Abstract
KSHV establishes characteristic latent infections in vitro, while RRV, a related macaque rhadinovirus, establishes characteristic permissive infections with virus replication. We identified cells that are not permissive for RRV replication and recapitulate the latent KSHV infection and reactivation processes. The RRV replication and transactivator (Rta) promoter was characterized in permissive and non-permissive cells and compared to the KSHV Rta promoter. Both promoters contained a critical Sp1 element, had equivalent activities in different cell types, and were inhibited by LANA. RRV and KSHV infections were non-permissive in cells with low Rta promoter activity. While RRV infections were permissive in cells with high basal promoter activity, KSHV infections remained non-permissive. Our studies suggest that RRV lacks the Rta-inducible LANA promoter that is responsible for LANA inhibition of the KSHV Rta promoter and induction of latency during KSHV infection. Instead, the outcome of RRV infection is determined by host factors, such as Spl.
Keywords: RRV, KSHV, Rta, replication, transactivator, promoter, Spl, macaque, rhadinovirus
Introduction
The human rhadinovirus Kaposi’s sarcoma herpesvirus (KSHV) has been associated with all forms of Kaposi’s sarcoma (KS) and two AIDS-related lymphoproliferative disorders (10, 57). In KS tumors in vivo and infected cultures in vitro the vast majority of cells are latently infected, with only a small percentage of cells undergoing active replication. Our understanding of the replicative program of infection is limited because a natural permissive system has not been identified for KSHV. In order to achieve high levels of replication in vitro, latent KSHV is commonly reactivated by treatment with histone deacetylase (HDAC) inhibitors like sodium butyrate or phorbol esters such as TPA. KSHV replication can also be induced by overexpression of the viral replication and transactivator (Rta), encoded by open reading frame (ORF) 50. Rta is the only viral gene that is necessary and sufficient for virus replication (42, 64). Because of its central role in the reactivation of KSHV, Rta is considered to be the master regulator of reactivation and its function has been extensively studied (23, 43, 64). Rta activates viral promoters through both direct DNA binding and interactions with cellular transcription factors (41, 56, 62). Numerous regulatory elements have been identified in the Rta promoter, including binding sites for the cellular transcription factors Octl, C/EBP, Spl, Apl, XBP-1, and YY1 (9, 39, 56, 67, 68, 70, 74, 75). Furthermore, the KSHV latency-associated nuclear antigen (LANA) and the virally encoded microRNAs negatively regulate Rta expression to promote the establishment and maintenance of latency (4, 35). Analysis of gene expression in latently infected cells following butyrate treatment demonstrated that Rta is one of the first genes induced after biochemical reactivation, with expression occurring within four hours (73). Studies have found that butyrate directly induces the KSHV Rta promoter, and the butyrate-responsive element has been mapped to an Spl-binding site 103 to 112 nucleotides upstream of the translational start site (39, 74).
Two studies of early gene expression kinetics during KSHV infection have shed light on the events that lead to the establishment of latency in infected cells. Both studies reported a discrete but transient increase in Rta expression early after infection (31, 36). Early Rta expression is thought to be regulated by the pre-existing balance of host factors present in the infected cell, as has been described for the transactivator genes of other herpesviruses (22). Additionally, Rta has been identified as a component of the KSHV virion, and the early surge in Rta expression may, in part, be due to promoter auto-activation by virion-associated Rta (3, 27). Instead of inducing active viral replication, however, Rta levels decline rapidly after KSHV infection and LANA levels steadily increase, peaking at 24 hours post-infection. At this time point LANA protein has readily accumulated in the nuclei of infected cells and viral latency is established. The steep decline in Rta levels after KSHV infection is thought to be a result of the inhibitory effects of LANA as it accumulates in the infected cell (35).
The early spike in Rta levels after infection prompted investigation into the role of Rta in the establishment of KSHV latency. A series of recent reports demonstrated that KSHV Rta induces LANA expression early in infection through an Rta-responsive promoter (LANApi) and that Rta-mediated activation of LANApi is critical for the establishment of KSHV latency (36, 40). LANApi is located in the intron region that is excised during maturation of the latent transcript and is distinct from the constitutive promoter (LANApc) that regulates LANA expression in latently infected cells (18, 45, 51, 58, 65). KSHV Rta activates LANApi through interaction with the cellular transcription factor RBP-Jkappa (RBP-Jk), a downstream effector of the Notch signaling pathway (49). Rta and RBP-Jk form a complex at LANApi, and DNA binding is mediated by an Rta-responsive element (RRE) and two RBP-Jk binding sites in the promoter region (36). Deletion of either RBP-Jk binding site abolishes LANApi activity and results in a diminished ability of the KSHV to establish latency (28, 40). These studies suggest that LANA and Rta together comprise a regulatory loop early in KSHV infection that leads to the establishment of viral latency: early expression of Rta activates the LANApi promoter and induces high-level expression of LANA. LANA shuts off Rta expression through transcriptional repression, ultimately resulting in the establishment of latency in the infected cell.
Two distinct lineages of rhadinoviruses have been identified in Old World primate species (25, 59). The RV1 rhadinovirus lineage consists KSHV and its close evolutionary homologs from different non-human primate species (24, 25, 32, 55). The RV2 rhadinovirus lineage is evolutionarily distinct from KSHV and the RV1 homologs. Members of the RV2 lineage have been detected in the same non-human primate species that are host to the RV1 rhadinoviruses (6,14, 25, 33, 34) and individual animals can be co-infected with both RV1 and RV2 viruses (59). Though RV2-lineage viruses have been found in numerous Old World primate species, an RV2-lineage rhadinovirus has not yet been identified in humans. Two variants of the rhesus rhadinovirus (RRV), the prototypical RV2-lineage virus, have been identified and full genome sequencing identified few strain differences. A comparison with the genome of KSHV showed that the RRV was highly homologous and co-linear with KSHV, although several differences were noted (14, 60). Major differences include the lack of the K3 and K5 immune evasion genes that are believed to be important for maintaining KSHV latency and the expansion of the viral interferon regulator factor (IRF) genes that control the host immune response to RRV infection (1, 54).
In vitro, tissue culture cells are permissive for RRV replication and produce high titers of infectious virus (14,17). In vivo, RRV is associated with a distinct group of T-cell and B-cell lymphomas in macaques that express markers of viral replication and contain high levels of RRV DNA (7). Because of this replicative phenotype, RRV represents a useful system to study the determinants of rhadinovirus replication. RRV encodes an Rta homolog that has considerable homology to KSHV Rta across functional domains including the DNA binding region (38). Studies have confirmed that RRV Rta is expressed early in infection (5,12,19, 38) and functions as a potent transactivator of viral promoters (12,19, 38). While ectopic expression of RRV LANA has been shown to decrease virus replication in permissive cells, no direct inhibitory effects of LANA on the RRV Rta promoter have been detected (15). The reasons that RRV and KSHV establish different programs of infection are not completely understood.
We have previously characterized RRV replication in rhesus primary fetal fibroblast (RPFF) and Vero cells using RV2-specific assays, and have shown that both cell lines are permissive for RRV infection, with lytic gene expression and production of infectious virus (5). Here, we have identified a novel natural infection system to study RRV latency in two orogastric epithelial cell lines that are non-permissive for RRV replication. We demonstrate that the RRV Rta promoter is highly active in cells that are permissive for virus replication, but only minimally active in non-permissive cells, in which RRV infection is latent. As in KSHV, an Sp1 response element was identified in the RRV Rta promoter that was critical for high-level Rta promoter activity. We further show that ectopic expression of RRV LANA inhibited Rta promoter activity and viral replication, as has been shown for KSHV LANA. However, RRV lacks a conserved homolog of the LANApi promoter required for the establishment of latency in KSHV, suggesting that the differential permissivity observed in RRV and KSHV infections may be due to differences in the induction and expression of LANA, rather than differences in Rta function. Our study demonstrates that RRV Rta promoter activity strongly correlates with cellular permissivity for viral replication and suggests an important role for host cell factors, such as Spl, in determining whether RRV infection results in latency or virus replication.
Materials and Methods
Plasmids
The pGL2-R-Rta plasmid containing 502 nucleotides of the RRV Rta promoter sequence was a gift from Dr. B. Damania. Truncated fragments of the Rta promoter were prepared from pGL2-R-Rta and inserted upstream of the firefly luciferase reporter gene in pGL2 basic (Promega) using Kpnl and Sacl restriction sites and the following forward primes: 487 (5’ ATAAGGTACCTCGCGTGATCTTTT 3’), 443 (5’ ATATGGTACCTAACAAACCTCACTCCCTGTAA 3’), 414 (5’ ATATGGTACCATAAGGTCCGTTCTTTCTATC 3’), 385 (5’ ATATGGTACWGAATCTATAGTTACATCTTTAAG 3’), 321 (5’ ATAAGGTACCTTAAAAAATCGCAAAAGCGAC 3’) 299 (5’ ATAAGGTACCGATGGCTCTATCCGCGTT 3’), 263 (5’ ATAAGGTACCTAGTCACGATGGATCTCCAGT 3’), 216 (5’ AATAGGTACCTTAACTGGAATGGAAACAGC 3’) 187 (5’ ATATGGTACCGTGAACTTCCTGATGTCTCCTA 3’), 153 (5’ ATAAGGTACCAAACAGAGCTAAATACCAATGAC, 136 (5’ AATAGGTACCAATGACTGTCACCCCTACCC 3’), 102 (5’ ATAAGGTACCGTACTATTAGACCAGGGGTGAG 3’), 73 (5’ ACTTGGTACCCTATCCTTTAAAAACCCATACG 3’). The same reverse primer was used for all RRV Rta promoter clones: (5’ ACTTGAGCTCTTTATGACAGGCGTC 3’). Mutagenesis of the Sp1 site at nucleotide −113 in pGL2-R-Rta was done by site directed mutagenesis (Mutagenex). pGL2-K-Rta, containing the KSHV Rta promoter upstream of the luciferase reporter gene, was constructed by amplifying the promoter sequence from KSHV-infected BCBL-1 cells using the following primers: forward (5’ ATTAGAGCTCGCTGTTGCCTGGCATTTTGC 3’) and reverse (5’ ATTAACGCGTTTTTGTGGCTGCCTGGACAG 3’). The KSHV Rta promoter sequence was inserted into pGL2 using the Sacl and Mlul sites. All constructs were verified by sequencing. The pRL-CMV plasmid, containing the CMV IE promoter driving a Renilla reporter gene, was obtained from Promega.
pCDNA N-Flag RRV LANA, containing the RRV 17577 strain ORF73 coding sequence with an N-terminal Flag tag, was constructed by inserting the RRV LANA sequence amplified from pRRV73GFP (8) into pCDNA3 using Hindlll and Xbal sites and the following primers: forward (5’ TAGCAAGCTTGCCACCATGGATTACAAGGATGACGACGATAAGTGGGGCAGCCGGCAA 3’, Flag coding sequence underlined) and reverse (5’ GCTATCTAGATTAGTGCTGAATTGGTAGTCCTCTG 3’). The pCDNA N-Flag KSHV LANA expression plasmid was constructed by ligating an insert with a Flag tag and the fourteen N-terminal amino acids of KSHV LANA (up to the Ascl site in the LANA coding sequence) and an insert containing the remaining 989 C-terminal amino acids (downstream of the Ascl site) into the pCDNA3 vector. Briefly, overlapping oligos encoding an initiating methionine, Flag sequence and the first fourteen amino acids of KSHV LANA were denatured at 95°C for 4 minutes in 100 mM potassium acetate, 2 mM magnesium acetate, and 20 mM HEPES and then annealed at 70°C for 10 minutes. The oligos were designed such that the annealed oligo insert had Kpnl and Ascl overhangs. The KSHV LANA coding sequence (minus the fourteen N terminal amino acids) was excised from pCDNA3.1 V5/HISA orf73 (53) with Ascl and Xhol. Both inserts were subcloned into pUC19 using Kpnl and Xho sites. The resulting pUC19/NFlag KSHV LANA plasmid was confirmed by diagnostic digest. The entire N-Flag KSHV LANA sequence was then excised and cloned into pCDNA3 using Kpnl and Xhol sites. LANA expression constructs were verified by sequencing.
Cell and virus stocks
Human salivary gland (HSG) epithelial cells (61) were a kind gift of Dr. K. Izutsu. Gastric adenocarcinoma (AGS) epithelial cells (52) were provided by Dr. N. Salama. Rhesus primary fetal fibroblast (RPFF) cells and RRV strain 17577 were kindly provided by Drs. M. Axthelm and S. Wong. HEK293 human embryonic kidney cells and African green monkey kidney epithelial (Vero) cells were obtained from the ATCC. All cells were maitained in Dulbecco’s modified Eagle’s medium supplemented with 10% cosmic calf serum, 1% penicillin-streptomycin and 1% HEPES. RRV virions were harvested by ultracentrifugation of supernatants from infected RPFF cell cultures as previously described (5). KSHV virions were gradient purified from filtered culture supernatants of TPA-treated BCBL-1 cells, as described (21).
Immunological reagents
The KSHV LANA rat monoclonal antibody (clone LN53) and ORF59 mouse monoclonal antibody were obtained from Advanced Biotechnologies Inc (ABI). The 425 rabbit polyclonal anti-RV2 ORF59 antisera was prepared by immunizing a rabbit with the coding sequences for amino acids 300–388 of ORF59 of the RRV (17577) and MneRV2 (442N), as described previously (5). M2 anti-Flag antibody (Sigma), Alexa488-conjugated goat anti-rabbit secondary antibody and Alexa594-conjugated goat anti-mouse secondary antibody (Molecular Probes), horse-radish peroxidase (HRP)-coupled goat anti-rat IgG (Biosource) and normal goat serum (Jackson) were purchased.
Immunofluorescence assays
To detect the lytic gene cascade characteristic of a permissive RRV infection, we developed an immunofluorescence assay to detect the nuclear accumulation of the early lytic gene ORF59, the DNA polymerase processivity factor, using the 425 rabbit anti-RV2 ORF59 antiserum, which is reactive with RRV ORF59, as described previously (5). ORF59 expression was visualized using an Alexa488-conjugated goat anti-rabbit secondary antibody (Molecular Probes). Flag-tagged RRV LANA expression was visualized using the M2 anti-Flag antibody (Sigma) and Alexa594-conjugated goat anti-mouse secondary antibody. KSHV LANA was visualized using the LN53 rat anti-LANA antibody and goat anti-rat IgG-coupled to HRP, as described (21). KSHV ORF59 was visualized using the monoclonal antibody reactive with KSHV ORF59. Cell nuclei were stained with TO-PRO. Images were acquired with a Zeiss LSM 5 Pascal confocal microscope.
Virus Infection
For RRV infections, cells were plated onto 17 mm spots, cultured overnight, and incubated with RRV for 3 hours at 37°C. To determine if the RRV infections were permissive, a low MOI (0.005) was used to allow local transmission of infection to adjacent cells. The infected cells were washed, cultured for 3 days and then evaluated for the presence of spreading ORF59-positive foci, using the 425 rabbit anti-RV2 ORF59 antiserum, as described previously (5). To determine whether the infection was productive for infectious virus, the cell supernatants were removed and incubated with permissive RPFF or Vero cells. The cells were evaluated after 2 days for the presence of ORF59-positive RRV-infected foci.
To determine if the RRV infections were latent, infected cultures that were ORF59-negative were treated 3 hours post-infection with 4 mM sodium butyrate (Millipore) to induce virus replication, and the cultures were evaluated 3 days later for the presence of ORF59-positive foci and infectious virus, as described above.
For comparison purposes, BCBL-1 cells, latently-infected with KSHV, were incubated with a monoclonal antibody reactive with KSHV ORF59 (Advance Biotechnologies) before and after treatment with 4mM sodium butyrate for 24 hours to determine the replicative state of the virus. For KSHV infections, AGS cells were incubated with gradient-purified KSHV, cultured for 24 hours and fixed in paraformaldehyde. KSHV LANA was visualized using the LN53 rat anti-LANA antibody and goat-anti-rat IgG-coupled to HRP, as described (21).
For LANA inhibition studies, cells were transfected with 200 ng pCDNA3-N-Flag RRV LANA and incubated with purified RRV virions eight hours post-transfection. Flag-tagged RRV LANA was visualized using the M2 anti-Flag antibody (Sigma) and Alexa594-conjugated goat anti-mouse secondary antibody. Cell nuclei were stained with TO-PRO. Images were acquired with a Zeiss LSM 5 Pascal confocal microscope.
Reporter assays
Cells were cultured on 6-well plates and transfected with 3ug of firefly reporter gene construct (either empty pGL2, pGL2-R-Rta or pGL2-K-Rta) and 5 ng of Renilla luciferase pRL-CMV as an internal control for transfection efficiency using LT1 reagent (Minis). For butyrate-treated wells, butyrate was added at 5 hours post-transfection to a final concentration of 4mM. Lysates were harvested 24 hours post-transfection using the Dual Luciferase kit (Promega). The mean luciferase activity from triplicate wells was determined and reported normalized to pRL-CMV activity as relative luciferase units (RLUs). In LANA inhibition studies, reporter plasmids and LANA expression plasmids were cotransfected using LT1 reagent (Mirus). The total volume of DNA was held constant using empty pCDNA3 vector and luciferase activity was measured at 24 hours.
Data analysis
The GraphPad Prism software was used for statistical analysis.
Results
Characterization of cell culture systems permissive and non-permissive for RRV replication
We and others have previously reported that rhesus fibroblasts and Vero kidney epithelial cells are permissive for RRV replication (5,14,17, 60) (Table 1). We have shown that RRV-infected rhesus primary fetal fibroblast (RPFF) and Vero cultures contain large numbers of cells expressing the ORF59 DNA polymerase processivity factor, an early marker of the lytic cycle, and produce high titers of infectious virus (5). In these permissive cell lines, ORF59 mRNA was expressed early after RRV infection and ORF59 protein accumulated in the nuclei of infected cells. Expression and nuclear accumulation of ORF59 increased over time, which strongly correlated with the production of infectious virions (5). Furthermore, expanding foci of ORF59-positive cells were detected, providing evidence of local transmission in the spreading infection in permissive cell cultures during the 3-day study.
Table 1.
Permissivity of cell lines to RRV infection
| Cell line | RPFF | Vero | HEK293 | HSG | HSG+NaButd | AGS | AGS+NaBut |
|---|---|---|---|---|---|---|---|
| ORF59 “early gene” expressiona | +e,g | +e,g | +g | −g | +g | −g | +g |
| Local virus transmissionb | +e,g | +e,g | +g | −g | +g | −g | +g |
| Production of infectious virionsc | +e,f | +e,g | +g | −g | +g | ndh | nd |
Positive nuclear immunofluorescence detected with rabbit anti-RV2 ORF59 antiserum
Expanding foci of ORF59-positive cells post infection
Infectious RRV virions in supernatent of post infection, titered on permissive RPFF cells
NaBut= sodium butyrate treated
Bruce et al., (2009)(5)
DesRosiers et al., 1997 (14), Searles et al., 1999 (60), DeWire et al., 2003(15)
This study
nd=not determined.
To identify natural infection systems to study RRV latency, we screened several cell lines for a non-permissive phenotype. Because a previous study reported that the human embryonic kidney cell line (HEK293) was not permissive for RRV replication, as measured by plaque assay (15), we incubated HEK293 cells with purified virions and evaluated for the induction of the lytic gene cascade examining the expression of RRV ORF59 3 days postinfection using the 425 anti-RV2 ORF59 antibody that reacts with RRV ORF59 (see Materials and Methods). RPFF and Vero cultures were also infected in parallel as positive controls for viral replication. As seen previously, the RRV-infected RPFF and Vero cell cultures contained expanding foci of ORF59-positive cells (Figure 1A and B), showing activation of the lytic gene cascade, production of infectious virions and local virus transmission to adjacent cells (Table 1). Unexpectedly, similar ORF59-positive foci were also detected in the RRV-infected HEK293 cell cultures (Figure 1C). By 3 days, the RRV infection in the HEK293 culture had spread from an initial focus of infection to adjacent cells, as seen previously with permissive RPFF and Vero cells (5). To test for the presence of newly replicated infectious RRV virions, supernatant from the RRV-infected HEK293 cultures was removed and used to infect naive RPFF cells. Supernatant from the RRV-infected RPFF cultures was used as a positive infection control. Three days post-infection similar high levels of ORF59-positive RRV-infected foci were detected in the naive RPFF cell cultures that had been treated with supernatants from either the RRV-infected HEK293 or RPFF cell cultures (Figure 2A and B). These results demonstrated that like RPFF and Vero cells, the HEK293 cells are permissive for RRV infection with expression of the ORF59 lytic gene cascade marker and production of infectious virions (Table 1).
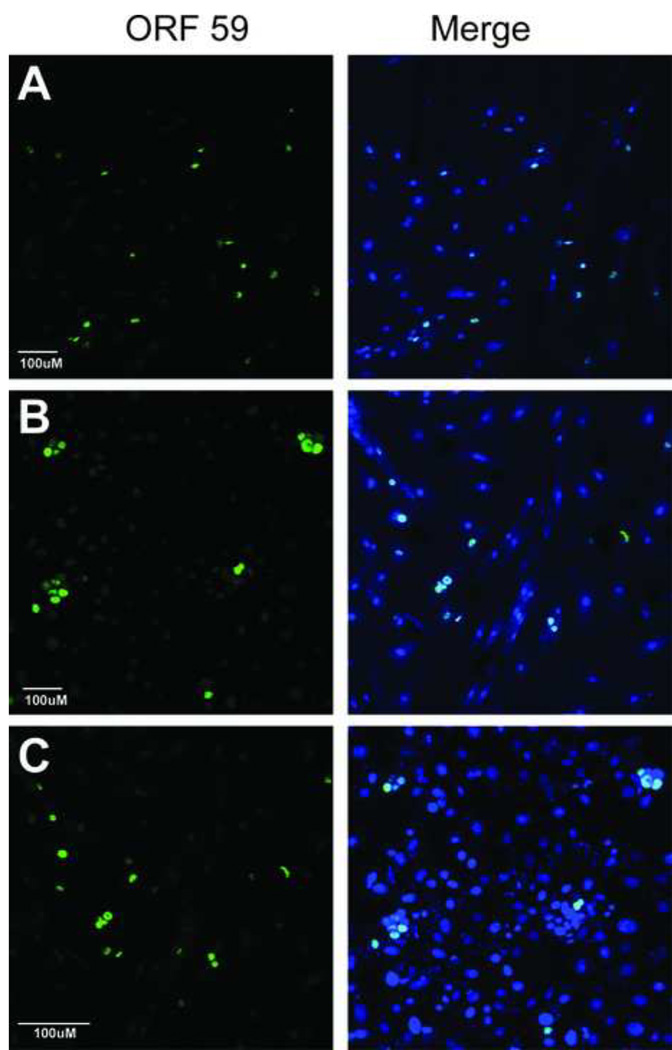
Figure 1. RRV infection induces lytic gene expression in a variety of cell lines from human and nonhuman Old World primate species.
(A) Rhesus primary fetal fibroblast (RPFF), (B) African green monkey kidney epithelial (Vero), and (C) human embryonic kidney epithelial (HEK293) cells were infected with RRV and evaluated three days post-infection for expression of the early lytic gene marker, the ORF59 DNA polymerase processivity factor, using rabbit anti-RV2 ORF59 antiserum (green), as described in Materials and Methods. The merged image of the ORF59 and Topro-3 (blue) nuclear staining is shown. Images were acquired by confocal microscopy and are shown as projections of the z-stack. The magnification scales are shown. Abundant RRV-infected ORF59-positive cell foci were detected in all three cell lines.
Figure 2. ORF59 positive RRV-infected cell cultures are permissive for virus replication and production of infectious virions.
Culture supernatant was removed from ORF59-positive RRV-infected RPFF (A), HEK293 (B) (see Fig. 1) and incubated with permissive RPFF cells to determine the presence of infectious virus. Similarly, supernatant from sodium butyrate-treated RRV-infected HSG cell cultures (see Fig. 3) was incubated with permissive Vero cells. The supernatant-treated cell cultures were screened for de novo infections using the anti-RV2 ORF59 antiserum (green) as described above. Similar titers of infectious RRV were detected in the supernatants of RRV-infected RPFF and HEK293 cell cultures and in the sodium-butyrate activated RRV-infected HSG cell cultures.
The permissivity of two additional epithelial cell lines, human salivary gland (HSG) and human gastric adenocarcinoma (AGS), was evaluated using ORF59 expression as a marker of RRV replication. The HSG epithelial cell line was originally isolated from an irradiated submandibular salivary gland and grows as an undifferentiated monolayer with cuboidal morphology under normal culture conditions (61). The AGS cell line was established from a surgically resected adenocarcinoma of the stomach and shown to be epithelial with expression of cytoplasmic mucin (2). HSG and AGS cell cultures were incubated with RRV for 3 hours and assayed for ORF59 expression 3 days post-infection (pi). Vero cells were infected in parallel as positive controls. While numerous ORF59-positive cells were detected in the permissive Vero cell culture 3 days pi, no ORF59 expression was detected in either the HSG or AGS cell cultures (Figure 3A and 3C). Even though the infected cells did not express ORF59, which is critical for virus replication, the culture medium from the infected HSG cells was assayed for the presence of infectious virions by incubating with permissive RPFF cells, as described above. As expected, the lack of ORF59 staining correlated with the absence of infectious virions, supporting the finding that HSG and AGS cells were not permissive for RRV replication (Table 1).
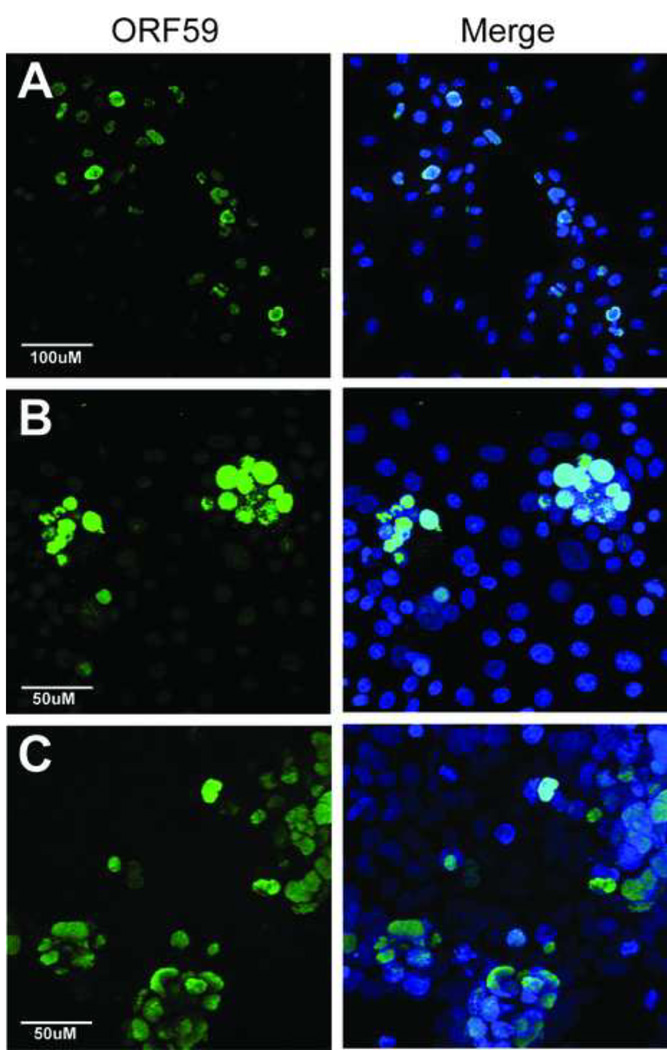
Figure 3. RRV latently infects orogastric epithelial cells, and activation of lytic gene expression and virus replication is induced by sodium butyrate.
Human salivary gland (HSG) and gastric adenocarcinoma (AGS) orogastric epithelial cell lines were incubated with RRV for 3 hours and then assayed 3 days post-infection for expression of RRV ORF59, the DNA polymerase processivity factor, using the 425 rabbit polyclonal anti-RV2 ORF59 antibody (green) to detect permissive RRV infections (A and C). Alternatively, HSG and AGS cells were incubated with RRV for 3 hours and then cultured for 3 days in the presence of sodium butyrate (NaBut) to activate latent RRV infections before evaluation of ORF59 expression (B, D). AGS cells are shown at lower magnification to visualize the presence of ORF59-positive nuclei throughout the culture, in addition to a higher magnification shown in panel D, insert). RRV lytic gene expression was detected in both cell lines after NaBut treatment indicating that the initial RRV infection was latent and that NaBut activated the RRV lytic cycle. The magnification scale bars are shown.
By definition, herpesvirus latency is a reversible state, and the ability to reactivate replication is considered a hallmark of latent infection (63). Because it is essential for viral replication, ORF59 expression and nuclear accumulation are commonly used as markers of reactivation of KSHV latency (29). Though little is known about latency in RRV, our ORF59 assay allowed us to evaluate whether viral replication could be reactivated in RRV-infected HSG and AGS cells. To determine if RRV had established a non-permissive but latent infection in HSG and AGS cells we evaluated ORF59 expression in RRV-infected cultures following treatment with sodium butyrate, an inhibitor of HDAC activity commonly used to reactivate latent herpesvirus infections. HSG and AGS cells were infected with RRV for 3 hours, extensively washed to remove free virus, and treated with sodium butyrate. Three days post-infection, the cells were assayed for ORF59 expression to detect reactivation of latent RRV infections. Numerous ORF59-positive nuclei were detected in RRV-infected HSG and AGS cell cultures following butyrate treatment (Fig. 3B and 3D), in contrast to the untreated cultures. Individual ORF59-positive nuclei were seen in butyrate-treated AGS cultures, shown at both low magnification (Figure 3D) and high magnification (Figure 3D, insert). In the butyrate-treated HSG cell cultures we detected foci of infection, which typically showed one cell strongly staining for ORF59 surrounded by several other cells showing weaker ORF59 staining (Figure 3B). The presence of ORF59-positive foci suggests that the infection had spread during the 3-day incubation from an initial infected cell to adjacent cells, which mirrors the spread of RRV infection in permissive RPFF and Vero cell cultures that we have previously described (5). To determine whether butyrate treatment resulted in the release of RRV virions, supernatant from the butyrate-induced HSG cells was incubated with naive Vero cell cultures. ORF59-positive Vero cells were detected 2 days post-infection (Figure 2C), indicating that butyrate treatment of the HSG cells had induced the production of infectious RRV virions (Table 1). The finding that sodium butyrate induced de novo ORF59 expression in RRV-infected AGS and HSG cells recapitulates the induction of ORF59 expression after butyrate reactivation of latent KSHV infections (Supplementary Figure S1). For both KSHV and RRV, butyrate-induced ORF59 expression correlates with replication and production of infectious virions, substantiating the similarities in reactivation of latency between these viruses.
The RRV Rta promoter is highly active in permissive cell lines
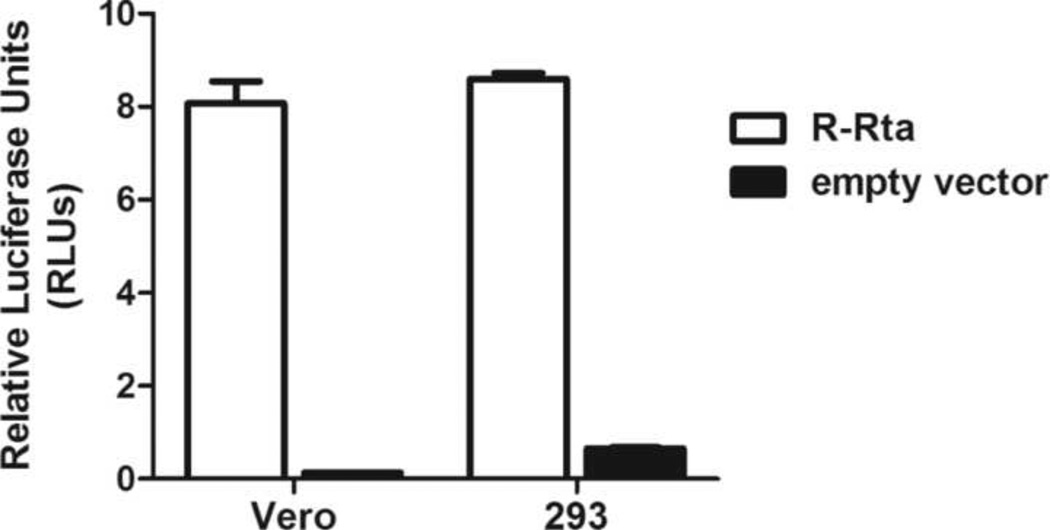
The availability of cell lines permissive for RRV replication allowed us to characterize the RRV Rta promoter in a natural cellular background that supports the complete lytic cascade, which has been difficult for KSHV. HEK293 and Vero cells were transfected with either the pGL2 R-Rta construct, containing 502 nucleotides of RRV Rta promoter sequence upstream of the firefly luciferase gene, or empty pGL2 vector and luciferase activity was measured at 24 hours. The pRL-CMV construct, in which the CMV immediate-early (IE) promoter drives expression of Renilla luciferase, was co-transfected and pGL2 firefly luciferase activity was normalized to pRL Renilla luciferase activity. The pGL2 R-Rta construct gave high levels of luciferase activity in both HEK293 and Vero cells (> 8 RLU) with a 45- and 19-fold increase in activity over empty pGL2 vector, respectively (Figure 4). These results demonstrated that the RRV Rta promoter was highly active in these permissive cell lines.
Figure 4. The RRV Rta promoter is highly active in cells that are permissive for RRV replication.
HEK293 and Vero cells were transfected with either the pGL2 R-Rta promoter construct, containing 502 nucleotides of RRV Rta promoter sequence driving luciferase expression, or empty pGL2 vector, and assayed for luciferase activity 24 hours post-transfection. Luciferase activity was normalized to the activity of pRL-CMV, containing the CMV IE promoter driving Renilla expression, which was co-transfected to control for transfection efficiency. Error bars represent the standard deviation from triplicate wells.
The activity of the RRV Rta promoter is low in non-permissive cell lines, but is highly induced by sodium butyrate treatment
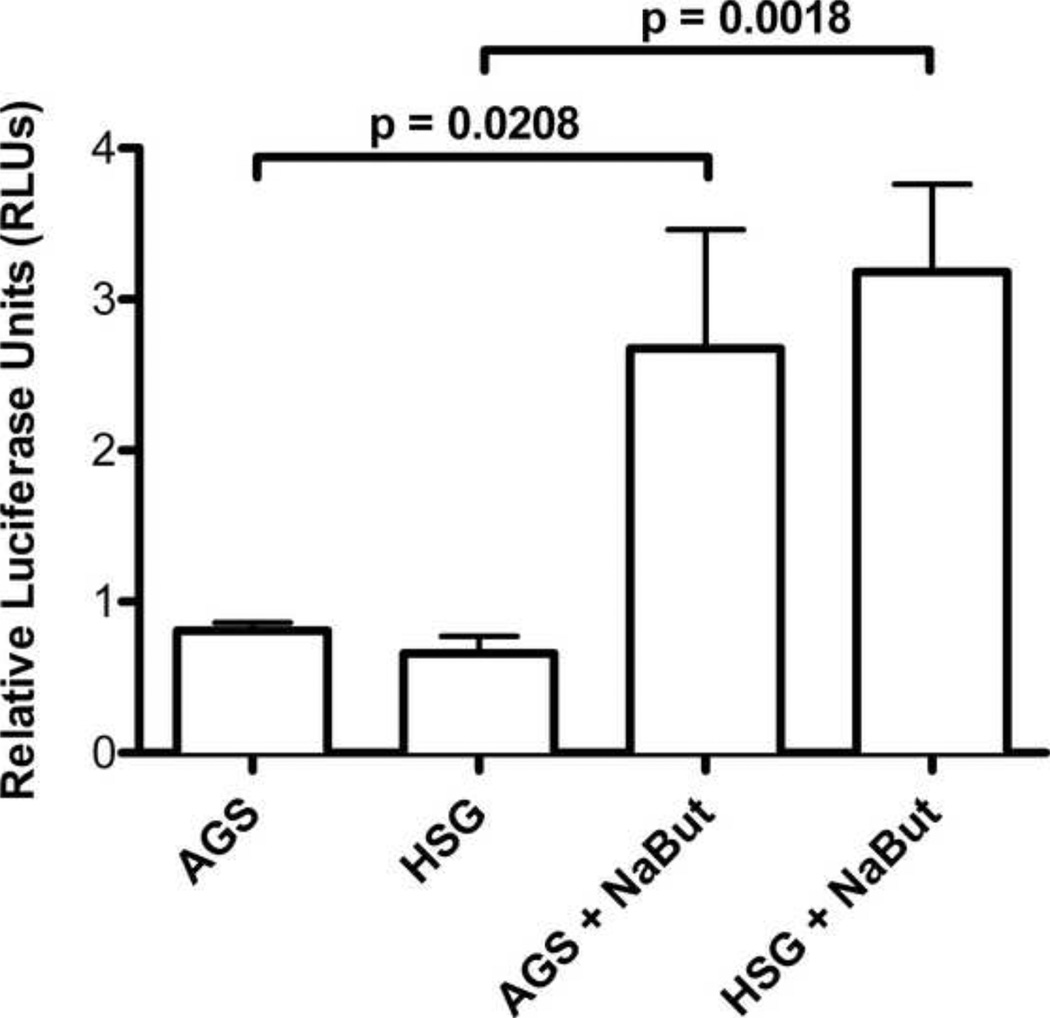
We next examined the activity of the RRV Rta promoter in AGS and HSG cells, which are non-permissive for RRV replication. Since butyrate treatment rendered AGS and HSG cells permissive for RRV replication, we also examined the effects of butyrate treatment on Rta promoter activity. AGS and HGS cells were transfected with pGL2 R-Rta and pRL-CMV in triplicate. In half of the cultures, sodium butyrate was added 5 hours post transfection, and luciferase activity was measured at 24 hours. Minimal luciferase activities of 0.66 and 0.81 RLU were detected in the non-permissive AGS and HSG cells, respectively (Figure 5). The transfection control, pRL-CMV, had comparable activity across all three cell lines (data not shown), indicating that the low activity of pGL2 R-Rta in AGS and HSG cells was not due to broad defects in promoter function in these cell lines. The butyrate-treated AGS and HSG cells had Rta promoter activities of 2.7 and 3.2 RLU (Figure 5). The Rta promoter activities in the butyrate-treated cells were significantly higher than the untreated cells, with 3.3 (p=0.0208) and 4.8 (p=0.0018) fold increased activity, respectively.
Figure 5. RRV Rta promoter activity is low in non-permissive cells and is activated by butyrate treatment.
Non-permissive AGS and HSG cells were transfected with the RRV Rta promoter construct, pGL2 R-Rta. Five hours post-transfection, butyrate was added to half of the cell cultures and luciferase activity was evaluated at 24 hours. Luciferase activity was normalized to the activity of pRL-CMV, as described in Figure 4. Error bars represent the standard deviation from triplicate wells. P values were calculated using the unpaired t test, two tailed.
The RRV and KSHV Rta promoters are similar in structure and share a conserved Sp1 element
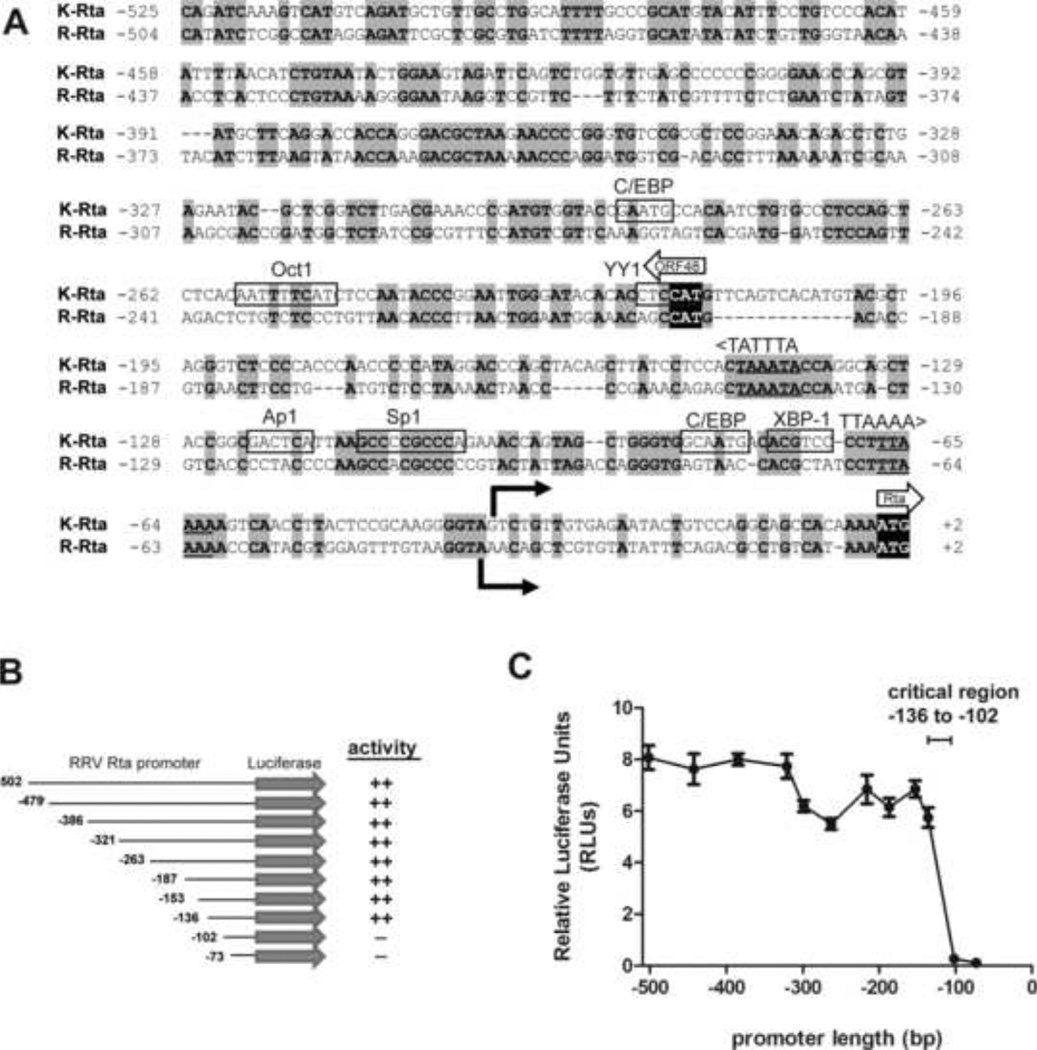
In order to identify regulatory elements in the RRV Rta promoter, the nucleotide sequence of the promoter region directly upstream of the RRV Rta translational start site was aligned with the corresponding region of the KSHV Rta promoter. Although the two promoter regions did not show a high level of sequence homology, an optimal alignment was obtained that showed a close relationship between the translational and transcriptional start sites that have been identified for KSHV and RRV Rta (16, 42). This alignment contained scattered regions of conservation extending to the upstream ORF48 translational start site and coding sequence (Figure 6A). A conserved TATA-like promoter motif (TTAAAA) that has been described upstream of the KSHV Rta transcriptional start site at nucleotides (nt) −67 to −62 (74) was conserved in the RRV Rta promoter region (nt −66 to −61). Similarly, another TATA-like motif (TATTTA, coding strand) was identified upstream of the ORF48 start site in both the KSHV and RRV sequences (nt −139 to −144). A number of binding sites for cellular transcription factors previously identified in the KSHV Rta promoter were mapped on the KSHV sequence, including a GC box containing nine consecutive guanines and cytosines (5’ GCCCCGCCC 3’; nt −112 to −103) that has been shown to be a binding site for the Sp1 transcription factor in the KSHV Rta promoter (39, 74). Similar proximal Spl-binding sites have been identified in a wide variety of eukaryotic promoters (37, 48). The transcription factor binding prediction program PROMO (47) identified a positionally conserved Sp1 site in the RRV Rta promoter (GCCACGCCC; nt −113 to −103). Eight of nine nucleotides in the GC box were identical between the KSHV and RRV Rta promoter sequences (Figure 6A). The binding sites for other transcription factors identified in the KSHV Rta promoter, including Oct1, Apl, XBP-1, and C/EBP, were not well conserved in the RRV Rta promoter.
Figure 6. Deletion analysis of the RRV Rta promoter reveals a critical regulatory region.
(A) Comparison of the promoter regions of K-Rta and R-Rta, the replication and transactivator genes for KSHV and RRV, respectively. Nucleotides 71170 to 71697 of KSHV (NC_009333.1) and 66858 to 67364 of RRV (NC_003401.1) genomic sequence were aligned. Nucleotide numbering indicates the position relative to the Rta translational start site. Identical nucleotides are highlighted in gray. Known transcription factor binding sites for C/EBP, Octl, YY1, Apl, Spl, and XBP-1 in the KSHV Rta promoter (9, 39, 56, 67, 68, 70, 74, 75) are indicated. Of these sites, only the Sp1 binding site was conserved in the RRV Rta promoter. Transcriptional start sites for KSHV Rta and RRV Rta (16, 42) are identified by solid arrows. Translational start sites for Rta and ORF48 are shown with block arrows with initiation codons shown in black. TATA-like elements upstream of ORF48 and Rta are shown. (B) 5’ serial truncations of the RRV Rta promoter region were constructed in the pGL2 firefly luciferase backbone. (C) Luciferase activity was measured in extracts from permissive Vero cells transfected with pGL2 R-Rta promoter deletion clones and normalized using pRL-CMV. Error bars represent the standard deviation of results from triplicate wells. The region from −136 to −102 containing the conserved Sp1 binding site was critical for Rta promoter activity.
The conserved Sp1 site in the RRV Rta promoter confers high-level activity in permissive cells and butyrate responsiveness in non-permissive cells
To identify specific regulatory elements in the RRV Rta promoter, we prepared 5’ truncations of the promoter region beginning 502 nucleotides upstream of the Rta translational start site. Serial truncations were cloned into the pGL2 firefly luciferase backbone, and the activity of each truncation construct was evaluated in Vero cells, which are permissive for RRV infection. Deletion of nucleotides −502 to −136 did not significantly impact promoter activity (Figure 6B). This region contains the upstream sequences encoding ORF48 and its putative TATA-like element. However, further deletion of nucleotides from −136 to −102 resulted in a dramatic decrease in promoter activity (Figure 6C). Similar results were obtained in HEK293 cells, which are also permissive for RRV infection (data not shown).
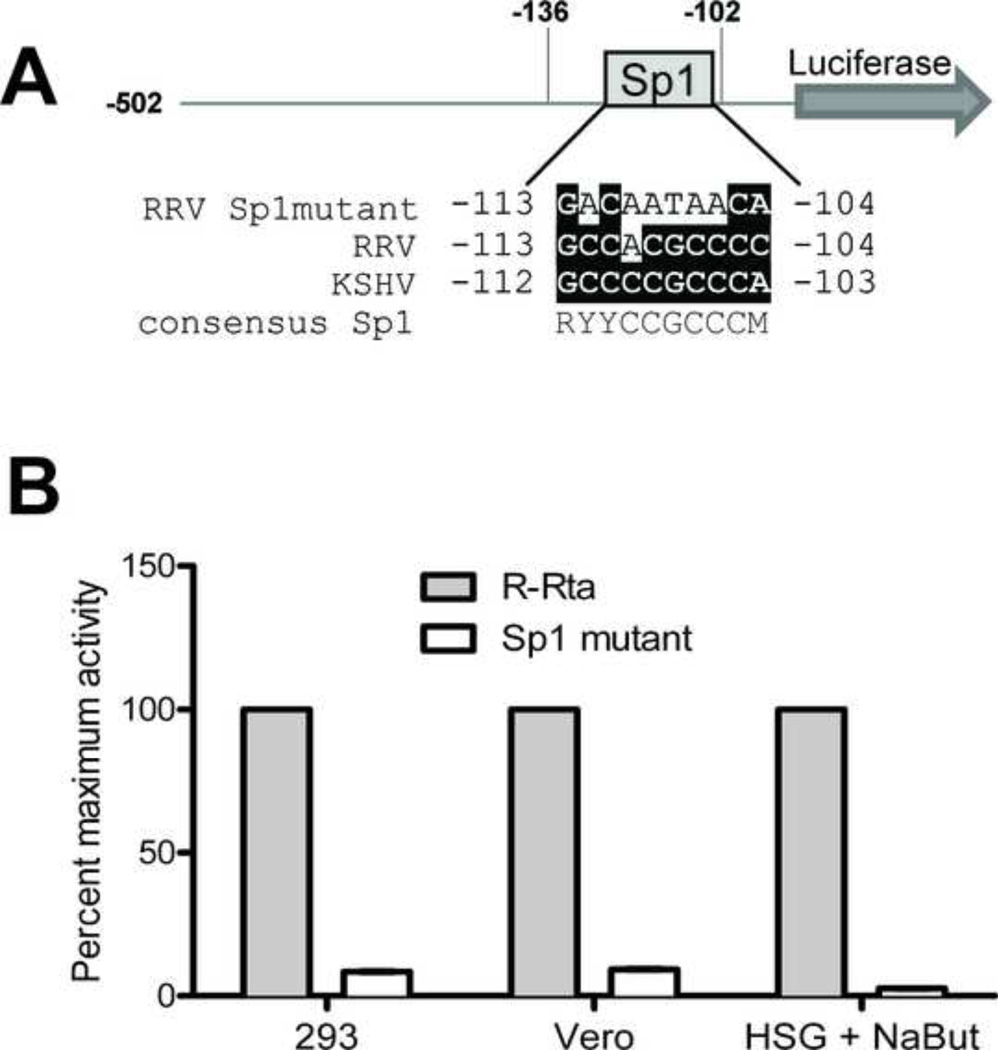
The truncation analysis suggested the presence of a critical regulatory element between nucleotides −136 and −102. Because this region contained the Sp1 element conserved between KSHV and RRV (Figure 7A), we tested whether the Sp1 site at −113 was required for RRV Rta promoter activity. The Sp1 element was eliminated in the full-length pGL2 R-Rta by site-directed mutagenesis (Figure 7A). The activity of the wild type and Sp1 mutant were analyzed in permissive Vero and HEK293 cells, which have high basal levels of Rta promoter activity, and in HSG cells that were treated with sodium butyrate to induce high-level Rta promoter activity. In HEK293 and Vero cells, the Sp1 mutation abolished RRV Rta promoter activity (Figure 7B), indicating that the Sp1 site at −113 was critical for high-level Rta promoter activity in permissive cells. A similar reduction in activity was seen in the butyrate-treated HSG cells (Figure 7B), indicating that the Sp1 site at −113 was also required for butyrate-responsiveness of the RRV Rta promoter in non-permissive cells. This result mirrors previous reports for the KSHV Rta promoter, which showed that the conserved Sp1 site was critical for butyrate induction in non-permissive cells (39, 74).
Figure 7. The conserved Sp1 element is critical for RRV Rta promoter activity.
(A) The Sp1 site in the critical RRV Rta promoter region was eliminated by site-directed mutagenesis in the full length pGL2 R-Rta construct. The conservation between the RRV and KSHV Sp1 sites are shown and the nucleotides matching the Sp1 consensus sequence (48) are highlighted. (B) Permissive Vero, HEK293 and HSG cells were transfected with either wild type pGL2 R-Rta or the Sp1 mutant and promoter activity was measured at 24 hours. pRL-CMV was co-transfected to control for transfection efficiency. The HSG cells were incubated with sodium butyrate 5 hours post transfection. Promoter activity, after normalization with pRL-CMV, is reported as percent of maximum activity. Mutagenesis of the RRV Sp1 site abrogated the RRV Rta promoter activity in all three permissive cell types. Error bars represent standard deviation from triplicate wells.
The KSHV and RRV Rta promoters have similar activities in a panel of cell lines
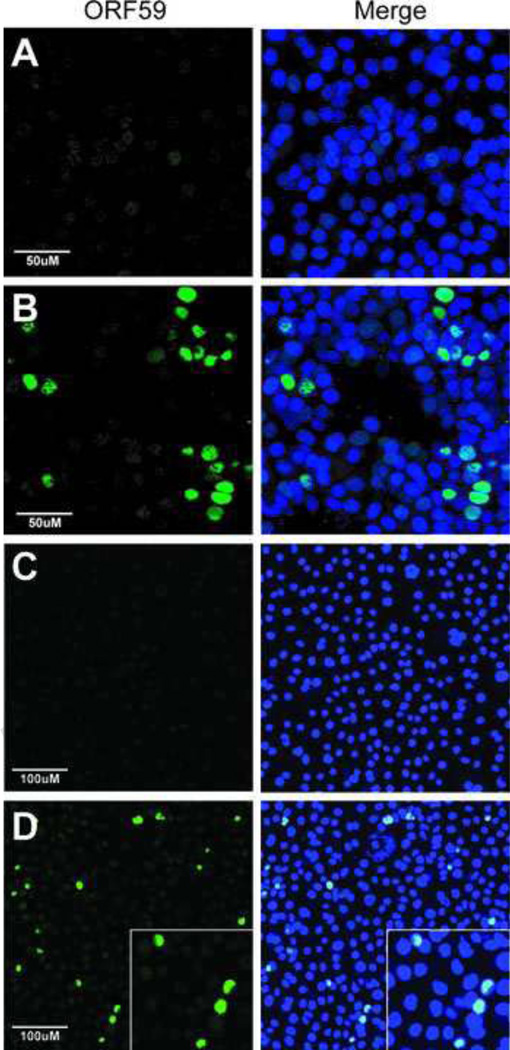
Our studies demonstrated that Vero and HEK293 kidney cell lines were permissive for RRV replication with expression of the early lytic gene, ORF59, and production of infectious virions. We also determined that HSG and AGS orogastric epithelial cell lines are non-permissive for RRV replication with no expression of the early lytic gene ORF59 or evidence of virus replication. We and others have shown that KSHV infection of Vero and HEK293 cells is non-permissive for viral replication with high numbers of latently infected cells expressing nuclear LANA. At most, only 1–5% of infected cells expressed ORF59, indicating minimal levels of viral replication. On the other hand, KSHV infection of AGS cells resulted in high numbers of latently infected cells expressing LANA (Supplementary Figure 2) with no ORF59-positive cells (data not shown). In other studies, we have shown that HSG cells are not susceptible to KSHV infection until reconstitution of the αvβ3 integrin receptor by transfection, which results in a characteristic latent KSHV infection (unpublished observations).
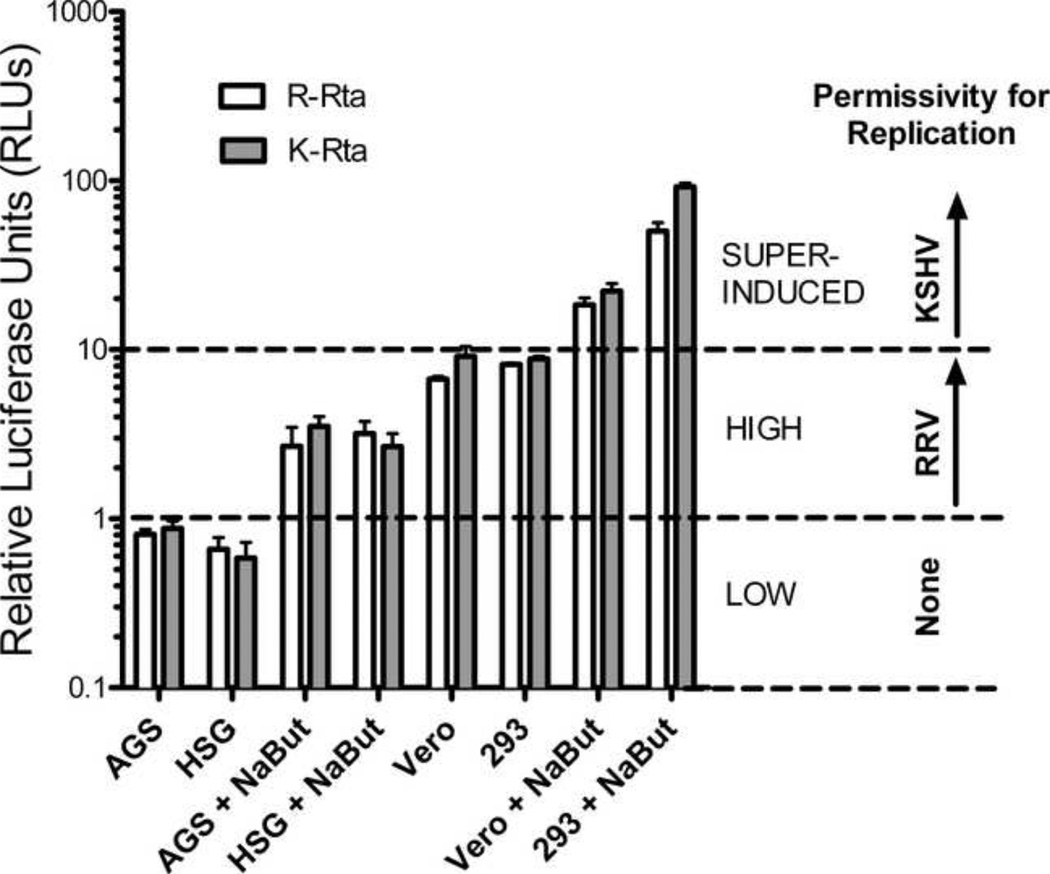
The availability of epithelial cell lines with different permissivity for KSHV and RRV replication allowed us to directly compare how the Rta promoters of these viruses function in the context of latency and replication. For this comparison, we cloned 503 nucleotides of KSHV Rta promoter sequence upstream of the firefly luciferase reporter gene to create pGL2 K-Rta, as described in Materials and Methods. The pGL2 R-Rta and pGL2 K-Rta constructs were transfected into AGS, HSG, Vero, and HEK293 cells and the luciferase activity was determined in the presence or absence of sodium butyrate trelfcnent. pRL-CMV was co-transfected to control for transfection efficiency. pGL2 luciferase activity was normalized using the pRL luciferase activity, as described previously. Both the RRV and KSHV Rta promoters had low Rta promoter activity (RLU <1) in untreated AGS and HSG cells (Figure 8). Butyrate treatment of AGS and HSG cells induced the activities of both promoters 3.3 to 4.8-fold, with levels of activity ranging from 2.7–3.5 RLU. Both promoters had similar levels of Rta activity in untreated Vero and HEK293 cells (RLU = 6.7 to 9.1). The KSHV and RRV Rta promoter activities were induced 2.4 and 2.7 fold by butyrate treatment of Vero cells, and 10.5 and 6.2 fold by butyrate treatment of HEK293 cells, respectively (Figure 8). Surprisingly, both promoters showed similar levels of activity in each cell line tested, suggesting that the promoters were similarly activated by host factors in these cells.
Figure 8. Comparison of KSHV and RRV Rta promoter activity.
pGL2 R-Rta and pGL2 K-Rta containing the RRV and KSHV Rta promoters directing luciferase expression, respectively, were transfected into AGS, HSG, Vero and HEK293 cells and luciferase activity was measured at 24 hours. pRL-CMV was cotransfected to control for transfection efficiency. Cells were treated with sodium butyrate as indicated. Luciferase activity was measured and normalized to pRL-CMV. Low levels of Rta promoter activity (RLU < 1) were associated with non-permissive infections with both KSHV and RRV. High levels of Rta promoter activity (RLU 1–10) were associated with non-permissive infections with KSHV and permissive infections with RRV leading to virus replication. Super-induced levels of Rta promoter activity (RLU >10) were required for permissive KSHV infections.
Rta promoter activity correlates with permissivity of RRV infection but not KSHV infection
As shown in Figure 8, low levels (RLU<1) of RRV and KSHV Rta promoter activity were associated with non-permissive infections of the AGS and HGS cells with both viruses, characterized by the complete lack of expression of the lytic replication marker ORF59 and no production of infectious virus. The high levels (RLU 1–10) of Rta promoter activity detected in butyrate-treated AGS and HSG cells and in untreated Vero and HEK293 cells were associated with RRV replication, suggesting that this level of Rta promoter activity was sufficient to induce the widespread expression of the lytic replication marker ORF59 and virus production in infected cells. However, the same levels of Rta promoter activity were insufficient to induce KSHV replication in the untreated Vero and HEK293 cells or butyrate treated AGS cells, as only minimal numbers of infected cells (1–5%) express the ORF59 lytic replication marker. Finally, super-induced levels (RLU 10–100) of Rta promoter activity were detected in butyrate-treated Vero and HEK293 cells, and were associated with widespread ORF59 expression and virion production in KSHV-infected cultures. Since there is sufficient Rta promoter activity in the untreated HEK293 and Vero cells and butyrate-treated AGS and HSG cells to induce the cascade of lytic replication of RRV, the inability of KSHV to replicate in these cells suggests that KSHV downregulates Rta promoter activity in a process that is not shared by RRV.
RRV LANA inhibits RRV replication in permissive cells
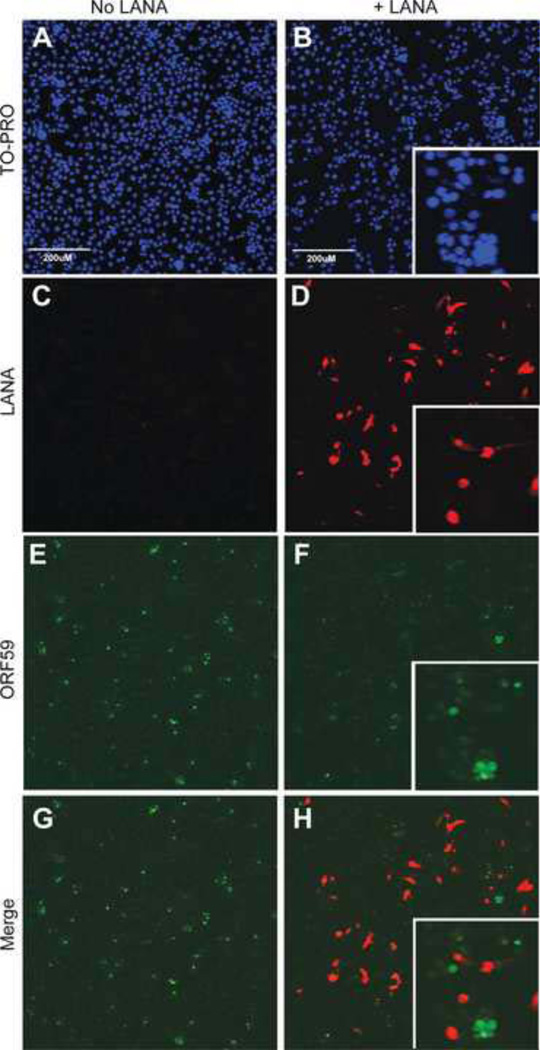
Previous studies have shown that KSHV LANA plays a major role in inhibiting the cascade of lytic gene expression and virus replication induced by Rta early after infection (35, 45). To determine whether RRV LANA can play a similar role in RRV infections, we transfected permissive Vero cells with pCDNA3 N-Flag R-LANA, a construct that expresses Flag-tagged RRV LANA from the highly active CMV IE promoter. Empty pCDNA3 was transfected as a negative control. LANA-transfected cell cultures were subsequently infected with RRV and evaluated 2 days post-infection for evidence of RRV replication using the anti-RV2 ORF59 antiserum. Nuclei were visualized with TO-PRO and expression of recombinant LANA was evaluated using an anti-Flag antibody. Cells transfected with empty pCDNA3 vector and infected with RRV yielded high numbers of ORF59-positive cells (Figure 9E; green), consistent with the permissive nature of these cells for RRV replication with widespread viral replication. Cells transfected with the LANA expression plasmid and infected with RRV showed abundant expression of Flag-tagged RRV LANA (Figure 9D; red). Analysis of the ORF59 expression in the LANA-transfected cell cultures revealed minimal numbers of ORF59-positive cells (Figure 9F; green), indicating that the LANA-transfected cell cultures were no longer permissive to widespread RRV replication. Analysis of the merged ORF59 (green) and LANA (red) images (Figure 9H) revealed that the few ORF59 positive cells detected in the transfected culture were LANA-negative, indicating that they had not been successfully transfected with the LANA expression plasmid. The LANA positive cells (red) showed no evidence of ORF59 (green) expression (Figure 9H and insert). Subsequent butyrate treatment of the LANA-transfected, RRV-infected cultures revealed strong ORF59 expression in the LANA-positive cells (data not shown). Thus, the LANA-expressing cells were susceptible to RRV infection, but were not permissive for expression of the ORF59 lytic replication marker. These data suggest that RRV LANA did not inhibit viral entry or infection, but instead blocked the lytic replication gene cascade. Sodium butyrate was able to overcome the LANA-mediated inhibition, allowing the replication gene cascade to proceed.
Figure 9. RRV latency associated nuclear antigen (LANA) inhibits RRV replication in Vero cells.
Cells were transfected with either pcDNA3 N-Flag R-LANA or empty pcDNA3, infected with RRV and evaluated 2 days post-infection for evidence of replication by detection of the ORF59 lytic cycle replication marker. (A–B) To-PRO stained cells show cell nuclei (blue). (C–D) Expression of the transfected RRV LANA was evaluated with an anti-Flag antibody (red). (E–F) ORF59 expression was evaluated with an anti-RV2 ORF59 antibody (green). (G) Merge of panels C and E. (H) Merge of panels D and F showing that only LANA-negative cells expressed the ORF59 marker of RRV replication.
RRV LANA is a potent inhibitor of Rta promoter activity
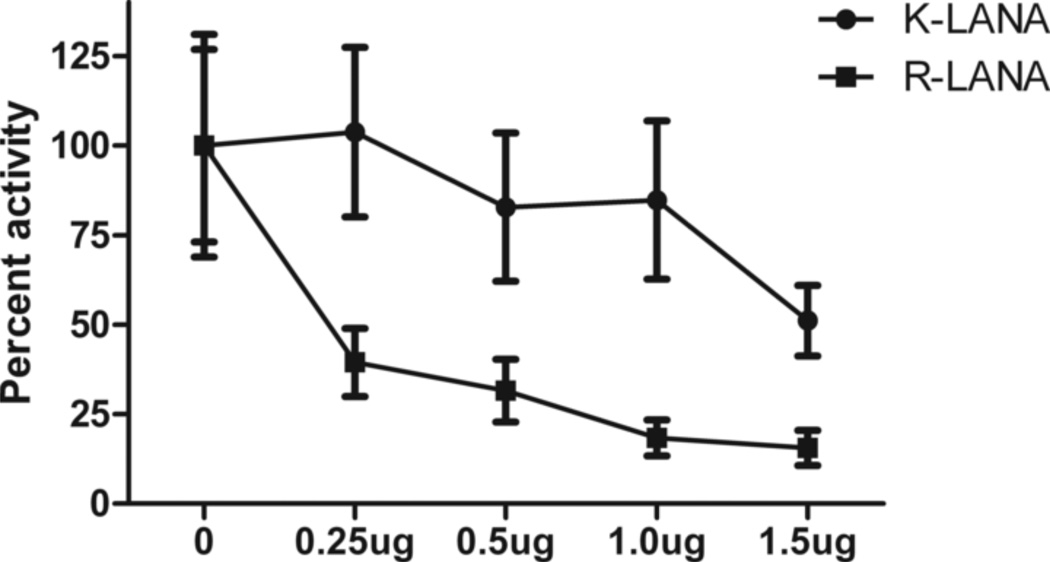
Previous studies have shown that RRV LANA inhibits the Rta-induced transactivation of viral promoters responsible for the cascade of lytic gene expression leading to virus replication (15, 69). To determine whether RRV LANA has a direct effect on the RRV Rta promoter, we performed luciferase reporter assays in permissive Vero cells expressing RRV LANA. The RRV Rta promoter construct pGL2-R-Rta was transfected with increasing amounts of pCDNA3 N-Flag R-LANA, and luciferase activity was measured at 24 hours. To compare with KSHV, we also evaluated the ability of KSHV LANA to inhibit the KSHV Rta promoter by transfecting Vero cells with pGL2-K-Rta and pCDNA3 N-Flag K-LANA, which expresses KSHV LANA with an N-terminal Flag tag. We observed a dose-dependent inhibition of KSHV Rta promoter activity by KSHV LANA, with a maximal inhibition of 50% at the highest plasmid concentration tested (Figure 10). This confirms previous results showing that KSHV LANA inhibits KSHV Rta promoter activity (35). RRV LANA strongly inhibited the RRV Rta promoter with greater than 60% inhibition at the lowest plasmid concentration tested. A dose-dependent response was detected with increasing inhibition to a maximal level of ~80%. These results demonstrate that the RRV Rta promoter can be strongly downregulated by RRV LANA and suggests that the inhibition of RRV replication by LANA that we detected in the transfected cell cultures was mediated through inhibition of the Rta promoter. Thus, the differential ability of KSHV and RRV to establish latency and replicative infection in cells that have high Rta activity, such as Vero and HEK293, may be due to differential expression of the Rta inhibitor, LANA.
Figure 10. RRV LANA inhibits RRV Rta promoter activity.
Vero cells were transfected with either the pGL2 K-Rta promoter construct and increasing amounts of pcDNA3 N-Flag K-LANA (triangles) or pGL2 R-Rta and increasing amounts of pcDNA3 N-Flag R-LANA (squares). pRL-CMV was used to control for transfection efficiency. Luciferase values were measured 24 hours post-transfection, normalized to pRL-CMV, and are shown as percent of maximal activity. The amount of total DNA in each well was held constant with empty pcDNA3 vector. While KSHV LANA modestly inhibited the KSHV Rta promoter, as previously shown (35), RRV LANA highly inhibited the RRV Rta promoter, in a dose-dependent fashion.
Discussion
Previous studies have identified and characterized in vitro culture systems to study latent infections of KSHV in non-permissive cells and replicative infections of RRV in permissive cells. Here we describe additional cell lines that are non-permissive for RRV replication and recapitulate the latent infection systems and virus reactivation processes that have been characterized for KSHV. This has allowed us to study the function of the RRV Rta promoter in natural permissive and non-permissive systems, something that has been difficult for KSHV. We have identified the human gastric epithelial (AGS) and the human salivary epithelial (HSG) cell lines as non-permissive for RRV infection, with no virus replication or production of infectious virions. The AGS and HSG cells were also non-permissive for KSHV replication, although the HSG cells lack the αvβ3 integrin receptor and are only susceptible to KSHV infection when αvβ3 is ectopically expressed (unpublished observations). Although these cell lines represent novel systems for studying KSHV and RRV latency, more study is needed to understand gene expression and viral persistence in these cells.
We have developed a polyclonal anti-RV2 ORF59 antiserum that is reactive with the RRV ORF59 DNA polymerase processivity factor. We have shown that rhesus fibroblasts and African green monkey Vero cells are permissive for RRV replication and produce infectious virus. RRV-infected fibroblast and Vero cells express ORF59 transcripts early after RRV infection with concomitant accumulation of ORF59 in the nucleus that is reactive with the anti-ORF59 antiserum (5). Thus, nuclear expression of RRV ORF59 is an early marker of virus replication. We did not detect any RRV-infected HSG and AGS cells expressing the ORF59 replication marker using the ORF59 antiserum. In contrast, butyrate treatment of RRV-infected HSG and AGS cells induced widespread ORF59 expression and virus replication, recapitulating the butyrate-induced reactivation of latent KSHV infections in pleural effusion lymphoma (PEL) cell lines, such as BCBL-1, and in KSHV-infected HEK293, Vero and other cell lines. Interestingly, while KSHV-infected AGS cells expressed significant levels of the latency antigen LANA, butyrate treatment did not induce widespread ORF59 expression (unpublished observations). AGS cells may be less efficiently reactivated because of low basal levels of Rta promoter activity.
It was previously reported that the HEK293 human embryonic kidney cell line is not fully permissive for RRV replication, based on the lack of plaque formation, and an RRV latency system was developed in which infected HEK293 cells were cultured long-term in ganciclovir, an inhibitor of viral replication that specifically targets the viral DNA polymerase (15). We found, however, that our ATCC HEK293 cells were permissive for RRV replication and showed strong expression of the ORF59 replication marker in expanding foci of infected cells with concomitant production of infectious virus, similar to that seen with RPFF cells. The different outcomes of RRV infection in HEK293 cells may be due to differences in the strain of RRV used for infection, as our studies used the 17577 RRV strain, while DeWire’s study used the 26–95 strain. Additionally, differences in the HEK293 cell lines or passages could contribute to the differences in permissivity.
Our studies demonstrated that the RRV Rta promoter was highly active in permissive cells, such as Vero and HEK293, in which RRV replicates, and minimally active in non-permissive cells, such as AGS and HSG, which are susceptible to infection but are not permissive for viral replication. In non-permissive cells, sodium butyrate treatment induced high-level RRV Rta promoter activity and RRV replication. An Sp1 element at −113 within the RRV Rta promoter was required for promoter activity in permissive cells and butyrate responsiveness in non-permissive cells. This Sp1 element is highly conserved with an Spl-binding site in KSHV that is critical for butyrate induction of the KSHV Rta promoter in non-permissive cells (39, 74). Our finding that high-level RRV Rta promoter activity correlates with cellular permissivity for RRV replication suggests that host factors, such as Spl, regulate RRV Rta expression and the downstream induction of the lytic gene cascade and virus replication in infected cells. Current studies are ongoing to examine the role of Sp1 in Rta regulation.
Our study adds to a growing number of reports linking butyrate treatment and Sp1 activity (30, 50, 71, 76). How sodium butyrate alters Sp1 binding and activity is not completely understood. Sp1 is widely expressed, and its activity is known to be highly regulated by multiple mechanisms, including transcriptional control, post-translational modification, and degradation (20, 44, 46, 72). A previous report by Ye et. al. showed that butyrate augments Sp1 binding to the KSHV Rta promoter, consistent with the model that butyrate enhances the functionality of Sp1 or Spl-containing complexes (74). While it has been suggested that the Sp1 site in the Rta promoter plays an important role in epigenetic silencing of Rta expression during KSHV latency (26, 39), the role of epigenetic regulation in RRV latency is unknown. However, our study suggests that the Sp1 site in the Rta promoter may play an important role in regulating RRV replication. Interestingly, we did not identify any sequence conservation with other transcription factor binding sites that have been characterized in the KSHV Rta promoter. While our studies suggest a role for Sp1 in the reactivation and replication of RRV, other host transcription factors may also play a role in these processes.
We found that the activities of the KSHV and RRV Rta promoters were essentially identical in all cell lines tested, indicating that the promoter structures and critical host factors regulating promoter activity were analogous. Both promoters had minimal activity in the HSG and AGS orogastric epithelial cells, which correlated with non-permissive phenotypes of these cells for both KSHV and RRV replication. Sodium butyrate treatment of the HSG and AGS cells increased the activity of both Rta promoters to the levels seen in untreated HEK293 and Vero cells. This level of Rta promoter activity was sufficient to induce the cascade of lytic gene expression leading to RRV replication in permissive HEK293 and Vero cells and butyrate-treated HSG and AGS cells but was unable to drive the lytic gene expression leading to KSHV replication. This suggests that the KSHV Rta promoter was down-regulated by viral factors present early after KSHV infection, leading to viral latency rather than replication. This is consistent with the current understanding that KSHV LANA mediates shutoff of KSHV Rta expression (35, 45). We found that sodium butyrate treatment of HEK293 and Vero cells increased the activity of both KSHV and RRV Rta promoters to very high (super-induced) levels, which correlated with reactivation of latent KSHV infection and induction of virus replication. This suggests that the super-induced levels of Rta promoter activity were sufficient to overcome inhibitory effects of LANA, allowing Rta induction of the lytic gene cascade and KSHV replication.
To understand why RRV replication in HEK293 and Vero cells was not similarly blocked by viral factors, such as LANA, we examined the ability of RRV LANA to inhibit RRV Rta promoter activity. It is known that KSHV LANA shuts off the viral replication cascade through direct repression of the Rta promoter (35, 45). We confirmed the inhibitory effects of KSHV LANA on the KSHV Rta promoter and demonstrated that RRV LANA can directly inhibit the RRV Rta promoter in a dose-dependent fashion. Our findings contrast with those from a previous study, which found no effect of RRV LANA on the RRV Rta promoter (15). This may be due to differences in approach, as we assayed for direct LANA inhibition of the RRV Rta promoter, while DeWire et al. normalized their data to detect LANA inhibition of Rta transactivation of the Rta promoter rather than direct Rta inhibition (15). We also showed that ectopic expression of RRV LANA in permissive cells, such as Vero, completely blocked expression of the early lytic gene ORF59 in cells expressing LANA, indicating that RRV replication was inhibited by LANA. Coupled with our finding that RRV LANA can inhibit the RRV Rta promoter, this indicates that the block in RRV replication by LANA was due to direct inhibition of the Rta promoter. This was substantiated by our finding that sodium butyrate, which activates the RRV Rta promoter, could relieve the LANA-induced inhibition of replication. The previous study by DeWire et al. detected a 50% decrease in RRV replication in RRV-infected rhesus fibroblasts ectopically expressing LANA. However, since they detected no direct effect of RRV LANA on the Rta promoter, they concluded that the inhibition of replication was due to LANA interference with Rta transactivation of lytic promotres. Our studies clearly show that RRV LANA can block RRV replication through direct inhibition of the RRV Rta promoter.
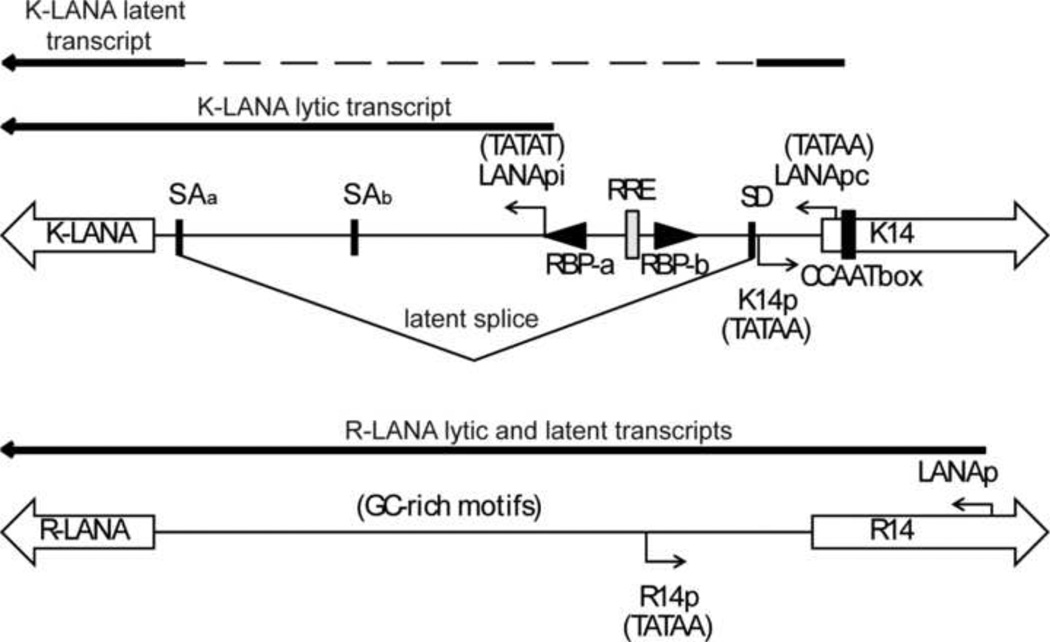
In cells, such as Vero and HEK293, that have high KSHV Rta promoter activity but are non-permissive for KSHV replication, it is believed that Rta expression is down-regulated by LANA. In KSHV, the constitutive LANApc promoter regulates expression of a spliced LANA transcript that originates from the adjacent K14 coding region (Figure 11). However, immediately after KSHV infection, Rta induces LANA expression through a second Rta-inducible LANApi promoter located within the intron region that is spliced out of the latent LANA transcript produced from the LANApc promoter (36, 40, 45). High-level expression of LANA coupled with the shut-off of Rta expression leads to the establishment of KSHV latency. The KSHV Rta-inducible LANApi promoter contains two RBP-Jk binding sites and an Rta-response element (RRE) adjacent to a TATA box that are critical for the Rta-mediated induction of KSHV LANA expression (Figure 11) (28). We have recently sequenced the complete genome of RFHV, the macaque homolog of KSHV, and have compared the RFHV and KSHV genome sequences. We identified a conserved RBP-Jk binding site adjacent to a conserved TATA box in the intron region of the RFHV latent LANA transcript, suggesting that RFHV also contains an Rta-inducible LANApi promoter (Bruce et al., In Press). Though the RRV LANA promoter region has not been fully characterized, the lytic and latent LANA transcriptional start sites have been co-localized to the same region within the R14 coding sequences (15), similar to the position of the latency-associated constitutive LANApc promoter of KSHV (Figure 11). Analysis of the RRV LANA sequence corresponding to the position of the Rta-inducible LANApi promoter within the spliced intron of KSHV failed to identify a comparable TATA-like promoter element or RBP-Jk binding sites, which are required for LANApi promoter activity in KSHV. Furthermore, none of the characterized RRV LANA transcripts initiate within this region. This analysis suggests that RRV lacks a conserved homolog of the KSHV Rta-inducible LANApi promoter and therefore lacks this mechanism for inducing the expression of sufficient levels of LANA to mediate the establishment of latency in cells expressing Rta. The lack of Rta-induced LANA expression could explain the ability of RRV to replicate in cells in which KSHV infections are latent. Previously, we have detected a very low level of RRV LANA transcripts in permissive RRV infections of Vero cells (5), suggesting that low levels of RRV LANA were not sufficient to overcome the Rta promoter activity induced by host factors like Spl. Interestingly, elimination of a critical RBP-Jk binding site within the KSHV LANApi promoter created a novel recombinant KSHV that induced lytic replication during primary infection, recreating the permissive types of infections seen with RRV (40).
Figure 11. Comparison of the promoter regions of the LANA homologs of KSHV and RRV.
(A) Schematic of the bi-directional promoter region for the LANA (leftward) and K14 (rightward) homologs in KSHV and RRV. The coding regions for the ORF73 homologs (K-LANA and R-LANA) and 0RF74 homologs (K14 and R14) are indicated as large open arrows. The homologous 0RF74 “TATAA” promoter elements (K14p and R14p) are shown. The latency-associated constitutive LANA promoter (LANApc) of KSHV and the Rta-inducible LANA promoter (LANApi) are shown. Splice acceptor (SAa, SAb) and splice donor (SD) sites utilized to produce the latent spliced LANA transcripts are indicated (18, 58, 65). RBP-Jkappa and RRE binding sites in LANApi are shown. RNA transcript and sequence analysis of the RRV LANA gene revealed no evidence for an inducible promoter analogous to the Rta-inducible KSHV LANApi promoter.
It is also possible that post-translational regulatory mechanisms, such as subcellular localization, could affect the ability of RRV LANA to inhibit Rta expression. Although antibodies specific for RRV LANA have not yet been developed, ectopic expression of RRV LANA fused to an enhanced green fluorescent protein (EGFP) revealed that RRV LANA has a different sub-nuclear localization than KSHV LANA and localizes to the nucleolus (15). We have confirmed these findings and have identified differences in the nuclear localization pathways utilized by KSHV and RRV LANAs that could account for these distinct localization patterns (unpublished results). Thus, nucleolar RRV LANA may be unavailable to inhibit the Rta promoter in the viral genome located in nuclear replication complexes.
Conclusions
Our studies provide an explanation for the differences observed in the ability of RRV and KSHV to establish lytic and latent infections in vitro. Both viruses establish non-permissive (latent) infections in cells in which Rta promoter activity is low or absent. The dependence of Rta promoter activity on the Sp1 site suggests that the low Rta promoter activity in these cells is linked to low levels of active Spl. In cells with sufficient levels of active Sp1 and Rta promoter activity to drive RRV Rta-induced lytic gene expression, RRV infections are permissive for virus replication. In the same cells, however, KSHV infections are latent as the Rta levels are not sufficient to overcome the inhibitory effects of LANA produced from the Rta-induced LANApi pathway. Our data suggest that the presence of the LANApi pathway in KSHV requires a super-induction of the KSHV Rta promoter to overcome the inhibitory effects of LANA and drive KSHV replication in an infected cell. Although we found that RRV LANA can directly inhibit the RRV Rta promoter, there was no evidence of a comparable Rta-inducible LANApi promoter that would induce latency in cells where the Rta promoter was active. Our study sheds light on the differential ability of RRV and KSHV to establish latent and replicative infections in different cell types and characterizes the interplay of Spl, Rta and LANA in determining the outcome of the infection.
Supplementary Material
Highlights.
Identified cell lines that are permissive and non-permissive for RRV infection
Characterized the RRV and KSHV Rta promoters in permissive and non-permissive cells
Identified a critical Sp1 element in the RRV Rta promoter that is conserved in KSHV
Although promoter activities were similar differences were noted in cell permissivity
Suggest that RRV lacks the Rta-inducible LANA promoter that induces latency in KSHV
Acknowledgements
We thank Blossom Damania for the full-length RRV Rta promoter construct, Ken Izutsu for the HSG cell line, and Nina Salama for the AGS cell line. We gratefully acknowledge Kellie Burnside for cloning the Flag-tagged LANA expression constructs and Jacques Garrigues for demonstrating the latent KSHV infections in AGS cells. This work was supported by the National Center for Research Resources (NCRR) and the Office of Research Infrastructure Programs (ORIP) of the National Institutes of Health (NIH) through Grant RR023343 (to. T.M.R.). L.K.D. was supported by a training grant from the University of Washington STD/AIDS Research Training Program T32AI007140-34.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Laura K. DeMaster, Email: demaster@mail.med.upenn.edu.
Timothy M. Rose, Email: timothy.rose@seattlechildrens.org.
References
- 1.Alexander L, Denekamp L, Knapp A, Auerbach MR, Damania B, Desrosiers RC. The primary sequence of rhesus monkey rhadinovirus isolate 26–95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J Virol. 2000;74:3388–3398. doi: 10.1128/jvi.74.7.3388-3398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barranco SC, Townsend CM, Jr., Casartelli C, Macik BG, Burger NL, Boerwinkle WR, Gourley WK. Establishment and characterization of an in vitro model system for human adenocarcinoma of the stomach. Cancer Res. 1983;43:1703–1709. [PubMed] [Google Scholar]
- 3.Bechtel JT, Winant RC, Ganem D. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. J Virol. 2005;79:4952–4964. doi: 10.1128/JVI.79.8.4952-4964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe. 2009;6:570–575. doi: 10.1016/j.chom.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruce AG, Bakke AM, Gravett CA, DeMaster LK, Bielefeldt-Ohmann H, Burnside KL, Rose TM. The ORF59 DNA polymerase processivity factor homologs of Old World primate RV2 rhadinoviruses are highly conserved nuclear antigens expressed in differentiated epithelium in infected macaques. Virol J. 2009;6:205. doi: 10.1186/1743-422X-6-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruce AG, Bakke AM, Thouless ME, Rose TM. Development of a real-time QPCR assay for the detection of RV2 lineage-specific rhadinoviruses in macaques and baboons. Virol J. 2005;2:2. doi: 10.1186/1743-422X-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruce AG, Bielefeldt-Ohmann H, Barcy S, Bakke AM, Lewis P, Tsai CC, Murnane RD, Rose TM. Macaque homologs of EBV and KSHV show uniquely different associations with simian AIDS-related lymphomas. PLoS Pathog. 2012;8:el002962. doi: 10.1371/journal.ppat.1002962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnside KL, Ryan JT, Bielefeldt-Ohmann H, Gregory Bruce A, Thouless ME, Tsai CC, Rose TM. RFHVMn ORF73 is structurally related to the KSHV ORF73 latency-associated nuclear antigen (LANA) and is expressed in retroperitoneal fibromatosis (RF) tumor cells. Virology. 2006;354:103–115. doi: 10.1016/j.virol.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 9.Chang PJ, Chen LW, Shih YC, Tsai PH, Liu AC, Hung CH, Liou JY, Wang SS. Role of the cellular transcription factor YY1 in the latent-lytic switch of Kaposi's sarcoma-associated herpesvirus. Virology. 2011;413:194–204. doi: 10.1016/j.virol.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Cohen A, Wolf DG, Guttman-Yassky E, Sarid R. Kaposi's sarcoma-associated herpesvirus: clinical, diagnostic, and epidemiological aspects. Crit Rev Clin Lab Sci. 2005;42:101–153. doi: 10.1080/10408360590913524. [DOI] [PubMed] [Google Scholar]
- 11.Condreay JP, Witherspoon SM, Clay WC, Kost TA. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. Proc Natl Acad Sci U S A. 1999;96:127–132. doi: 10.1073/pnas.96.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damania B, Jeong JH, Bowser BS, DeWire SM, Staudt MR, Dittmer DP. Comparison of the Rta/Orf50 transactivator proteins of gamma-2-herpesviruses. J Virol. 2004;78:5491–5499. doi: 10.1128/JVI.78.10.5491-5499.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Poorter JJ, Lipinski KS, Nelissen RG, Huizinga TW, Hoeben RC. Optimization of short-term transgene expression by sodium butyrate and ubiquitous chromatin opening elements (UCOEs) J Gene Med. 2007;9:639–648. doi: 10.1002/jgm.1057. [DOI] [PubMed] [Google Scholar]
- 14.Desrosiers RC, Sasseville VG, Czajak SC, Zhang X, Mansfield KG, Kaur A, Johnson RP, Lackner AA, Jung JU. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeWire SM, Damania B. The latency-associated nuclear antigen of rhesus monkey rhadinovirus inhibits viral replication through repression of Orf50/Rta transcriptional activation. J Virol. 2005;79:3127–3138. doi: 10.1128/JVI.79.5.3127-3138.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeWire SM, McVoy MA, Damania B. Kinetics of expression of rhesus monkey rhadinovirus (RRV) and identification and characterization of a polycistronic transcript encoding the RRV Orf50/Rta, RRV R8, and R8.1 genes. J Virol. 2002;76:9819–9831. doi: 10.1128/JVI.76.19.9819-9831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeWire SM, Money ES, Krall SP, Damania B. Rhesus monkey rhadinovirus (RRV): construction of a RRV-GFP recombinant virus and development of assays to assess viral replication. Virology. 2003;312:122–134. doi: 10.1016/s0042-6822(03)00195-8. [DOI] [PubMed] [Google Scholar]
- 18.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dittmer DP, Gonzalez CM, Vahrson W, DeWire SM, Hines-Boykin R, Damania B. Whole-genome transcription profiling of rhesus monkey rhadinovirus. J Virol. 2005;79:8637–8650. doi: 10.1128/JVI.79.13.8637-8650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fojas de Borja P, Collins NK, Azizkhan-Clifford Du J, Mudryj M. Cyclin A-CDK phosphorylates Sp1 and enhances Spl-mediated transcription. Embo J. 2001;20:5737–5747. doi: 10.1093/emboj/20.20.5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrigues HJ, Rubinchikova YE, Dipersio CM, Rose TM. Integrin alphaVbeta3 Binds to the RGD motif of glycoprotein B of Kaposi's sarcoma-associated herpesvirus and functions as an RGD-dependent entry receptor. J Virol. 2008;82:1570–1580. doi: 10.1128/JVI.01673-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gelman IH, Silverstein S. Herpes simplex virus immediate-early promoters are responsive to virus and cell trans-acting factors. J Virol. 1987;61:2286–2296. doi: 10.1128/jvi.61.7.2286-2296.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gradoville L, Gerlach J, Grogan E, Shedd D, Nikiforow S, Metroka C, Miller G. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J Virol. 2000;74:6207–6212. doi: 10.1128/jvi.74.13.6207-6212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greensill J, Sheldon JA, Murthy KK, Bessonette JS, Beer BE, Schulz TF. A chimpanzee rhadinovirus sequence related to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8: increased detection after HIV-1 infection in the absence of disease. AIDS (London, England) 2000;14:F129–F135. doi: 10.1097/00002030-200012010-00001. [DOI] [PubMed] [Google Scholar]
- 25.Greensill J, Sheldon JA, Renwick NM, Beer BE, Norley S, Goudsmit J, Schulz TF. Two distinct gamma-2 herpesviruses in African green monkeys: a second gamma-2 herpesvirus lineage among old world primates? J Virol. 2000;74:1572–1577. doi: 10.1128/jvi.74.3.1572-1577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunther T, Grundhoff A. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 2010;6:el000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison SM, Whitehouse A. Kaposi's sarcoma-associated herpesvirus (KSHV) Rta and cellular HMGB1 proteins synergistically transactivate the KSHV ORF50 promoter. FEBS Lett. 2008;582:3080–3084. doi: 10.1016/j.febslet.2008.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilton IB, Dittmer DP. Quantitative analysis of the bidirectional viral G-protein-coupled receptor and lytic latency-associated nuclear antigen promoter of Kaposi's sarcoma-associated herpesvirus. J Virol. 2012;86:9683–9695. doi: 10.1128/JVI.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katano H, Sata T, Suda T, Nakamura T, Tachikawa N, Nishizumi H, Sakurada S, Hayashi Y, Koike M, Iwamoto A, Kurata T, Mori S. Expression and antigenicity of human herpesvirus 8 encoded ORF59 protein in AIDS-associated Kaposi's sarcoma. J Med Virol. 1999;59:346–355. doi: 10.1002/(sici)1096-9071(199911)59:3<346::aid-jmv15>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 30.Kim YH, Park JW, Lee JY, Kwon TK. Sodium butyrate sensitizes TRAIL-mediated apoptosis by induction of transcription from the DR5 gene promoter through Sp1 sites in colon cancer cells. Carcinogenesis. 2004;25:1813–1820. doi: 10.1093/carcin/bgh188. [DOI] [PubMed] [Google Scholar]
- 31.Krishnan HH, Naranatt PP, Smith MS, Zeng L, Bloomer C, Chandran B. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J Virol. 2004;78:3601–3620. doi: 10.1128/JVI.78.7.3601-3620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lacoste V, Mauclere P, Dubreuil G, Lewis J, Georges-Courbot MC, Gessain A. KSHV-like herpesviruses in chimps and gorillas. Nature. 2000;407:151–152. doi: 10.1038/35025145. [DOI] [PubMed] [Google Scholar]
- 33.Lacoste V, Mauclere P, Dubreuil G, Lewis J, Georges-Courbot MC, Gessain A. A novel gamma 2-herpesvirus of the Rhadinovirus 2 lineage in chimpanzees. Genome Res. 2001;11:1511–1519. doi: 10.1101/gr.158601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lacoste V, Mauclere P, Dubreuil G, Lewis J, Georges-Courbot MC, Rigoulet J, Petit T, Gessain A. Simian homologues of human gamma-2 and betaherpesviruses in mandrill and drill monkeys. J Virol. 2000;74:11993–11999. doi: 10.1128/jvi.74.24.11993-11999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lan K, Kuppers DA, Verma SC, Robertson ES. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J Virol. 2004;78:6585–6594. doi: 10.1128/JVI.78.12.6585-6594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lan K, Kuppers DA, Verma SC, Sharma N, Murakami M, Robertson ES. Induction of Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen by the lytic transactivator RTA: a novel mechanism for establishment of latency. J Virol. 2005;79:7453–7465. doi: 10.1128/JVI.79.12.7453-7465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lania L, Majello B, De Luca P. Transcriptional regulation by the Sp family proteins. Int J Biochem Cell Biol. 1997;29:1313–1323. doi: 10.1016/s1357-2725(97)00094-0. [DOI] [PubMed] [Google Scholar]
- 38.Lin SF, Robinson DR, Oh J, Jung JU, Luciw PA, Kung HJ. Identification of the bZIP and Rta homologues in the genome of rhesus monkey rhadinovirus. Virology. 2002;298:181–188. doi: 10.1006/viro.2002.1490. [DOI] [PubMed] [Google Scholar]
- 39.Lu F, Zhou J, Wiedmer A, Madden K, Yuan Y, Lieberman PM. Chromatin remodeling of the Kaposi's sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency. J Virol. 2003;77:11425–11435. doi: 10.1128/JVI.77.21.11425-11435.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J, Verma Q, Cai Q, Robertson ES. The single RBP-Jkappa site within the LANA promoter is crucial for establishing Kaposi's sarcoma-associated herpesvirus latency during primary infection. J Virol. 2011;85:6148–6161. doi: 10.1128/JVI.02608-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukac DM, Garibyan L, Kirshner JR, Palmeri D, Ganem D. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J Virol. 2001;75:6786–6799. doi: 10.1128/JVI.75.15.6786-6799.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lukac DM, Kirshner JR, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukac DM, Renne R, Kirshner JR, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 44.Majumdar G, Harmon A, Candelaria R, Martinez-Hernandez A, Raghow R, Solomon SS. O-glycosylation of Sp1 and transcriptional regulation of the calmodulin gene by insulin and glucagon. Am J Physiol Endocrinol Metab. 2003;285:E584–E591. doi: 10.1152/ajpendo.00140.2003. [DOI] [PubMed] [Google Scholar]
- 45.Matsumura S, Fujita Y, Gomez E, Tanese N, Wilson AC. Activation of the Kaposi's sarcoma-associated herpesvirus major latency locus by the lytic switch protein RTA (ORF50) J Virol. 2005;79:8493–8505. doi: 10.1128/JVI.79.13.8493-8505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merchant JL, Du M, Todisco A. Spl phosphorylation by Erk 2 stimulates DNA binding. Biochem Biophys Res Commun. 1999;254:454–461. doi: 10.1006/bbrc.1998.9964. [DOI] [PubMed] [Google Scholar]
- 47.Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 48.Nagaoka M, Shiraishi Y, Sugiura Y. Selected base sequence outside the target binding site of zinc finger protein Spl. Nucleic Acids Res. 2001;29:4920–4929. doi: 10.1093/nar/29.24.4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nellesen DT, Lai EC, Posakony JW. Discrete enhancer elements mediate selective responsiveness of enhancer of split complex genes to common transcriptional activators. Dev Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- 50.Omotehara F, Kawamata H, Uchida D, Hino S, Nakashiro K, Fujimori T. Vesnarinone, a differentiation inducing drug, directly activates p21(wafl) gene promoter via Sp1 sites in a human salivary gland cancer cell line. Br J Cancer. 2002;87:1042–1046. doi: 10.1038/sj.bjc.6600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pearce M, Matsumura S, Wilson AC. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi's sarcoma-associated herpesvirus originate from a common promoter. J Virol. 2005;79:14457–14464. doi: 10.1128/JVI.79.22.14457-14464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinto-Santini DM, Salama NR. Cag3 is a novel essential component of the Helicobacter pylori Cag type IV secretion system outer membrane subcomplex. J Bacteriol. 2009;191:7343–7352. doi: 10.1128/JB.00946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Renne R, Barry C, Dittmer D, Compitello N, Brown PO, Ganem D. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J Virol. 2001;75:458–468. doi: 10.1128/JVI.75.1.458-468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson BA, O'Connor MA, Li H, Engelmann F, Poland B, Grant R, DeFilippis V, Estep RD, Axthelm MK, Messaoudi I, Wong SW. Viral interferon regulatory factors are critical for delay of the host immune response against rhesus macaque rhadinovirus infection. J Virol. 2012;86:2769–2779. doi: 10.1128/JVI.05657-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rose TM, Strand KB, Schultz ER, Schaefer G, Rankin GW, Jr., Thouless ME, Tsai CC, Bosch ML. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakakibara S, Ueda K, Chen J, Okuno T, Yamanishi K. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J Virol. 2001;75:6894–6900. doi: 10.1128/JVI.75.15.6894-6900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sarid R, Olsen SJ, Moore PS. Kaposi's sarcoma-associated herpesvirus: epidemiology, virology, and molecular biology. Adv Virus Res. 1999;52:139–232. doi: 10.1016/s0065-3527(08)60299-7. [DOI] [PubMed] [Google Scholar]
- 58.Sarid R, Wiezorek JS, Moore PS, Chang Y. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol. 1999;73:1438–1446. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schultz ER, Rankin GW, Jr., Blanc MP, Raden BW, Tsai CC, Rose TM. Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi's sarcoma-associated herpesvirus. J Virol. 2000;74:4919–4928. doi: 10.1128/jvi.74.10.4919-4928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Searles RP, Bergquam EP, Axthelm MK, Wong SW. Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shirasuna K, Sato M, Miyazaki T. A neoplastic epithelial duct cell line established from an irradiated human salivary gland. Cancer. 1981;48:745–752. doi: 10.1002/1097-0142(19810801)48:3<745::aid-cncr2820480314>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 62.Song MJ, Li X, Brown HJ, Sun R. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 2002;76:5000–5013. doi: 10.1128/JVI.76.10.5000-5013.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Speck SH, Ganem D. Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe. 2010;8:100–115. doi: 10.1016/j.chom.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Talbot SJ, Weiss RA, Kellam P, Boshoff C. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka J, Sadanari H, Sato H, Fukuda S. Sodium butyrate-inducible replication of human cytomegalovirus in a human epithelial cell line. Virology. 1991;185:271–280. doi: 10.1016/0042-6822(91)90774-6. [DOI] [PubMed] [Google Scholar]
- 67.Wang SE, Wu FY, Chen H, Shamay M, Zheng Q, Hayward GS. Early activation of the Kaposi's sarcoma-associated herpesvirus RTA, RAP, and MTA promoters by the tetradecanoyl phorbol acetate-induced AP1 pathway. J Virol. 2004;78:4248–4267. doi: 10.1128/JVI.78.8.4248-4267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang SE, Wu FY, Yu Y, Hayward GS. CCAAT/enhancer-binding protein-alpha is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J Virol. 2003;77:9590–9612. doi: 10.1128/JVI.77.17.9590-9612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wen KW, Dittmer DP, Damania B. Disruption of LANA in rhesus rhadinovirus generates a highly lytic recombinant virus. J Virol. 2009;83:9786–9802. doi: 10.1128/JVI.00704-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson SJ, Tsao EH, Webb BL, Ye H, Dalton-Griffin L, Tsantoulas C, Gale CV, Du MQ, Whitehouse A, Kellam P. X box binding protein XBP-ls transactivates the Kaposi's sarcoma-associated herpesvirus (KSHV) ORF50 promoter, linking plasma cell differentiation to KSHV reactivation from latency. J Virol. 2007;81:13578–13586. doi: 10.1128/JVI.01663-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang J, Kawai Y, Hanson RW, Arinze IJ. Sodium butyrate induces transcription from the G alpha(i2) gene promoter through multiple Sp1 sites in the promoter and by activating the MEK-ERK signal transduction pathway. J Biol Chem. 2001;276:25742–25752. doi: 10.1074/jbc.M102821200. [DOI] [PubMed] [Google Scholar]
- 72.Yang X, Su K, Roos MD, Chang Q, Paterson AJ, Kudlow JE. 0-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc Natl Acad Sci U S A. 2001;98:6611–6616. doi: 10.1073/pnas.111099998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ye J, Gradoville L, Daigle D, Miller G. De novo protein synthesis is required for lytic cycle reactivation of Epstein-Barr virus, but not Kaposi's sarcoma-associated herpesvirus, in response to histone deacetylase inhibitors and protein kinase C agonists. J Virol. 2007;81:9279–9291. doi: 10.1128/JVI.00982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ye J, Shedd D, Miller G. An Sp1 response element in the Kaposi's sarcoma-associated herpesvirus open reading frame 50 promoter mediates lytic cycle induction by butyrate. J Virol. 2005;79:1397–1408. doi: 10.1128/JVI.79.3.1397-1408.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu F, Feng J, Harada JN, Chanda SK, Kenney SC, Sun R. B cell terminal differentiation factor XBP-1 induces reactivation of Kaposi's sarcoma-associated herpesvirus. FEBS Lett. 2007;581:3485–3488. doi: 10.1016/j.febslet.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 76.Zhang C, Wang Y, Zhou Z, Zhang J, Tian Z. Sodium butyrate upregulates expression of NKG2D ligand MICA/B in HeLa and HepG2 cell lines and increases their susceptibility to NK lysis. Cancer Immunol Immunother. 2009;58:1275–1285. doi: 10.1007/s00262-008-0645-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.