 3000 g, to obtain the nuclear pellets. DNA was isolated by CHCl3/phenol extraction (Protocol S-1). Alternatively, the nuclear pellet was processed with the ZR FFPE DNA MiniPrep™ kit (Zymo Research), using the equivalent of 20 – 25 mg of dry weight deparaffinized tissue. The protocol followed the manufacturer’s instructions with minor modifications (Supporting Information, Protocol S-2).
Isolation of DNA from Freshly Frozen Human Kidney
Genomic DNA was isolated by CHCl3/phenol extraction36 with modifications as described in Supporting Information (Protocol S-3).
Deparaffinzation, Rehydration and Isolation of DNA from FFPE Human Kidney Tissue-Sections
Two tissue-section cuts of 10 µm-thickness each containing a piece of kidney occupying a surface area of 0.9 – 1.8 cm2 were combined in an Eppendorf tube, and washed with p-xylene (1.5 mL), followed by ethanol/water (95%/5%). The tissue was air-dried for 30 min and then resuspended in TE buffer (1 mL). The samples were digested with proteinase K for 16 h at 55 °C, employing the lysis buffer of the ZR FFPE DNA MiniPrep™ kit. DNA was isolated according to the manufacturer’s instructions with minor modifications as described in Supporting Information (Protocol S-2).
Assessing the Efficacy of Removal of DNA Crosslinks and Recovery of AL-DNA Adducts
The efficacy of removal of DNA crosslinks in mouse and human FFPE tissues was determined by monitoring the recovery of deoxyribnucleosides, following enzymatic digestion of the DNA. DNA hydrolysis was performed with a cocktail of enzymes containing DNase I, nuclease P1, alkaline phosphatase, and phosphodiesterase as previously described.37 Deoxyribonucleosides were monitored by HPLC with UV detection, as previously described.38
Measurement of AL-DNA Adducts by On-Line Solid Phase Extraction Coupled with UPLC-ESI/MS3
Adduct quantification was done by the stable isotope dilution method and employed [15N5]-dA-AL-I as the internal standard at a level of 2 to 5 adducts per 108 DNA bases.31 Analyses were performed with a NanoAcquity UPLC system (Waters Corporation, Milford, MA) interfaced with an Advance CaptiveSpray source from Michrom Bioresource Inc. (Auburn, CA) and an ion-trap mass spectrometer (LTQ Velos, Thermo Fisher, San Jose, CA). A Waters Symmetry trap column (180 µm × 20 mm, 5 µm particle size) was employed for online solid phase enrichment of the DNA adducts. A C18 AQ (0.3 × 150 mm, 3 µm particle size, Michrom Bioresources Inc.) was used for chromatography.31 The DNA adducts were measured in the positive ionization mode at the MS3 scan stage. The chromatographic and mass spectra acquisition parameters were previously described.31 The method is summarized in Supporting Information (Protocol S-4).
Enzymatic Digestion of AL-DNA and 32P-Postlabeling/PAGE Analysis of Fresh Frozen Samples
Human DNA digests (5 or 20 µg) were prepared, then incubated with 20 µCi of [γ-32P]-ATP (6000 Ci/mmol) and 10 units of 3′-phosphatase-free OptiKinase at 37 °C, as described previously.39,40 32P-labeled adducts were subjected to electrophoresis for 5 h on a nondenaturing 30% polyacrylamide gel, and AL-DNA adducts were measured using a ß-phosphorimager with Image QuaNT v5.2 software (Molecular Dynamics Inc. Piscataway, NJ).36,39,40
Statistical Analyses
Differences between AL-DNA adduct levels in freshly frozen and FFPE tissue as a function of dose were determined with the unpaired 2-tailed t-test, and differences in recovery of AL-DNA adducts by different retrieval methods were assessed by one-way ANOVA with Dunnett’s multiple comparison post-test, using Graphpad Prism® 4 Software (San Diego, CA).
Results and Discussion
Elevated temperature and/or alkaline pH conditions have been used to reverse crosslinks formed between formaldehyde with DNA and protein in FFPE tissues.9–11,13,14 The macromolecules retrieved have been employed with varying degrees of success in genomics, proteomics, and immunohistochemistry applications.6,13,14,41 Genomic studies strive to optimize the recovery of high-quality, uncrosslinked DNA from FFPE tissue for usage as templates for DNA amplification by PCR. In contrast, our goal is to recover chemically modified DNA that is fully digestible to form single DNA bases containing an adducted mutagen/carcinogen for analysis by mass spectrometry.
We examined the effect of temperature on the reversal of DNA crosslinks and evaluated different methods by which DNA could be retrieved from archived FFPE tissue so as to optimize the recovery of AL-DNA adducts. Incomplete removal of DNA crosslinks results in the formation of oligomers containing dA-AL-I; consequently, decreasing the recovery of the dA-AL-I adduct. Furthermore, AL-DNA adducts must be stable to both the conditions employed to reverse the DNA crosslinks and the retrieval of DNA from FFPE tissue; otherwise, levels dA-AL-I will be underestimated.
Effect of Temperature and Methods of DNA Retrieval on Reversal of DNA Crosslinks and Recovery of AL-DNA Adducts in FFPE Tissue
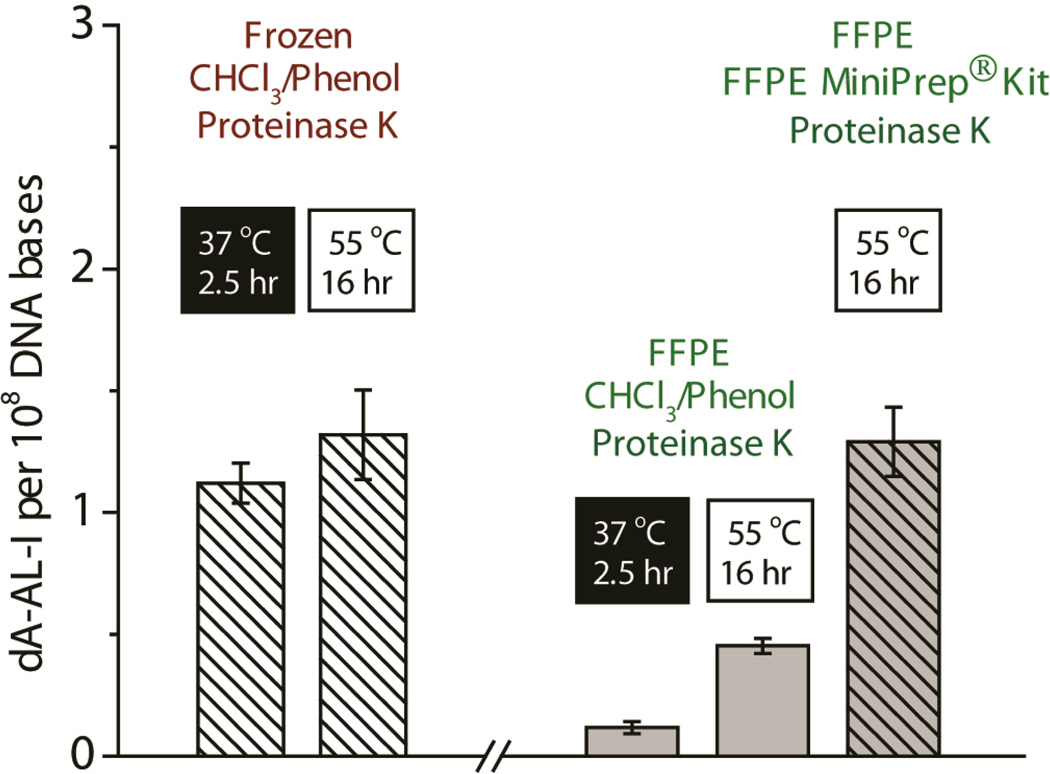
Different temperatures and times of heat treatment were examined to determine the optimal condition for retrieval of AL-DNA adducts from freshly frozen and FFPE liver tissues of mice exposed to AA-I (0.1 mg/kg body weight). The results are depicted in Figure 2. Levels of dA-AL-I adducts were similar in freshly frozen tissue retrieved by the CHCl3/phenol extraction method, irrespective of the length of time or temperature (2.5 h at 37 °C or 16 h at 55 °C) of proteinase K digestion of the nuclear extract. However, the amounts of dA-AL-I recovered from DNA of FFPE tissue processed by the same conditions were much lower. The yield of dA-AL-I adducts was significantly increased when the rehydrated FFPE tissue was processed with the MiniPrep™ kit (Zymo Research), and levels of dA-AL-I adduct were comparable to those measured in freshly frozen tissue (Figure 2).
Figure 2.
The effect of proteolysis and extraction methods on the recovery of dA-AL-I adducts from the freshly frozen and FFPE liver tissue from mice treated with AA-I (0.1 mg/kg body weight). Mean adduct levels are shown (N = 4 and SD). In freshly frozen tissue, high recovery of AL-DNA is achieved by proteinase K treatment of the nuclear pellet at 37 or 55 °C, followed by retrieval of DNA by the CHCl3/phenol method. Both elevated temperature (55 °C) with proteinase K treatment and processing of FFPE tissue with the ZR MiniPrep™ kit are required to efficiently remove crosslinks in DNA and recover dA-AL-I in high yield. One-way ANOVA, P < 0.0001; Dunnett’s multiple comparison post-test reveals that dA-AL-I adduct levels in the nuclear pellet of FFPE tissue heated at 37 or 55 °C with proteinase K and processed with CHCl3/phenol are significantly different from the level of adducts in the nuclear pellet of freshly frozen tissue heated at 37 °C with proteinase K, followed by processing of DNA with CHCl3/phenol (P < 0.01). The level of dA-AL-I in FFPE tissue processed with the ZR MiniPrep™ kit is not significantly different from the level of dA-AL-I in freshly frozen tissue (P > 0.05).
The DNA from freshly frozen tissue samples was completely digested to deoxyribonucleosides, when assayed by HPLC (data not shown). However the DNA of FFPE tissue, processed by the same CHCl3/phenol method, was poorly digested: the amount of deoxyribonucleosides recovered was low, and the DNA was present as large oligomers. The samples also were contaminated with appreciable amounts of RNA, and high levels of guanosine and uracil were detected (Supporting Information, Figure S-1). High levels of RNA contamination were previously detected in DNA isolated from FFPE tissues across different species,42 indicating that RNase inefficiently digests RNA present in the nuclear extract of FFPE tissue processed by heat treatment at 55 °C. However, the DNA of FFPE mouse liver tissue processed with the MiniPrep™ kit was quantitatively digested to deoxyribonucleosides, and there was no contamination with RNA (Supporting Information, Figure S-1). Therefore, proprietary reagents in the MiniPrep™ kit are critical for efficient proteolysis, the quantitative removal of the DNA crosslinks, the efficient enzymatic digestion of DNA, and the recovery of dA-AL-I in high yield from DNA of FFPE tissue. dA-AL-I was stable in the different buffers and digestion conditions employed with proteinase K: there was no evidence of depurination or oxidation of the dA-AL-I adduct (Supporting Information, Figure S-2).
Estimates of AL-DNA Adduct Formation as a Function of Dose in Mouse Liver and Kidney: Freshly Frozen versus FFPE Tissues
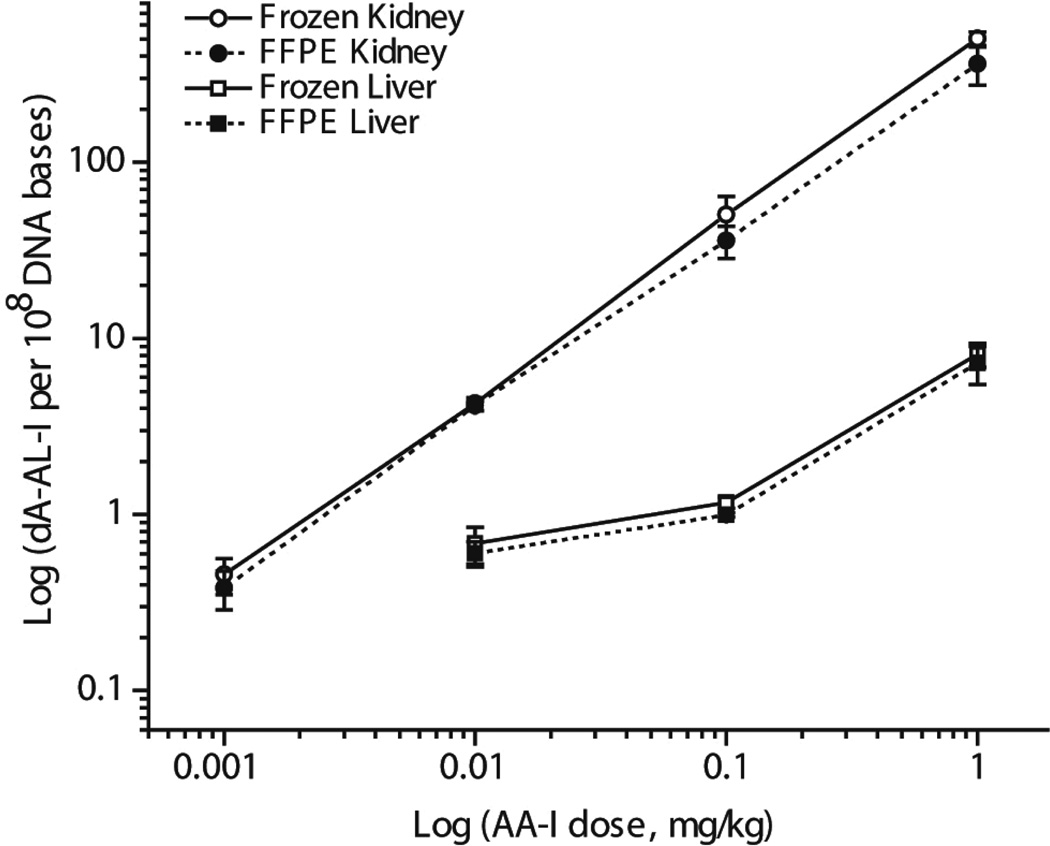
The levels of dA-AL-I adduct formation were measured in animals exposed to AA-I at dosages ranging from 0.001 to 1 mg/kg body weight. The mean level of adducts per group (N = 4 animals per dose, quadruplicate measurements per animal) are plotted as a function of dose (Figure 3). The overall mean difference between dA-AL-I adduct levels in DNA of freshly frozen and FFPE kidney and liver tissues across all doses was 21 ± 14% (mean ± SD). For the most part, the mean and median levels of adducts in freshly frozen and FFPE tissues of the same treatment groups were not statistically significantly different. AL-DNA adduct formation in kidney, a target organ of AA-I,43,44 was 7 to 15-fold greater than the adduct levels formed in liver for all dose treatments. The level of dA-AL-I adduct, at the lowest dose treatment (0.001 mg/kg body wt), in DNA of freshly frozen and FFPE kidney was, respectively, 4.5 ± 0.5 and 3.8 ± 0.5 adducts per 109 DNA bases (mean ± SEM, N = 4 animals per group). These levels of adducts were above the limit of quantification (LOQ),45 which was 3 adducts per 109 DNA bases. Adduct formation in the liver was below the limit of detection (1 adduct per 109 DNA bases) at this dose treatment. Representative mass chromatograms and product ion spectra of the dA-AL-I adduct recovered from a freshly frozen and FFPE kidney tissue of a mouse treated with the lowest dose of AA-I (0.001 mg/kg) are shown in Supporting Information (Figure S-3).
Figure 3.
Mean level of dA-AL-I adducts present in mouse kidney and liver following treatment with AA-I (0.001 to 1 mg/kg body weight). Adduct levels measured in freshly frozen and FFPE mouse kidney (open and filled circle) and liver (open and filled square) (mean adduct level, SD, N = 4 animals per dose, quadruplicate measurements per animal) were plotted as a function of dose. The overall mean difference in adduct levels between freshly frozen and FFEP kidney and liver tissues across all doses was 21 ± 14% (mean ± SD). dA-AL-I adduct formation was below the limit of detection in liver of mice dosed with AA-I at 0.001 mg/kg body weight. Mean levels of dA-AL-I adducts were significantly statistically different between freshly frozen and FFPE kidney or liver at the following dose treatments of AA-I: kidney, 1 mg/kg, P = 0.03; liver, 0.1 mg/kg, P = 0.01, unpaired two-tailed t-test.
Identification and Quantitation of dA-AL-I in Freshly Frozen and FFPE Kidney of Patients with UUC
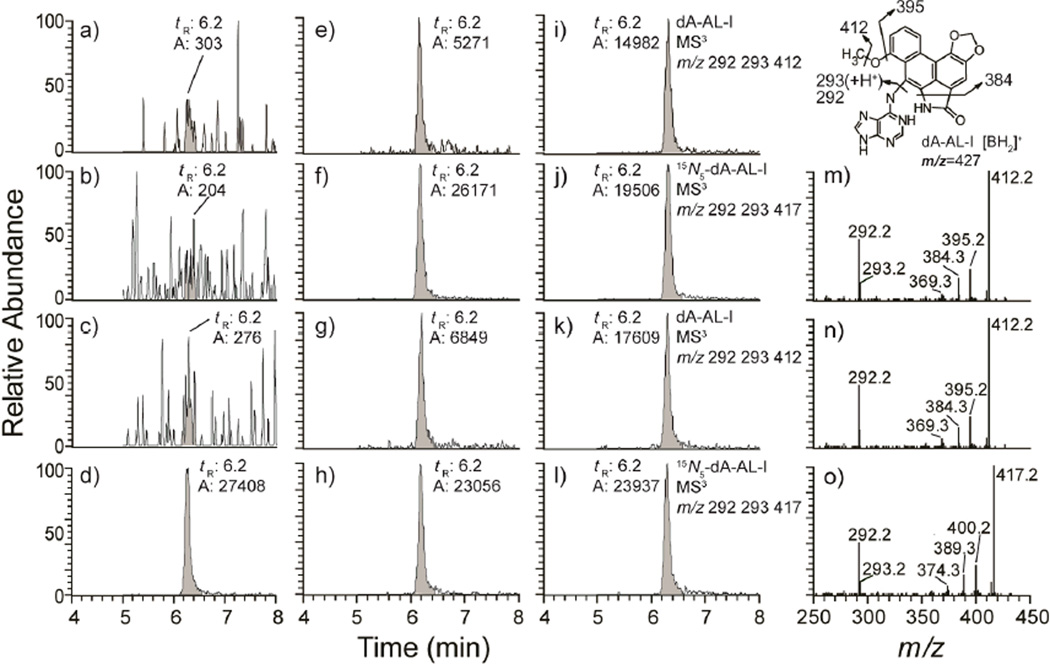
The levels of dA-AL-I adducts present in freshly frozen and FFPE kidney are tabulated in Table 1. The recovery of DNA from 2-combined tissue-section cuts from 5 different subjects ranged between 3.0 and 8.0 µg. These amounts of DNA were assayed for AL-DNA adducts, whereas 5 µg of DNA was assayed for adducts in the freshly frozen tissue sample. Representative mass chromatograms and product ion spectra of the dA-AL-I adducts present in two human kidney specimens, both freshly frozen and FFPE, along with calf thymus DNA which served as the negative control, are shown in Figure 4. The levels of dA-AL-I in DNA from four sets of two 10-µm tissue-section cuts of kidney from subject 1 ranged between 4.5 to 6.3 adducts per 108 bases, whereas the estimate of dA-AL-I in DNA from the freshly frozen biospecimen ranged between 6.6 and 7.2 adducts per 108 bases. These values are similar to the level determined by the 32P-postabeling method,39 at 6.5 adducts per 108 bases in DNA from the same freshly frozen tissue. dA-AL-I adduct levels in freshly frozen tissue samples from subjects 2 and 3 obtained by 32P-postlabeling and UPLC-ESI/MS3, and from FFPE tissue assayed by UPLC-ESI/MS3 are in good agreement. It is worthy of note that 32P-postlabeling failed to detect dA-AL-I in freshly frozen tissue of subjects 4 and 5; the levels were below the limit of detection by this method (~ 1 adduct per 108 bases). However, dA-AL-I was detected by UPLC-ESI/MS3. These findings are consistent with our previous results, which showed UPLC-ESI/MS3 to be several-fold more sensitive than 32P-postlabeling for monitoring dA-AL-I.31 Thus, biomonitoring of dA-AL-I adducts in freshly frozen and FFPE tissues by UPLC-ESI/MS3 , using our optimized conditions of DNA isolation, is highly robust.
Table 1.
dA-AL-I adduct levels in human freshly frozen and FFPE kidney tissue
| Subject | Tissue |
32P-postlabeling dA-AL-I / 108 bases |
LC-MS/MS3 dA-AL-I /108 bases |
Surface area of tissue in block (cm2) |
DNA yield, µg per 2 FFPE 10 µm-section cuts |
|---|---|---|---|---|---|
| 1 | FR | 6.5 | 7.0 ± 0.2a | - | |
| FFPE | n.a. | 6.1 | 1.8 | 3.0 | |
| FFPE | n.a. | 4.6 | 1.8 | 4.8 | |
| FFPE | n.a. | 4.7 | 1.8 | 4.0 | |
| FFPE | n.a. | 6.2 | 1.8 | 7.2 | |
| 2 | FR | 3.2 | 2.7 ± 0.1b | - | |
| FFPE | n.a. | 1.8 | 1.6 | 2.5 | |
| 3 | FR | 2.2 | 3.9 ± 0.1b | - | |
| FFPE | n.a. | 3.8 | 1.5 | 7.5 | |
| FFPE | n.a. | 3.7 | 1.5 | 8.0 | |
| 4 | FR | n.d. | 1.0 ± 0.1b | - | |
| FFPE | n.a. | 1.5 | 0.9 | 5.3 | |
| FFPE | n.a. | 1.6 | 0.9 | 4.9 | |
| 5 | FR | n.d. | 0.13 ± 0.02b,c | - | |
| FFPE | n.a. | 0.48 | 1.0 | 3.3 | |
Independent triplicate measurements;
independent duplicate measurements; other samples were measured in singlicate;
above the limit of detection (~1 adduct per 109 bases by UPLC-ESI-MS/MS3);
n.a. = not analyzed; n.d. = not detected, below the limit of detection of ~1 adduct per 108 bases by 32P-postlabeling; FR= freshly frozen tissue; FFPE = formalin fixed paraffin embedded tissue.
Figure 4.
Representative mass chromatograms at the MS3 scan stage and product ion spectra of dA-AL-I in freshly frozen and FFPE human kidney DNA (5 µg) spiked with 15N5-labeled internal standard at a level of 5 adducts in 108 bases (d, f, h, j, and l). The chromatograms were reconstructed from the extracted ions of dA-AL-I (MS3 at m/z 292, 293, 412) and [15N5]-dA-AL-I (MS3 at m/z 292, 293, 417). Three untreated calf thymus DNA samples served as the negative controls (a–c); freshly frozen human kidney tissue (e, i) and FFPE kidney tissue (g, k) from 2 human patients with UUC. Product ion spectra were acquired at MS3 scan stage for dA-AL-I in frozen (m) and FFPE tissue (n), and for the internal standard [15N5]-dA-AL-I (o). The structure of the aglycone [BH2]+ adduct of dA-AL-I and proposed mechanisms of fragmentation of the adduct at the MS3 scan stage are shown in the top right corner.
Potential and Limitations of DNA Adduct Biomarkers in FFPE Tissue for Molecular Epidemiology Studies
The ability to examine, retrospectively, FFPE tissue for which there is clinical diagnosis of disease opens a previously untapped source of biospecimens for measurement of DNA adducts in molecular epidemiology studies that seek to assess the causal role of environmental carcinogens. The usage of FFPE tissue represents an important advance in DNA adducts biomarker research as freshly frozen tissues are often unavailable for analysis. Our findings on the measurement of AL-DNA adducts in FFPE tissue are highly promising; however, further validation of the FFPE method is required before it can be routinely employed in molecular epidemiology studies. For example, the temperature and/or duration of formalin fixation can adversely affect the quality of DNA and up to 30% of nucleic acids may be lost during fixation.8 Therefore, the ability to quantitatively recover DNA and measure carcinogen DNA adducts in tissues stored in formalin under variable temperature and/or time prior to paraffin embedding must be shown. In our study, a time period of 24 h was used for formalin fixation of tissues, because 24 h is an optimal period of tissue preservation for histological and immunohistochemical analyses.6,7 However, the time interval between tissue procurement and formalin fixation may vary, and different temperatures and times may be employed for formalin fixation in the clinical setting. Thus, a prolonged time period of formalin fixation may adversely affect the recovery of high quality, fully digestible DNA, thereby impacting the reliability of measurement of carcinogen DNA adducts. Our preliminary data reveal that the extraction method can recover AL-DNA adducts in high yield from mouse liver and kidney tissue preserved with formalin for different times up to 7 days, prior to paraffin embedding (unpublished observations, BH Yun).
Biomonitoring DNA Adducts in Human Tissues: 32P-postlabeling versus Mass Spectrometry
Historically, many studies involving the biomonitoring of DNA adducts employed various modifications of the original 32P-postlabeling method,46,47 including measurements of AL-DNA adducts.22–24,29,30 Postlabeling methods are highly sensitive but suffer from several drawbacks.31 The labeling efficiencies of DNA adducts are highly variable,48 which leads to uncertain estimates of adduct levels. Furthermore, postlabeling methods fail to provide spectral data, and the identities of adducts depend on co-chromatography with standards. Thus, mass spectrometry is the sole method that can be employed in molecular epidemiology studies to quantify DNA adducts, while providing spectral data to support the identity of the lesion. Current MS instruments have the requisite sensitivity to measure various DNA adducts in human specimens at levels below 1 adduct per 108 bases, a level of sensitivity that is comparable to 32P-postlabeling methods.38,49,50 The linear quadrupole ion trap MS employed in our study is particularly powerful, since product ion mass spectra of the aglycone adducts [BH2]+ acquired at the MS3 scan stage provide rich structural information about the structure of the adduct and corroborate the identity of the lesion (Figure 4).31,37
Biological Significance of DNA Adducts
The identification of DNA adducts in clinical samples is frequently due to recent exposures, and the interpretation of negative findings must be done with caution. Ideally, biomonitoring of DNA adducts should be more relevant when measured earlier in time, when the multistage process of malignant tumor formation and progression has begun, rather than many years later when the cancer has been diagnosed. However, tobacco smoking, diet, and environmental pollution often represent long-term exposures, and current adduct levels of carcinogens from these exposures are likely to correlate with adduct levels that existed during the time of tumor formation and progression. In the Balkans, dietary exposure to AA occurred through daily ingestion of bread prepared from flour derived from contaminated grain, whereas in Asia, the exposure to AA occurs through herbal remedies containing Aristolochia sp.51 AA is unusual among many environmental carcinogens in its persistence: the dA-AL-I adduct is resistant to global genomic repair,52 and the adduct can be detected decades after ingestion of Aristolochia herbs has ceased.22,29–31 Thus, the measurement of dA-AL-I in FFPE kidney likely represents an accumulation of long-term chronic exposure to AA-I.
Conclusions
Our study represents an important advance in the application of DNA adduct biomarkers for human cancer research. The high level of sensitivity provided by multistage scan mass spectrometry, employing UPLC-ESI/MS3, has allowed us to reliably quantify AL-DNA adducts in rodents exposed to AA-I at very low single dosage of 1 µg /kg body weight, and also in kidney tissue of humans exposed to AA-I in their diet. A second major advance is the ability to assay for AL-DNA adducts in archived FFPE specimens. Our method of DNA retrieval from archival FFPE tissues efficiently removes formalin-induced crosslinks, producing DNA that is fully digestible and amenable for quantitative measurement of AL-DNA adducts. The dA-AL-I adduct can be measured in FFPE tissue at a level of sensitivity that is comparable to freshly frozen tissue, using only 2.5 – 5 µg DNA for mass spectrometric analysis. To our knowledge, this study is the first report on the identification and quantitation of a carcinogen DNA adduct, by mass spectrometry, in human FFPE tissue.
Traditional Chinese medicines, some of which may still contain AA, are used worldwide, most commonly in Asia.51,53 There has also been inadvertent dietary exposure to AA in certain parts of the world, such as in endemic nephropathy in the Balkans,51 or in a cluster of cases in Belgian, where Aristolochia herbs were used as part of a slimming regime.54 Indeed, a recent commentary55 reinforced the notion that AA-associated cancer may have global proportions.51 Given the widespread use of Aristolochia sp. in traditional herbal remedies, all of which contain AA-I, the biomonitoring of dA-AL-I adducts can be used to identify exposure to AA-I and to assess the etiologic role of this toxin in chronic renal disease and/or UUC world-wide.22–24,29 The use of FFPE tissue as a biospecimen for biomonitoring DNA adducts of other environmental, dietary, and endogenous genotoxicants, by mass spectrometric techniques, warrants investigation. The identification of carcinogen DNA adducts in FFPE tissue may provide clues to the origin of human cancers for which an environmental cause is suspected.
Supplementary Material
Acknowledgements
Technical support provided by the Stony Brook University Research Histology Core Laboratory is gratefully acknowledged. The technical assistance of Mr. Steven Wong, Senior Research Associate, Zymo Research Corporation (Irvine, CA), is greatly appreciated.
Funding: This research was supported by grants R01 ES019564 (to R.J.T.) and ES04068 (to A.P.G.) from the National Institute of Environmental Health Sciences
Footnotes
Conflict of Interest Disclosure. “The authors declare no competing financial interest.”
Reference List
- 1.Miller JA. Cancer Res. 1970;30:559–576. [PubMed] [Google Scholar]
- 2.Miller EC. Cancer Res. 1978;38:1479–1496. [PubMed] [Google Scholar]
- 3.Himmelstein MW, Boogaard PJ, Cadet J, Farmer PB, Kim JH, Martin EA, Persaud R, Shuker DE. Crit Rev. Toxicol. 2009;39:679–694. doi: 10.1080/10408440903164163. [DOI] [PubMed] [Google Scholar]
- 4.Jarabek AM, Pottenger LH, Andrews LS, Casciano D, Embry MR, Kim JH, Preston RJ, Reddy MV, Schoeny R, Shuker D, Skare J, Swenberg J, Williams GM, Zeiger E. Crit. Rev. Toxicol. 2009;39:659–678. doi: 10.1080/10408440903164155. [DOI] [PubMed] [Google Scholar]
- 5.Loeb LA, Harris CC. Cancer Res. 2008;68:6863–6872. doi: 10.1158/0008-5472.CAN-08-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayat MA. Microscopy, Immunohistochemistry, and Antigen Retrieval Methods for Light and Electron Microscopy. New York: Kluwer Academic / Plenum Publishers; 2002. [Google Scholar]
- 7.Nirmalan NJ, Harnden P, Selby PJ, Banks RE. Mol. Biosyst. 2008;4:712–720. doi: 10.1039/b800098k. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasan M, Sedmak D, Jewell S. Am. J. Pathol. 2002;161:1961–1971. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valera VA, Walter BA, Linehan WM, Roberts DD, Merino MJ. Current Proteomics. 2009;6:122–139. [Google Scholar]
- 10.Shi SR, Cote RJ, Wu L, Liu C, Datar R, Shi Y, Liu D, Lim H, Taylor CR. J. Histochem. Cytochem. 2002;50:1005–1011. doi: 10.1177/002215540205000802. [DOI] [PubMed] [Google Scholar]
- 11.Shi SR, Datar R, Liu C, Wu L, Zhang Z, Cote RJ, Taylor CR. Histochem. Cell Biol. 2004;122:211–218. doi: 10.1007/s00418-004-0693-x. [DOI] [PubMed] [Google Scholar]
- 12.Goelz SE, Hamilton SR, Vogelstein B. Biochem. Biophys. Res. Commun. 1985;130:118–126. doi: 10.1016/0006-291x(85)90390-0. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert MT, Haselkorn T, Bunce M, Sanchez JJ, Lucas SB, Jewell LD, Van ME, Worobey M. PLoS. One. 2007;2:e537. doi: 10.1371/journal.pone.0000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hood BL, Conrads TP, Veenstra TD. Proteomics. 2006;6:4106–4114. doi: 10.1002/pmic.200600016. [DOI] [PubMed] [Google Scholar]
- 15.Santella RM. Cancer Epidemiol. Biomarkers Prev. 1999;8:733–739. [PubMed] [Google Scholar]
- 16.Poirier MC, Santella RM, Weston A. Carcinogenesis. 2000;21:353–359. doi: 10.1093/carcin/21.3.353. [DOI] [PubMed] [Google Scholar]
- 17.Zhu J, Chang P, Bondy ML, Sahin AA, Singletary SE, Takahashi S, Shirai T, Li D. Cancer Epidemiol. Biomarkers Prev. 2003;12:830–837. [PubMed] [Google Scholar]
- 18.Faraglia B, Chen SY, Gammon MD, Zhang Y, Teitelbaum SL, Neugut AI, Ahsan H, Garbowski GC, Hibshoosh H, Lin D, Kadlubar FF, Santella RM. Carcinogenesis. 2003;24:719–725. doi: 10.1093/carcin/bgg013. [DOI] [PubMed] [Google Scholar]
- 19.Johnson F, Huang CY, Yu PL. Environ. Health Perspect. 1994;102(Suppl 6):143–149. doi: 10.1289/ehp.94102s6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindahl T. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 21.Hewer A, Phillips DH. IARC Sci. Publ. 1993:211–214. [PubMed] [Google Scholar]
- 22.Grollman AP, Shibutani S, Moriya M, Miller F, Wu L, Moll U, Suzuki N, Fernandes A, Rosenquist T, Medverec Z, Jakovina K, Brdar B, Slade N, Turesky RJ, Goodenough AK, Rieger R, Vukelic M, Jelakovic B. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CH, Dickman KG, Moriya M, Zavadil J, Sidorenko VS, Edwards KL, Gnatenko DV, Wu L, Turesky RJ, Wu XR, Pu YS, Grollman AP. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8241–8246. doi: 10.1073/pnas.1119920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jelakovic B, Karanovic S, Vukovic-Lela I, Miller F, Edwards KL, Nikolic J, Tomic K, Slade N, Brdar B, Turesky RJ, Stipancic Z, Dittrich D, Grollman AP, Dickman KG. Kidney Int. 2012;81:559–567. doi: 10.1038/ki.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanherweghem JL, Debelle FD, Muniz Martinez MC, Nortier JL. In: Clinical Nephrotoxins. 2nd Edition. De Broe ME, Porter GA, Bennett WM, Verpooten GA, editors. Dordrecht: Kluwer; 2003. [Google Scholar]
- 26.Pfau W, Schmeiser HH, Wiessler M. Carcinogenesis. 1990;11:1627–1633. doi: 10.1093/carcin/11.9.1627. [DOI] [PubMed] [Google Scholar]
- 27.Pfau W, Schmeiser HH, Wiessler M. Carcinogenesis. 1990;11:313–319. doi: 10.1093/carcin/11.2.313. [DOI] [PubMed] [Google Scholar]
- 28.Arlt VM, Stiborova M, vom BJ, Simoes ML, Lord GM, Nortier JL, Hollstein M, Phillips DH, Schmeiser HH. Carcinogenesis. 2007;28:2253–2261. doi: 10.1093/carcin/bgm082. [DOI] [PubMed] [Google Scholar]
- 29.Schmeiser HH, Bieler CA, Wiessler M, van Ypersele De SC, Cosyns JP. Cancer Res. 1996;56:2025–2028. [PubMed] [Google Scholar]
- 30.Bieler CA, Stiborova M, Wiessler M, Cosyns JP, van Ypersele De SC, Schmeiser HH. Carcinogenesis. 1997;18:1063–1067. doi: 10.1093/carcin/18.5.1063. [DOI] [PubMed] [Google Scholar]
- 31.Yun BH, Rosenquist TA, Sidorenko V, Iden CR, Chen CH, Pu YS, Bonala R, Johnson F, Dickman KG, Grollman AP, Turesky RJ. Chem. Res. Toxicol. 2012;25:1119–1131. doi: 10.1021/tx3000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nedelko T, Arlt VM, Phillips DH, Hollstein M. Int. J. Cancer. 2009;124:987–990. doi: 10.1002/ijc.24006. [DOI] [PubMed] [Google Scholar]
- 33.Schmeiser HH, Janssen JW, Lyons J, Scherf HR, Pfau W, Buchmann A, Bartram CR, Wiessler M. Cancer Res. 1990;50:5464–5469. [PubMed] [Google Scholar]
- 34.Attaluri S, Bonala RR, Yang IY, Lukin MA, Wen Y, Grollman AP, Moriya M, Iden CR, Johnson F. Nucleic Acids Res. 2010;38:339–352. doi: 10.1093/nar/gkp815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmeiser HH, Frei E, Wiessler M, Stiborova M. Carcinogenesis. 1997;18:1055–1062. doi: 10.1093/carcin/18.5.1055. [DOI] [PubMed] [Google Scholar]
- 36.Gupta RC. IARC Sci. Publ. 1993;124:11–23. [PubMed] [Google Scholar]
- 37.Goodenough AK, Schut HA, Turesky RJ. Chem. Res. Toxicol. 2007;20:263–276. doi: 10.1021/tx0601713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu D, Turesky RJ, Tao Y, Langouet SA, Nauwelaers GC, Yuan JM, Yee D, Yu MC. Carcinogenesis. 2012;33:124–130. doi: 10.1093/carcin/bgr252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong H, Suzuki N, Torres MC, Bonala RR, Johnson F, Grollman AP, Shibutani S. Drug Metab Dispos. 2006;34:1122–1127. doi: 10.1124/dmd.105.008706. [DOI] [PubMed] [Google Scholar]
- 40.Shibutani S, Kim SY, Suzuki N. In: DNA Repair Protocols: Eukaryotic systems. second edition. Henderson DS, editor. Totawa, NJ: Humana Press Inc.; 2005. [Google Scholar]
- 41.Holley T, Lenkiewicz E, Evers L, Tembe W, Ruiz C, Gsponer JR, Rentsch CA, Bubendorf L, Stapleton M, Amorese D, Legendre C, Cunliffe HE, McCullough AE, Pockaj B, Craig D, Carpten J, Von HD, Iacobuzio-Donahue C, Barrett MT. PLoS. One. 2012;7:e50586. doi: 10.1371/journal.pone.0050586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmermann J, Hajibabaei M, Blackburn DC, Hanken J, Cantin E, Posfai J, Evans TC., Jr Front Zool. 2008;5:18. doi: 10.1186/1742-9994-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shibutani S, Dong H, Suzuki N, Ueda S, Miller F, Grollman AP. Drug Metab Dispos. 2007;35:1217–1222. doi: 10.1124/dmd.107.014688. [DOI] [PubMed] [Google Scholar]
- 44.Arlt VM, Levova K, Barta F, Shi Z, Evans JD, Frei E, Schmeiser HH, Nebert DW, Phillips DH, Stiborova M. Chem. Res. Toxicol. 2011;24:1710–1719. doi: 10.1021/tx200259y. [DOI] [PubMed] [Google Scholar]
- 45.MacDougall D, Amore FJ, Cox GV, Crosby DG, Estes FL, Freeman DH, Gibbs WE, Gordon GE, Keith LH, Lal J, Langner RR, McClelland NI, Phillips WF, Pojasek RB, Sievers RE. Anal. Chem. 1980;52:2242–2249. [Google Scholar]
- 46.Randerath K, Reddy MV, Gupta RC. Proc. Natl. Acad. Sci U. S. A. 1981;78:6162–6129. doi: 10.1073/pnas.78.10.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phillips DH. Mutat. Res. 2005;577:284–292. doi: 10.1016/j.mrfmmm.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 48.Phillips DH, Farmer PB, Beland FA, Nath RG, Poirier MC, Reddy MV, Turteltaub KW. Environ Mol. Mutagen. 2000;35:222–233. doi: 10.1002/(sici)1098-2280(2000)35:3<222::aid-em9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 49.Singh R, Farmer PB. Carcinogenesis. 2006;27:178–196. doi: 10.1093/carcin/bgi260. [DOI] [PubMed] [Google Scholar]
- 50.Tretyakova N, Goggin M, Sangaraju D, Janis G. Chem Res. Toxicol. 2012;25:2007–2035. doi: 10.1021/tx3002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grollman AP. Environ. Mol. Mutagen. 2012 doi: 10.1002/em.21756. In Press. [DOI] [PubMed] [Google Scholar]
- 52.Sidorenko VS, Yeo JE, Bonala RR, Johnson F, Scharer OD, Grollman AP. Nucleic Acids Res. 2012;40:2494–2505. doi: 10.1093/nar/gkr1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Debelle FD, Vanherweghem JL, Nortier JL. Kidney Int. 2008;74:158–169. doi: 10.1038/ki.2008.129. [DOI] [PubMed] [Google Scholar]
- 54.Nortier JL, Martinez MC, Schmeiser HH, Arlt VM, Bieler CA, Petein M, Depierreux MF, De PL, Abramowicz D, Vereerstraeten P, Vanherweghem JL. N. Engl. J Med. 2000;342:1686–1692. doi: 10.1056/NEJM200006083422301. [DOI] [PubMed] [Google Scholar]
- 55.Olivier M, Hollstein M, Schmeiser HH, Straif K, Wild CP. Nat. Rev. Cancer. 2012;12:503–504. doi: 10.1038/nrc3311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.