Abstract
Bladder cancer is a relatively common and strikingly costly malignancy. Here, we will focus on recent advances in our understanding of the molecular pathogenesis of metastatic bladder cancer, a stage of this disease curable in only a minority of patients. Our group has recently investigated the role of a class of small G-proteins known as the Ras-like or Ral GTPases and their role in this disease. These signaling proteins, regulated by the Ras pathway and other mechanisms, have been shown to be necessary for key cellular phenotypes associated with transformation or cancer progression in diverse cancer systems. In bladder cancer we have observed that these GTPases are overexpressed, are necessary for key phenotypes in models of bladder cancer progression, and finally, are essential for the regulation of expression of key molecules, including the prognostic marker and cell surface GPI-linked glycoprotein, CD24. These findings are reviewed here and suggest that Ral GTPases and their downstream pathways constitute key mediators of bladder cancer progression and may include targets for future therapeutic strategies.
Introduction
An early review of the role of Ral GTPases in cellular biology suggested that, consistent with pop-artist Andy Warhol's immortal words, the time might be coming for these molecules to have their “15 minutes of fame” [1]. That the time of Ral GTPases has come appears now to be quite indisputable, with multiple reports finding important and general functions of these GTPases in diverse cellular processes and phenotypes. Herein, we hope to review key findings that provide understanding to the role of Ral GTPases in the pathogenesis and potential future treatment of human bladder cancer.
Bladder cancer is the 5th most commonly diagnosed cancer in the United States and most expensive cancer to treat per patient. Recent reviews have estimated the societal cost of bladder cancer at 3.7 billion dollars in the U.S. alone, with a cost per patient ranging from 96,000-187,000 dollars [2]. Cost aside, studies show that bladder cancer is the fourth most common cause of deaths from cancer. Many of these deaths are of patients initially diagnosed with muscle invasive disease but also arise in patients previously diagnosed with refractory superficial bladder cancer [3]. Most patients diagnosed with metastatic disease die within two years of diagnosis [4]. Because of the substantial mortality associated with invasive and metastatic bladder cancer, our group has focused our research at this stage of disease. We have uncovered cell regulators that must be lost for progression to metastatic stage disease, including the metastasis-suppressor gene RhoGDI2 [5-9], but we have also studied cell regulators that drive cancer progression, converging on the Ral GTPase pathway [10, 11].
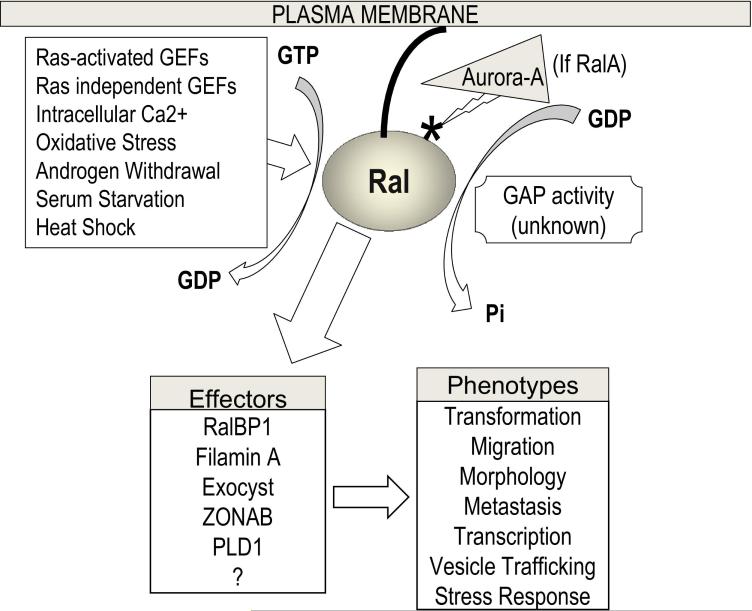
Ral GTPases were originally cloned through their homology to the Ras oncogene [1]. These two GTPase paralogs constitute a distinct subfamily of the Ras family of small G proteins, and exhibit >90% similarity and >80% identity to each other on the level of amino acid sequence. As is the case with other G-proteins, or GTP binding proteins, RalA and RalB function as GTPase switches and possess intrinsic GTPase activity. Their signaling is stimulated by GTP exchange for GDP, commonly by molecules known as GEFs or guanidine nucleotide exchange factors, recruited by activated upstream Ras or other means. Once activated, Ral GTPases mediate downstream signaling through diverse effectors, including RalBP1, exocyst components (Sec5 and Exo84), Filamin A, ZONAB, and PLD1 (reviewed in [1]). Through these and other uncharacterized effectors, Ral signaling has been shown to affect cellular processes as diverse as exocytosis and endocytosis, as well as cellular motility, proliferation, cytoskeletal organization, and transformation (Figure 1).
Figure 1. The regulation of Ral and downstream signaling.
RalGTPases may be activated by a variety of stimuli, including guanidine nucleotide exchange factors (RalGEFs) activated by upstream activation of Ras or independent of Ras activation. Also, several other stimuli have been identified that activate Ral, perhaps independently of either of these factors. These include processes known to be hyperactive in cancer cells, including oxidative damage, calcium signaling, and androgen withdrawal [1]. Once activated by exchange of GDP for GTP, Ral GTPases are able to associate physically with several known (and likely other uncharacterized) effector proteins, to mediate diverse cellular effects.
Proceeding from observations of association of overexpression of EGFR in invasive stage bladder cancer [12], we initially screened for what downstream pathways impinged on EGFR-stimulated motility of the invasive but non-metastatic T24 bladder cancer cell line [13]. We observed that interruption of the Ral pathway using dominant negative mutants, in contrast to other pathways downstream of the EGFR, reduced cellular motility, for the first time implicating these GTPases in bladder cancer invasion and progression [14]. Concurrently, other groups discovered that the Ral pathway is indispensable for mediating Ras-dependent transformation of human cells of various tissues, including the observation that RalA appears to be the paralog in this family necessary for such function [15]. Finally, a growing body of literature has implicated Ral in regulating several transcription factor pathways, some known to be essential in cancer. These include through NFκB, AP-1, c-Jun, forkhead family members, HSF-1, ZONAB and RREB1 (reviewed in [1]).
Given this foundation, recent reports have started to answer key questions regarding the role for Ral in cancer progression and metastasis. Our group has focused on bladder cancer, with the general approach of using molecular analyses of human bladder tumor tissues and cell lines to guide laboratory investigation. In so doing, we have uncovered intriguing aspects of the role of Ral in the biology of bladder cancer, progression, and metastasis. Here, we summarize these observations and those of other groups that make the case that Ral may be an important new target for future therapeutic modalities.
Status of the Ral Pathway in human tumors, including bladder cancer
Despite the observations of roles for RalA and RalB downstream of the Ras oncogene in the transformation of human cell lines, until recently, very little was known of the expression or mutation state of these key molecules in actual human tissues. Indeed, though reports have shown that Ral GTPases may be activated by oncogenic Ras, such mutations occur quite rarely in tumors, including bladder cancer, begging the question of how this pathway might be dysregulated in vivo. To this end, we surveyed the status of this pathway in human bladder cancer, on the level of mutation, activation, and expression in human bladder tumor tissues and cell lines [16].
First, we sequenced RalA and RalB coding regions from 20 different human bladder cancer-derived cell lines, including many known to be invasive and metastatic by virtue or derivation or xenograft studies. However, we only found a single mutation of RalA in these cell lines: an E97Q substitution mutation in the cell line, UM-UC-6, that did not affect the intrinsic GTP-binding activity of this protein. Though this observation of a lack of mutation of Ral in bladder cancer cell lines does not exclude that they may occur at some frequency in vivo in human tumors, it does tend to suggest that mutation does not occur at a high frequency. Moreover, it begs the question of how Ral GTPase signaling may be increased in vivo, as when we examined the activation state of RalA and RalB in 10 different bladder cancer cell lines, we found substantial levels of activated Ral in all of them.
When we examined the mRNA and protein expression of RalA and RalB in bladder cancer tissues, we were surprised to discover that overexpression of these GTPases may constitute an important part of their dysregulation. We observed in a microarray series containing 80 different human bladder cancers of differing stage and grade, that RalA mRNA is overexpressed, as a function of transformation (cancer versus normal) and stage (T2+ cancer versus superficial cancer). In contrast, it does not appear that RalB mRNA is substantially overexpressed in bladder cancer. On the other hand, to determine the reliability of Ral mRNA measures as a surrogate of protein expression, we correlated RalB mRNA expression to RalB protein expression across 28 cell lines and found a very low degree of concordance. Consistent with this disconnect, we found that both RalA and RalB protein were overexpressed in invasive stage disease as determined by immunoblotting human bladder tumors.
Finally, we examined Ral effector proteins and found that two canonical direct Ral effectors, RalBP1 and Filamin A, were strongly differentially expressed in invasive disease. RalBP1 mRNA was substantially overexpressed, while RalBP1 protein expression, detected by immunoblotting of homogenized tissues, went from nearly undetectable to strongly expressed. In contrast, Filamin A mRNA and protein expression was substantially downregulated in tumors.
Scanning the Oncomine database of published microarray studies [17], we found other tumor types also exhibit RalA overexpression, including seminoma, glioblastoma, hepatoma, pancreatic adenocarcinoma, and prostate adenocarcinoma. Finally, another recent report, which performed an integrative genomic and proteomic analysis of prostate cancer progression, found that RalA was overexpressed at both the RNA and protein level in metastatic prostate cancer [18]. Taken together, these results suggest that one mechanism for increasing Ral signaling in the cancer milieu is overexpression of these proteins in tumor tissue themselves.
A Role for Ral in Bladder Cancer Cellular Migration and Metastasis
Two reports initially implicated Ral in cellular migration. First, Suzuki et al found that activation of the Ral pathway by either Ral pathway-specific mutants of activated mutant Ras, constitutively active mutants of a RalGEF, or constitutively activated mutant Ral stimulated increased motility of skeletal myoblasts in migration assays [19]. Additionally, another group found that through a direct interaction with the cytoskeletal protein, Filamin A, RalA could trigger filopodial formation [20]. Specific to bladder cancer, we initially implicated Ral in motility of human bladder cancer by finding that transfection of dominant negative mutant Ral constructs results in substantial abrogation of T24 bladder cancer cell motility [14]. However, overexpression of these dominant negative mutants disrupts both RalA and RalB function by forming extensive unproductive complexes with RalGEFs that are necessary for GTPase loading, suggesting that that this finding is unlikely to be specific to RalA or RalB function [1]. With this limitation in mind, we determined that RalA and RalB have different functions in serum-stimulated motility of bladder cancer cells, finding that siRNA-mediated depletion of RalB, as well as transfection of constitutively-active mutant RalA, inhibit bladder cancer cellular migration [11]. The observation of this indispensable role for RalB in cellular motility was confirmed by another group, who found in rodent cells that RalB is necessary for wound closure migration, and that it is associated with formation of a RalB-Sec5 complex and resultant direction of exocyst components to the leading edge of migrating cells [21].
A Role for Ral in metastasis
Recent reports suggest that these intriguing results from in vitro surrogate models for metastasis may translate into a real functional role for RalA, and particularly, RalB in metastasis. First, Yin et al. determined that the activation of the Ral pathway by either Ral pathway-specific mutants of oncogenic Ras or a constitutively active RalGEF (both of which cause strong activation of Ral) were sufficient to produce bone metastasis of the prostate cancer cell line, DU145 in nude mice [22]. By contrast, stable depletion of RalA or RalB using RNA interference or by expression of a dominant negative RalB construct resulted in substantial abrogation of metastasis of these cells. Moreover, using pancreatic cancer cell lines stably expressing RNA interference for RalA and RalB, Lim et al. elegantly showed that both RalA and RalB have key roles in metastasis [23]. RalA is necessary for tumor growth because it is necessary for transformed and tumorigenic growth of pancreatic cell lines in any setting. RalB, though apparently not essential for tumorigenic growth, is nonetheless indispensable for invasion of most pancreatic cell lines and for pulmonary metastasis from tail vein injected cells. Given our prior results for RalB in human bladder cancer cellular phenotypes of migration and proliferation, we are currently examining the relative roles for activated RalA and RalB in a metastatic model of human bladder cancer. The ultimate question remains how RalB mediates such a function, as well as determination of what effector(s) are necessary for RalB to exhibit this function.
A Role for Ral-dependent Transcriptional Targets in Bladder Cancer?
A growing body of literature suggests that a key mechanism for Ral function may be through regulation of transcription. Indeed, transcriptional signaling pathways converging on NFκB, AP-1, c-Jun, forkhead family members, HSF-1, and ZONAB, have been shown to be regulated by Ral [1, 24]. Interestingly, though none were performed in bladder cancer cell lines, some of these studies involved, targets that are already known to be key players in bladder cancer. For example, Henry et al. found that activation of Ral causes activation and nuclear accumulation of NFκB and resultant induction of cyclin D1, known to be overexpressed in bladder cancer [25]. Okan et al found that activation of RalA was sufficient to induce expression of the urokinase plasminogen activator receptor (uPAR) in an AP-1 site dependent manner [26], while uPAR itself has been associated with invasion of bladder cancer. Finally, one group had found that activation of RalA results in transcriptional induction of VEGF-C, itself a prognostic factor for bladder cancer progression [27].
In an effort to determine more globally what quantity and types of transcripts are regulated by the expression of Ral GTPases, we evaluated human bladder cancer cells, depleted by siRNAs of RalA, RalB, or both GTPases, and analyzed these by oligonucleotide microarray [28]. From the standpoint of probe sets regulated uniquely by depletion of one Ral paralog or another, we found that approximately 80 genes were regulated uniquely by RalA or RalB, while a many more were regulated by both RalA and RalB deletion or by simultaneous depletion of both GTPases. Interestingly, these targets included several Ral-regulated targets identified in unrelated cell culture systems in the aforementioned papers, including cyclin D1, uPAR, and VEGF-C, suggesting that Ral's role in the regulation of these key molecules may be a general phenomenon.
One important Ral-dependent transcriptional target that we have examined is the cell surface GPI-linked glycoprotein, CD24, a molecule whose overexpression has been associated with invasion, metastasis, poor prognosis in immunohistochemical studies of diverse tumor types [29]. Functionally CD24 has been implicated in cellular invasion and metastasis in vivo [30], and recent reports suggest that CD24 may prove to be a marker for highly tumorigenic stem cell-like populations in human cancer [31]. We found that depletion of RalB, or particularly of both RalA and RalB in bladder and other cancer cells resulted in downregulation of CD24 [32]. Moreover, expression of CD24 itself was necessary for proliferation, anchorage independence, and survival of cancer cells, making a case that this Ral target may be essential across tumor types.
Finally, by integrating microarray expression studies of human bladder tumor tissue with the Ral-dependent transcriptional targets empirically discovered by the siRNA-microarray study, we were able to examine the extent to which Ral-dependent targets appear to be implicated in the global transcriptional dysregulation of human bladder cancer. Comparing genes significantly regulated by RalA, RalB, or both RalA and RalB, to genes highly significantly dysregulated in human bladder cancer microarray studies, we found in all cases a highly significant enrichment of Ral-dependent targets in genes dysregulated in bladder cancer. Nearly 1 in 7 probe sets differentially expressed in human bladder cancer were RalA or RalB-dependent transcriptional targets. This highly significant enrichment of RalA and RalB targets strongly suggests that transcriptional pathways downstream from these GTPases are essential in the pathogenesis of bladder cancer [28].
Targeting the Ral Pathway in Human Bladder Cancer?
Though the future might hold many novel strategies for targeting these molecules, at present we identify several mechanisms that might prove to be efficacious in targeting Ral function. First, RalA-dependent transformation of human cells has been shown to be dependent on phosphorylation of RalA on serine 194 [33], and perhaps additionally, serine 183 [34]. Aurora-A kinase, itself overexpressed in bladder cancer [16] has been shown to phosphorylate RalA on serine 194, though it remains unclear whether other kinases might also perform this function [33]. Pharmacologic inhibitors of Aurora kinases have been developed and thus might be able to abrogate RalA function [35].
More promising at present appears to be the potential to target Ral signaling function through targeting CD24 expressing cells. As CD24 has been shown to be a cell surface marker for poor prognosis and stem cell-like properties in tumor cells, this molecule appears to be an attractive therapeutic target. Several monoclonal antibodies against CD24 have been developed and have even been employed successfully in preclinical and clinical trials to treat the CD24-expressing cells of a post-transplant lymphoproliferative disorder [36]. Preliminarily, we have also had some success in treating nude mice inoculated subcutaneously with highly CD24+ variant cells of the UM-UC-3 cell line with anti-CD24 monoclonal antibodies (unpublished observations). If future experiments extend the utility of these experiments to other cell types and metastatic models of bladder or other cancers, this may prove to be an important line of investigation.
Conclusions
Taken together, these data suggest that the Ral pathway may mediate key functions in bladder cancer, particularly in progression and metastasis. At a very minimum, recent findings regarding the overexpression of RalA and RalB in bladder and other cancers serves to anchor prior phenotypic findings from model systems in actual pathologic samples. While our observation of a pervasive role for Ral in the dysregulation of the bladder cancer transcriptome speaks to a central role for these GTPases in signaling to the gene expression changes that define cells as cancer. One key area of future investigation will be to determine what aspect of RalA versus RalB function, whether it is the scant differences in sequence between these proteins or higher order differences such as differential post-translational modifications or subcellular localization, suffice to mediate the paralog specific functions of these proteins. Another important area will be to determine which downstream pathways—which effectors—are necessary for Ral to mediate these functions. Finally, though our understanding of Ral function in cancer is still young and incomplete, it is never too early to start identifying tractable therapeutic targets. While targeting a pathway output, like CD24, may seem promising, future strategies must likely target both RalA and RalB to disable both arms of this crucial cancer pathway at the source.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–25. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 2.Botteman MF, Pashos CL, Redaelli A, Laskin B, Hauser R. The health economics of bladder cancer: a comprehensive review of the published literature. Pharmacoeconomics. 2003;21:1315–30. doi: 10.1007/BF03262330. [DOI] [PubMed] [Google Scholar]
- 3.Malkowicz SB, van Poppel H, Mickisch G, et al. Muscle-invasive urothelial carcinoma of the bladder. Urology. 2007;69:3–16. doi: 10.1016/j.urology.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 4.Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer. 2005;5:713–25. doi: 10.1038/nrc1697. [DOI] [PubMed] [Google Scholar]
- 5.Gildea JJ, Seraj MJ, Oxford G, et al. RhoGDI2 is an invasion and metastasis suppressor gene in human cancer. Cancer Res. 2002;62:6418–23. [PubMed] [Google Scholar]
- 6.Theodorescu D, Sapinoso LM, Conaway MR, Oxford G, Hampton GM, Frierson HF., Jr. Reduced expression of metastasis suppressor RhoGDI2 is associated with decreased survival for patients with bladder cancer. Clin Cancer Res. 2004;10:3800–6. doi: 10.1158/1078-0432.CCR-03-0653. [DOI] [PubMed] [Google Scholar]
- 7.Titus B, Frierson HF, Jr., Conaway M, et al. Endothelin axis is a target of the lung metastasis suppressor gene RhoGDI2. Cancer Res. 2005;65:7320–7. doi: 10.1158/0008-5472.CAN-05-1403. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y, McRoberts K, Berr SS, Frierson HF, Jr., Conaway M, Theodorescu D. Neuromedin U is regulated by the metastasis suppressor RhoGDI2 and is a novel promoter of tumor formation, lung metastasis and cancer cachexia. Oncogene. 2006 doi: 10.1038/sj.onc.1209835. [DOI] [PubMed] [Google Scholar]
- 9.Nicholson BE, Frierson HF, Conaway MR, et al. Profiling the evolution of human metastatic bladder cancer. Cancer Res. 2004;64:7813–21. doi: 10.1158/0008-5472.CAN-04-0826. [DOI] [PubMed] [Google Scholar]
- 10.Harding MA, Arden KC, Gildea JW, et al. Functional genomic comparison of lineage-related human bladder cancer cell lines with differing tumorigenic and metastatic potentials by spectral karyotyping, comparative genomic hybridization, and a novel method of positional expression profiling. Cancer Res. 2002;62:6981–9. [PubMed] [Google Scholar]
- 11.Oxford G, Owens CR, Titus BJ, et al. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–20. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- 12.Theodorescu D, Laderoute KR, Gulding KM. Epidermal growth factor receptor-regulated human bladder cancer motility is in part a phosphatidylinositol 3-kinase-mediated process. Cell Growth Differ. 1998;9:919–28. [PubMed] [Google Scholar]
- 13.Gildea JJ, Golden WL, Harding MA, Theodorescu D. Genetic and phenotypic changes associated with the acquisition of tumorigenicity in human bladder cancer. Genes Chromosomes Cancer. 2000;27:252–63. doi: 10.1002/(sici)1098-2264(200003)27:3<252::aid-gcc5>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 14.Gildea JJ, Harding MA, Seraj MJ, Gulding KM, Theodorescu D. The role of Ral A in epidermal growth factor receptor-regulated cell motility. Cancer Res. 2002;62:982–5. [PubMed] [Google Scholar]
- 15.Lim KH, Baines AT, Fiordalisi JJ, et al. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–45. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 16.Smith SC, Oxford G, Baras AS, et al. Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin Cancer Res. 2007;13:3803–13. doi: 10.1158/1078-0432.CCR-06-2419. [DOI] [PubMed] [Google Scholar]
- 17.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki J, Yamazaki Y, Li G, Kaziro Y, Koide H. Involvement of Ras and Ral in chemotactic migration of skeletal myoblasts. Mol Cell Biol. 2000;20:4658–65. doi: 10.1128/mcb.20.13.4658-4665.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP. The small GTPase RalA targets filamin to induce filopodia. Proc Natl Acad Sci U S A. 1999;96:2122–8. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosse C, Hatzoglou A, Parrini MC, White MA, Chavrier P, Camonis J. RalB mobilizes the exocyst to drive cell migration. Mol Cell Biol. 2006;26:727–34. doi: 10.1128/MCB.26.2.727-734.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin J, Pollock C, Tracy K, et al. Activation of the RalGEF/Ral pathway promotes prostate cancer metastasis to bone. Mol Cell Biol. 2007;27:7538–50. doi: 10.1128/MCB.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim KH, O'Hayer K, Adam SJ, et al. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–94. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 24.Frankel P, Aronheim A, Kavanagh E, et al. RalA interacts with ZONAB in a cell density-dependent manner and regulates its transcriptional activity. Embo J. 2005;24:54–62. doi: 10.1038/sj.emboj.7600497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry DO, Moskalenko SA, Kaur KJ, et al. Ral GTPases contribute to regulation of cyclin D1 through activation of NF-kappaB. Mol Cell Biol. 2000;20:8084–92. doi: 10.1128/mcb.20.21.8084-8092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okan E, Drewett V, Shaw PE, Jones P. The small-GTPase RalA activates transcription of the urokinase plasminogen activator receptor (uPAR) gene via an AP1-dependent mechanism. Oncogene. 2001;20:1816–24. doi: 10.1038/sj.onc.1204260. [DOI] [PubMed] [Google Scholar]
- 27.Rinaldo F, Li J, Wang E, Muders M, Datta K. RalA regulates vascular endothelial growth factor-C (VEGF-C) synthesis in prostate cancer cells during androgen ablation. Oncogene. 2006 doi: 10.1038/sj.onc.1209971. [DOI] [PubMed] [Google Scholar]
- 28.Oxford G, Smith SC, Hampton G, Theodorescu D. Expression profiling of Ral-depleted bladder cancer cells identifies RREB-1 as a novel transcriptional Ral effector. Oncogene. 2007 doi: 10.1038/sj.onc.1210521. [DOI] [PubMed] [Google Scholar]
- 29.Kristiansen G, Sammar M, Altevogt P. Tumour biological aspects of CD24, a mucin-like adhesion molecule. J Mol Histol. 2004;35:255–62. doi: 10.1023/b:hijo.0000032357.16261.c5. [DOI] [PubMed] [Google Scholar]
- 30.Baumann P, Cremers N, Kroese F, et al. CD24 expression causes the acquisition of multiple cellular properties associated with tumor growth and metastasis. Cancer Res. 2005;65:10783–93. doi: 10.1158/0008-5472.CAN-05-0619. [DOI] [PubMed] [Google Scholar]
- 31.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 32.Smith SC, Oxford G, Wu Z, et al. The Metastasis-Associated Gene CD24 Is Regulated by Ral GTPase and Is a Mediator of Cell Proliferation and Survival in Human Cancer. Cancer Res. 2006;66:1917–22. doi: 10.1158/0008-5472.CAN-05-3855. [DOI] [PubMed] [Google Scholar]
- 33.Wu JC, Chen TY, Yu CT, et al. Identification of V23RalA-Ser194 as a critical mediator for Aurora-A-induced cellular motility and transformation by small pool expression screening. J Biol Chem. 2005;280:9013–22. doi: 10.1074/jbc.M411068200. [DOI] [PubMed] [Google Scholar]
- 34.Sablina AA, Chen W, Arroyo JD, et al. The tumor suppressor PP2A Abeta regulates the RalA GTPase. Cell. 2007;129:969–82. doi: 10.1016/j.cell.2007.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katayama H, Brinkley WR, Sen S. The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 2003;22:451–64. doi: 10.1023/a:1023789416385. [DOI] [PubMed] [Google Scholar]
- 36.Benkerrou M, Jais JP, Leblond V, et al. Anti-B-cell monoclonal antibody treatment of severe posttransplant B-lymphoproliferative disorder: prognostic factors and long-term outcome. Blood. 1998;92:3137–47. [PubMed] [Google Scholar]