Abstract
MicroRNAs (miRNAs) are evolutionary conserved small non-coding RNAs that regulate gene expression. Early studies have shown that miRNA expression is deregulated in cancer, and experimental data indicate that cancer phenotypes can be modified by targeting miRNA expression. Based on these observations, miRNA-based anticancer therapies are being developed either alone or in combination with current targeted therapies, with the goal to improve disease response and increase cure rate. The advantage of using miRNA approaches is based on the ability to concurrently target multiple effectors of pathways involved in cell differentiation, proliferation and survival. In this review, we describe the role of miRNAs in tumorigenesis, and critically discuss the rationale, strategies and challenges for therapeutic targeting of miRNAs in cancer.
The rising new world of microRNAs
Rescued from the forgotten landscape of dark genomic matter, microRNAs (miRNAs) have become rising stars in cancer genetics. MiRNAs are small non-coding RNAs of 18-25 nucleotides in length that act as expression regulators of genes involved in fundamental cell processes such as development, differentiation, proliferation, survival and death1.
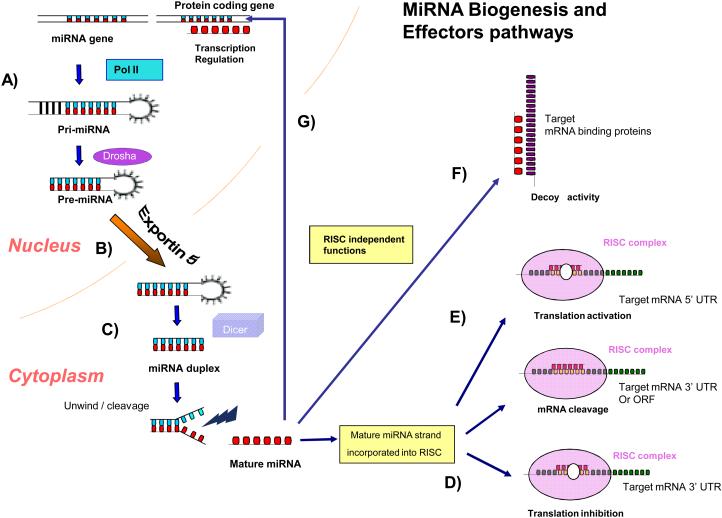
MiRNAs are mostly transcribed from intragenic or intergenic regions by RNA polymerase II into primary transcripts of variable length (usually between 1 to 3 kb), called pri-miRNAs (Figure 1)2-3. The primary transcripts undergo further processing by the ribonucleases Drosha and DiGeorge syndrome critical region gene 8 (DGCR8) complex in the nucleus, thereby resulting in a hairpin intermediate of about 70-100 nucleotides, called pre-miRNA4-5. The latter is then transported out of the nucleus to the cytoplasm by Exportin 56. In the cytoplasm, the pre-miRNA is processed by another ribonuclease, Dicer, into a mature double strand miRNA of variable length (~18-25 nucleotides)7. After strand separation, the guide strand or mature miRNA is incorporated into an RNA-induced silencing complex (RISC), while the passenger strand called “miRNA* is commonly degraded8-10. The RISC is the effector complex of the miRNA pathway and is comprised by miRNA, Argonaute (AGO) proteins (AGO 1-4) and other protein factors8-10. AGO proteins play critical role in the miRNA biogenesis, maturation and microRNA effector functions8-10. The mature strand is critical for target recognition and incorporation of specific target mRNAs to the RISC complex (Figure 1)8-10.
Figure 1. MicroRNA biogenesis and effectors pathways.
(A) miRNAs are transcribed by RNA polymerase II (pol II) into long primary miRNA transcripts of variable size (pri-miRNA), which are recognized and cleaved in the nucleus by the RNase III enzyme Drosha, resulting in a hairpin precursor form called pre-miRNA1-3. (B) Pre-miRNA is exported from the nucleus to the cytoplasm by exportin 5 and is further processed by another RNase enzyme called Dicer (C), which produces a transient 19–24-nt duplex4-7. Only one strand of the miRNA duplex (mature miRNA) is incorporated into a large protein complex called RISC (RNA-induced silencing complex)8-10. D) The mature miRNA leads RISC to cleave the mRNA or induce translational repression, depending on the degree of complementarity between the miRNA and its target8-10. While the most frequent site of interaction is the 3' UTR of the target mRNA, miRNAs have been described that bind to the open reading frame sequences as well as the 5' UTR16-17(E). This last interaction has been associated with activation rather repression17. (F) MiRNAs can also bind directly to proteins, in particular RNA binding proteins in a sequence dependent manner and prevent these proteins to bind their RNA targets. These “decoy activities” of miRNAs are RISC independent18. G) MiRNAs can also regulate gene transcription by binding directly or by modulating methylation patterns at the target gene promoter level20-22.
The specificity of miRNA targeting is defined by Watson-Crick complementarities between position 2-8 from the 5' miRNA (also known as “the seed”), with the 3' untranslated region (3' UTR) of their target mRNAs10. When miRNA and its target messenger RNA (mRNA) sequence show perfect complementarities, the RISC complex induces mRNA degradation. Should an imperfect miRNA-mRNA target pairing occur, translation into protein is blocked10. Regardless of which of these two events occur, the net result is a decrease in the amount of the proteins encoded by the mRNA targets.
Each miRNA has the potential to target a large number of genes (on average about 500 for each miRNA family)11-14. Conversely, an estimated 60% of the mRNAs have one or more evolutionary conserved sequences that are predicted to interact with miRNAs11-14. Bioinformatic analysis predicts that the 3' UTR of a single gene is frequently targeted by several different miRNAs11-12. Many of these predictions have been validated experimentally, suggesting that miRNAs might cooperate to regulate gene expression15.
Besides the aforementioned canonical mechanisms of miRNAs gene regulation through 3' UTR interactions, other “non canonical” miRNA-mediated mechanisms of mRNA expression modulation are emerging 16-21. Some miRNAs have been shown to bind to the open reading frame or the 5' UTR of the target genes and, in a few cases, they have been shown to activate rather than to inhibit gene expression16-17. Our group has recently reported that miRNAs can bind to ribonucleoproteins in a seed sequence and RISC-independent manner and then interfere with their RNA binding functions (decoy activity)18-19. Three studies have reported that miRNAs can also regulate gene expression at the transcriptional level by binding directly to the DNA (Figure 1)20-22. Overall, these data demonstrate the complexity and widespread regulation of gene expression by miRNAs that should be taken into consideration when developing miRNA-based therapies.
MicroRNAs meet cancer: a paradigm shift
Following our initial demonstration of deletion/down-regulation of miR-15a/miR-16-1 in B-cells of patients with chronic lymphocytic leukemia (CLL)23, other studies established that malignant tissues in human cancer patients exhibited distinctive miRNA expression signatures 24-25. Genome-wide profiling established that such miRNA expression signatures allowed different cancer types to be discriminated with high accuracy24-25 and the tissue of origin of poorly differentiated tumors to be identified. Messenger RNA profiles, in contrast, were highly inaccurate indicators of tissue or cancer type24.
Supporting the mechanistic involvement of miRNAs in the cancer, it was reported that selected groups of distinct miRNAs were commonly up- or down-regulated concurrently in distinct types of human neoplasia and often associated with distinct cytogenetic abnormalities25. miR-17 and miR-21 were found to be consistently up-regulated in colon, lung, stomach, prostate and pancreatic tumors, and miR-155 up-regulated in breast, lung and colon cancer25. These results have been validated over time in different cohorts of patients and similar results were found in other types of cancer as well (Table 1)26-38. In contrast, miR-29 was down-regulated in CLL, acute myeloid leukemia (AML), lung and breast cancer, rhabdomyosarcoma, cholangiocarcinoma, liver cancer and mantle cell lymphoma30,32,28,31,40-43, miR-15-a/miR-16-1 was down-regulated in CLL, prostate and pituitary adenomas23,44-45 and members of the let-7 family in lung, colon, breast, ovarian and stomach cancer28,33,31,46-51.
Table 1.
MiRNAs involved in cancer
| microRNA | Genomic Location |
Expression in patients | Deregulation Mechanism |
Function | Targets | Experimental data | Therapeutic Strategy |
|---|---|---|---|---|---|---|---|
| miR-15a/miR-16-1 | 13q31 | Down in CLL24 , prostate Cancer44 and pituitary adenomas45 |
Genomic loss24
Mutations30 Positive reg. by p5358-59 |
TS |
BCL-261
MCMCL-161 |
In vitro over-expression induces apoptosis in CLL and prostate cancer cells24,44 In vivo silencing causes CLL in mice62 |
Mimics Vector-based (viral) Drugs |
| Let-7a-2 | 11q24 | Down in lung46 ,colon33
breast31 , ovarian50 and stomach cancer27 |
Negative reg. by MYC70 | TS |
K-RAS, N-RAS46
CDK6, CDC25A123 HMGA2124 MYC64 |
In vitro over-expression reduces cell growth in lung, breast and colon cancer cells46, 48-49 In vivo over-expression reduces breast and lung tumor burden in mice 48,123 |
Mimics Vector-based (viral) Drugs |
| miR-29b-1/miR-29a miR-29b-2/miR-29c |
7q32 1q30 |
Down in NPM1 wt AML39, CLL30
Lung28 and breast cancer31, cholangiocarcinoma41, lymphoma43, hepatocarcinoma42 and rhabdomyosarcoma40 |
Genomic loss63
Negative reg. by MYC60 Positive reg. by p5359 |
TS |
MCL-1, CDK641,63
TCL-1, DNMT1125-118 DNMT3a and b119 |
In vitro over-expression induces apoptosis , inhibits cell proliferation and induces DNA hypomethylation in several cancers41,63,118-119 In vivo over-expression inhibits tumorigenicity in AML, liver and lung cancer42,63,119 |
Mimics Vector-based (viral) Drugs |
| miR-34a miR-34b and c |
1p36 11q23 |
Down in colon, lung, breast kidney, bladder cancer and melanoma cell lines126 Down in neuroblastoma34 |
Methylation reg. 65,126
Positive reg. by p53 58-59 Deletion |
TS |
CDK4, CDK6,65,127
CCNE2,-D1127-128 MET, MYC,127,129 CREB, E2F3130,131 BCl-2131 |
In vitro over-expression induce cell cycle arrest apoptosis and inhibits cell proliferation61-62 |
Mimics Vector-based (viral) Drugs |
| miR-26a | 3p22 | Down in Liver cancer93 | Negative reg. by MYC60 | TS | CCND2, CCNE293 | Restoration of miR-26 inhibits MYC-induced liver cancer93 |
Vector-based (viral) |
| miR-155 | 21q21 | Up in high risk CLL30, AML32,39
Lung28, colon33, breast cancer31 and lymphomas37-38 |
Positive reg. by NFκB118 | OG |
SHIP-1 and CEBPb(71,73) |
Over-expression in HSC induce myeloid pro- liferation and block erythropoiesis in mice72 In vivo over-expression in lymphocytes induce pre-B lymphoma/Leukemia71 |
Antisense oligos miR-MASK Sponges, Drugs |
| miR-17-92 | 13q22 | Up in lung28, breast31, colon33, and stomach cancer27 , myeloma36 and t(11q23) AML132 |
Amplification23
Transcription (E2F and MYC133) |
OG |
BIM, PTEN27,70
CDKN1A27 |
Cooperate with C-MYC to induce lymphoma73
In vivo over-expression in lymphocytes induce lymphoid proliferation and autoimmmunity72 |
Antisense oligos miR-MASK Sponges, Drugs |
| miR-21 | 17q23 | Up in pancreas33, breast31, Lung28, prostate, and stomach cancer 23 , CLL30, AML32, myeloma39 and gliobastoma34 |
Positive reg, by IL-6
and Gf1a134-135 |
OG |
PDCD4, PTEN67-68
TPM1136 |
In vitro silencing enhances apoptosis in glioblastoma, lung, breast and heaptocarcinoma cell lines66-70 |
Antisense oligos miR-MASK Sponges, Drugs |
| miR-372 miR-373 |
19q13 | Up in Testicular germ cell tumors and in breast cancer31,130 |
? | OG | LATS2137 | Neutralize p53 pathway in vitro137
In vivo over-expression stimulated cancer cell invasion130 |
Antisense oligos miR-MASK Sponges, Drugs |
TS: tumor suppressor
OG: oncogene
Reg: regulated
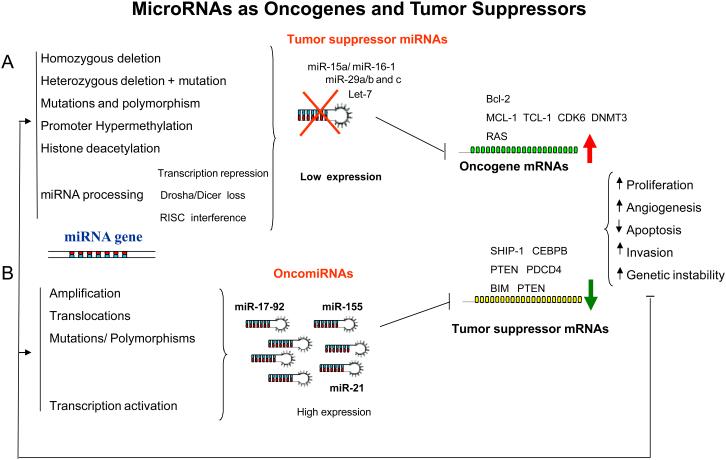
These commonalities in miRNA expression patterns suggested that deregulation of these miRNAs was unlikely to be a random event in cancer and led to the hypothesis that up-regulated miRNAs may act as oncogenes and lost miRNAs as tumor suppressors (Figure 2). Consequently, similar to coding genes involved in cancer, it was postulated that genes encoding miRNAs can be subjected to genomic alteration leading to expression up-regulation (e.g, translocations, amplification,) or loss-of-function (e.g., deletions, insertions, mutations) (Figure 2).
Figure 2. MicroRNAs as oncogenes and tumor suppressors.
(A) In this model, we propose that a miRNA that normally downregulates an oncogene can be defined as a tumor suppressor gene, and is often lost in tumor cells. The loss of function of this miRNA by mutation, deletion, promoter methylation or any abnormalities in the miRNA biogenesis might result in an abnormal expression of the target oncogene, which subsequently contributes to tumor formation by inducing cell proliferation, invasion, angiogenesis and decreased cell death. Some of the proposed mechanisms for inactivation of miRNAs in cancer are experimentally proven, such as the down-regulation of miR-15a/miR-16-1 expression in CLL patients that harbor homozygous and heterozygous deletions at 13q14.3, where the miR-15a/miR-16–1 cluster is located23 and the loss of miR-29b-1/miR-29a cluster in AML patients with 7q- (This cluster is located in 7q32)63. In addition, germ-line mutations were found in the miR-15a/miR-16–1 precursor that resulted in lower miR-15a and miR-16-1 expression levels. Overall, the loss of both miR-15a/miR-16-1 and miR-29b-1/miR-29a cluster results in up-regulation of target oncogenes like BCL-2, MCL-1, TCL-1, CDK6 and DNMT3a61,41,63,119,125. (B) The amplification or overexpression of a miRNA that downregulates a tumor suppressor or other important genes involved in differentiation might contribute to tumor formation by stimulating proliferation, angiogenesis and invasion and preventing apoptosis and increasing genetic instability. For example, amplifications of the oncogenic miRNAs, miR-17–92 cluster, miR-21 and miR-155 have been clearly associated with tumor initiation and progression by repressing the expression of tumor suppressor genes like PTEN, BIM and PDCD427,67-68,70. The impact of the aberrant miRNA expression on the transcriptome and proteome will result in increased cell proliferation, angiogenesis, invasion, anti-apoptosis and genomic instability, which in turn will damage further the genome, perpetuating a dangerous cycle. For example, increased genomic instability may predispose for more mutations that may induce cancer progression or refractoriness to treatment.
Consistent with this hypothesis, we found that miRNAs are indeed frequently located inside or close to fragile sites and in minimal regions of loss of heterozygosity, minimal regions of amplification, and common breakpoints associated with cancer52. The miR-17-92 cluster, which comprises six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92-1), is located within 800 base pairs region within the non coding gene C13orf25, a genomic region commonly found to be amplified in lymphomas. The miR-17-92 cluster has frequently been found to be over-expressed in solid tumours or hematological malignancies26-46. In contrast, the expression of the miR-15a/miR-16-1 cluster, which is located in the chromosome 13q14 region (mapping to the 30-kb deleted region between exons 2 and 5 of the non coding gene LEU2), is frequently down-regulated in patients with CLL harboring a genomic deletion of this region (Figure 2)23,30.
In addition to structural genetic alterations, silencing of structurally normal miRNAs genes by DNA promoter hypermethylation and/or histone hypoacetylation has been described in solid tumors and hematological malignancies53-55. Saito and colleagues, first demonstrated that miR-127 is silenced by promoter DNA hypermethylation and down-regulated in human bladder cancer. It is re-expressed in response to treatment with hypomethylating agents; this coincided with a down-regulation of the oncogene BCL6, which is a bona fide target of miR-127 53.
Aberrant miRNA expression in cancer may also result from downstream miRNA processing (Figure 2). Kumar et al showed that global repression of miRNA expression can be induced by short hairpin RNAs against Dicer and Drosha (the two ribonucleases involved in miRNA processing), and that this treatment promotes cellular transformation and tumorigenesis in vivo56. Furthermore, the conditional loss of Dicer 1 in mice lung tissues enhances the development of lung tumors in a K-Ras mouse model56. Loss of Dicer and Drosha has also been correlated inversely with outcome in epithelial ovarian cancers57.
Finally, a deregulation of miRNA expression can result from increased or decreased transcription from their respective miRNA genes by aberrant transcription factor activity. For example, the miR-34a, -b, and –c family of miRNAs is induced directly by the tumour suppressor TP53, and it was suggested that some of the TP53 effects could be mediated through transcriptional activation of miRNAs58-59. Using different models, the authors compared miRNA expression in cells with high or low TP53 expression and found that miR-34 expression is increased in cells with high TP53 levels58-59. Chromatin immunoprecipitation experiments revealed that TP53 binds to the promoter of miR-34 58-59. Recent work also suggests that the oncogene c-MYC negatively regulates transcription of tumor suppressor miRNAs, such as let-7 (let-7a- ,let-7f-1, let-7d, let-7c and let-7g) and miR-29 family members (-a, -b, and -c)60. Chromatin inmunoprecipitation experiments showed that c-MYC binds to conserved sequences of the miRNA promoter that it represses. Functionally, it was shown that c-MYC -induced repression of miRNAs contributes to lymphomagenesis, since the restoration of the silenced miRNAs decreases the tumorigenic potential of the lymphoma cells 60.
Nevertheless, despite the advances in our understanding of the mechanisms causing miRNA deregulation, the daunting task is to elucidate the biological role of miRNAs in the initiation and development in cancer.
Functional investigations of miRNAs in cancer
Gain- and loss-of-function experiments in combination with target prediction analyses have provided insights into the role of miRNAs in carcinogenesis. For example, miRNAs that are frequently lost in cancer, such as miR-15/miR-16 in CLL23, were ectopically expressed in leukemic cells and biological effects such as apoptosis and proliferation were investigated. These experiments revealed that the miR-15a/miR-16-1 cluster over-expression resulted in apoptosis of leukemic cells61. Target prediction programs identified BCL-2, a known anti-apoptotic gene which is up-regulated in a subset of CLL patients albeit by unknown mechanisms, as the target of miR-15-a/miR-16-161. Further work by our group showed that miR-15a/miR-16-1 directly interact with BCL-2 3'-UTR and inhibit its protein translation61. A negative correlation was also found between miR-15-a/miR-16-1 and Bcl-2 protein expression in CLL patients, supporting this interaction. Thus, the loss of miR-15-a/miR-16-1 in CLL patients by genomic deletion and mutations results in unblocking BCL-2 transcription in CLL cells (Figure 2)61. It was recently reported that miR-15-a/miR-16-1 knock out mice developed a CLL-like disease and lymphomas, further supporting a tumor suppressor role in CLL62. Other examples of miRNAs that act as tumour suppressors are listed in Table 1. Tumour suppressor miRNAs have now been shown to target oncogenes that play critical roles in various cancer pathways, such RAS (let-7)46, MCL-1 (miR-29)41,63 and MYC (let-7 and miR-34)64-65 (Table 1 and Figure 2).
In contrast, to assess the biological effects of miRNAs found to be overexpressed in cancer cells, in vitro experiments were carried out to block their expression using antisense oligonucleotides. For example, miR-21 expression has been reported at high levels in glioblastomas34, pancreas35, breast31 and colon cancer33 among others (Table 1). Chan and colleagues blocked miR-21 expression in glioblastoma cell lines and found an increased activation of caspases and apoptosis66. Further studies revealed that miR-21 exerts its antiapoptotic effects by targeting the tumor suppressors phosphatase and tensin homolog (PTEN) and programmed cell death 4 (PDCD4)67-68.
The oncogenic activity of the miR-17-92 cluster and of miR-155, both found to be over-expressed in lymphoproliferative disorders including lymphomas and leukemia69,32, were demonstrated in animal models. (Table 1 and Figure 2). Infection of murine hematopoietic stem cells with a retrovirus carrying the miR-17-92 cluster accelerated the development of lymphomas in MYC transgenic mice69. Transgenic mice that overexpressed miR-17-92 in B-cells were found to develop lymphoproliferative disease and autoimmunity70.The higher proliferation and less activation-induced cell death of lymphocytes in these mice was partially attributed to the direct targeting of the antiapoptotic genes BIM and PTEN by the miR-17-92 cluster70. Ventura and colleagues showed that mice deficient for miR-17-92 die shortly after birth with lung hypoplasia and a ventricular septal defect71. The miR-17-92 cluster is also essential for B cell development since the loss of miR-17-92 inhibits B cell development at pro-B to pre-B transition71. The contributing role of each of the six miRNAs within the miR-17-92 cluster to the oncogenic function was investigated by two different groups. Mu and colleagues found that deletion of the complete miR-17-92 cluster slows c-Myc-induced oncogenesis72. This phenotype was rescued by reintroduction of the full cluster, but not by the cluster lacking miR-19a and miR-19b, thereby suggesting miR-19 as the most important miRNA of the cluster72. Using a different approach, Olive and colleagues overexpressed individual miRNAs in the Eμ-MYC mice model. The authors found that overexpressing the whole cluster, the cluster without miR-92, but not the cluster lacking miR-19a or miR-19b promotes oncogenesis73. Further studies by both groups identified PTEN as the major target for miR-1972-73. Altogether, both studies indicate that miR-19 is critical for the oncogenic activities of this cluster.
In contrast to the miR-17-92 cluster, overexpression of miR-155 alone in the lymphoid compartment was sufficient to cause cancer and did not require any other cooperative mutation or the expression of MYC. miR-155 TG mice developed polyclonal lymphoid proliferation followed by acute lymphoblastic lymphoma/leukemia74 (Table 1 and Figure 2). To our knowledge, this was the first demonstration that the dysregulation of a single miRNA can lead to malignancy. Another group reported that the ectopic over-expression of miR-155 in hematopoietic stem cells via infection with retroviral constructs caused a myeloproliferative disorder (MPD)75. Recently, the mechanisms for these effects were discovered. Elegant experiments performed independently by two groups have shown that the Src homology-2 domain-containing inositol 5-phosphatase 1 (SHIP-1) is the target of miR-15576-77. SHIP-1 is expressed in the hematopoietic system, and by blocking the AKT pathway, it plays an important role in the differentiation of macrophages and lymphocytes78. SHIP-1 deficient mice develop a MPD characterized by increased granulocyte-monocyte populations, and decreased B-lymphocyte numbers, similar to the phenotype observed for the miR-155 over-expressing mice76. Thus, SHIP-1 repression by miR-155 seems to be one of the critical events for miR-155 induced leukemogenesis.
In addition to classical tumor suppressor or oncogene functions, miRNAs have been recently implicated in cell migration and metastasis. Ma and colleagues reported that miR-10a, which is highly expressed in metastatic breast cancer, positively regulates cell migration and invasion79. Elegant experiments confirmed that overexpressing miR-10a in non metastatic breast cancer cells initiates invasion and metastasis79. The authors showed that these effects are mediated by direct targeting of HOXD10 by miR-10a, enabling the overexpression of the well known pro-metastatic gene RhoC79. Furthermore, a recent study reported on silencing of miR-10b to inhibit metastasis in a mouse breast tumor model using a 2'-O-methyl antagomir oligonucleotide, thereby highlighting the therapeutic potential of targeting metastasis-associated80. Tavazoie and colleagues have recently reported on two miRNAs (miR-126 and miR-335) which act as negative regulators of tumor invasion and metastasis in breast and lung cancer81.
In figure 2, we present a simplified model of miRNAs acting as oncogenes and tumor suppressors. It should be stressed that the function of miRNAs depends on the expression of their critical targets. Some miRNAs could function as oncogenes in some cell types and as suppressors in others. Thus their definition of miRNAs as oncogenes or tumor suppressor genes requires an indication of the type of cells in which they act. It is anticipated that in this model will need to be refined in the near future as other potentially key aspects of miRNA biology are uncovered. It is unlikely that miRNAs will be responsible for a specific phenotype only by aiming at a single target. It is instead expected that miRNAs activity results through complex interactions with the machinery that controls the transcriptome and the concurrent targeting of multiple mRNAs.
Establishing the rationale for targeting miRNAs in cancer
The rationale for using miRNAs as anticancer drugs is based on two major findings: miRNA expression is deregulated in cancer compared to normal tissues and the cancer phenotype can be changed by targeting miRNA expression23-40,47-51. But why can miRNA-based therapeutic approaches offer an advantage over other strategies such as targeting protein expression?
Unus Pro Omnibus: "One for all”
One of the most appealing properties of miRNAs as therapeutic agents is their ability to target multiple genes, frequently within the context of a network, making them very efficient in regulating distinct biological cell processes relevant to normal and malignant cell homeostasis 59,63,70,82-84 (Figure 3). This concept was elegantly demonstrated by the Chen group, while studying T cell receptor (TCR) biology84. It is known that TCR signaling and antigen recognition are controlled by sequential phosphorylation and dephosphorylation events by more than 40 different kinases and phosphatases 84. Li and colleagues showed that miR-181 has a critical role in the regulation of TCR sensitivity and signaling strength at the posttranscriptional level by targeting multiple phosphatases. More importantly, the authors demonstrated that this task can be carried out very efficiently by miR-181a alone, but not by a single shRNA, which is designed to target individual genes84. The ability of miRNAs to regulate multiple genes within a molecular pathway makes them excellent candidates for novel molecular targeting for treatment.
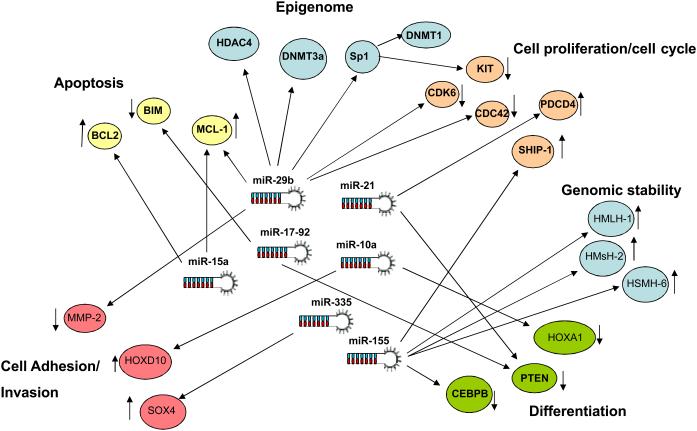
Figure 3. Transcriptome-miRNA networks in cancer.
In thiscartoon we graphically represent the relationship between critical oncogenic transcriptome networks and the miRNome. Target mRNAs for each major pathway are represented by circles with a unique color. MiRNAs are represented as hairpins structures in the center. The arrows connecting miRNAs and mRNAs indicate validated mRNA-miRNA interactions. The small arrow next to the circles indicate the biological effects on the pathway by the miRNA action on its target (i.e. miR-15a induces apoptosis by targeting BCL-2 or miR-29b suppresses cell proliferation by blocking CDK6. Some miRNAs like miR-29b, coordinately regulate multiple targets within different pathways. As shown in the cartoon, miR-29b modulates target mRNAs involved in apoptosis, cell proliferation, DNA methylation, histone acetylation and cell adhesion.
Cancer networks are miRNA wired
In cancer, as a result of multiple genetic and epigenetic events, perturbation of critical gene and protein networks occurs, resulting in malignant transformation. Apoptosis, cell cycle, cell adhesion, chromosome stability and DNA repair networks are frequently affected in carcinogenesis (Figure 3)85. Since miRNAs regulate many different pathways and orchestrate integrated responses in normal “healthy” cells and tissues, it is reasonable to think that they also play key roles in coordinating cancerous networks. One can envision miRNAs as a ‘power grid’ that keeps all these genes and protein networks connected (Figure 3). The degree of miRNA perturbation in cancer could be measured and compared to normal tissue patterns. This way, it might be possible to obtain a miRNA snapshot map, the ‘core of the cancer connectivity grid’. Restoring normal miRNA programs in the cancer cell may rewire the cell connectivity map and reverse cancer phenotypes. Developing therapeutic strategies to restore homeostasis by modifying miRNA expression may prove to be more comprehensive and successful than targeting individual genes or protein, since there are only a few miRNAs deregulated in cancer, compared to the large perturbations of the transcriptome and proteome in cancer cells.
Strategies for miRNA-based therapeutics
There are two main strategies to target miRNA expression in cancer. Direct strategies involve the use of oligonucleotides or virus-based constructs to either block the expression of an oncogenic miRNA or to substitute for the loss of expression of a tumor suppressor miRNA. The indirect strategy involves the use of drugs to modulate miRNA expression by targeting their transcription and processing (Figure 4).
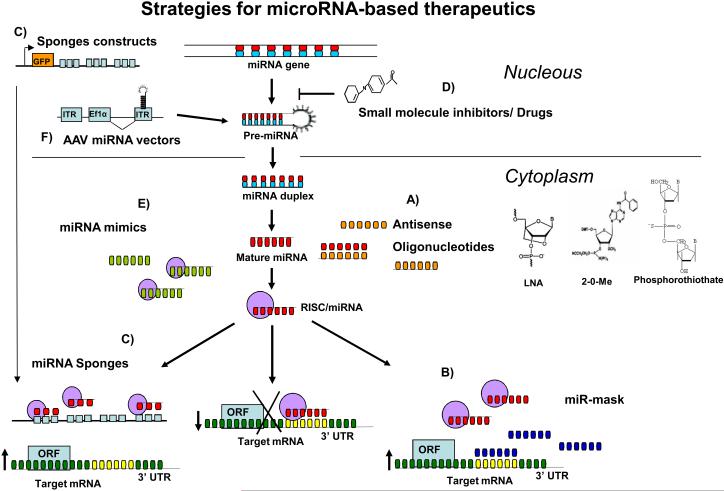
Figure 4. Strategies for miRNA-based therapies.
Blocking oncogenic miRNAs can be achieved by the use of antisense oligonucleotides, miRNA sponges, miRNA-mask and small RNA inhibitors91-99. (A) Antisense oligonucleotides can bind to the target miRNAs following the Watson and Crick complementarities and induces either degradation or duplex formation101,107. The three most common oligonucleotide modification structures are shown; Locked nucleic acid (LNA), 2-0-methyl (2-0-ME) and phosphorothiolate (PS)101,107. (B) The miR-mask oligonucleotides are synthetic oligonucleotides complementary to the 3' UTR target mRNA that compete with endogenous miRNAs for its target98. Therefore, miR-mask is able to block oncogenic miRNA deleterious functions at the target level. (C) The miRNA sponges are oligonucleotide constructs with multiple complementary miRNA binding sites (in tandem) to the target miRNA97. When introduced to the cell, sponges will “soak” endogenous miRNAs (Red oligos), decreasing the expression levels of an oncogenic miRNA. (D) Small molecule miRNA inhibitors regulate miRNA expression at transcriptional level99. Restoring down-regulated miRNA expression could be achieved by (E) using synthetic miRNAs (miRNA mimics) or (F) by inserting genes coding for miRNAs into viral constructs, such as the adeno associated viral vectors 100-101.
Blocking oncogenic miRNAs using antisense oligonucleotides
The demonstration that oncogenic miRNAs were up-regulated in cancer (Table 1) provided a rationale to investigate the use of antisense oligonucleotides to block their expression. Antisense oligonucleotides work as competitive inhibitors of miRNAs, presumably by annealing to the mature miRNA guide strand and inducing degradation or stoichiometric duplex formation. While this mechanism was demonstrated successfully by injecting complementary double strand sequences to miRNAs into Drosophila embryos86, further experiments using naked or unmodified antisense DNA oligonucleotides were ineffective in C. Elegans87. Researchers overcame these difficulties by introducing modifications to the chemical structure of the oligonucleotides to increase stability, binding affinity and specificity (Figure 4)88. Among these modifications, the introduction of 2'-O- methyl groups contributes to nuclease resistance and improved binding affinities to RNA (reviewed by Weiler J, et al)87. Oligonucleotides with 2'-O-methyl groups have proven to be effective inhibitors of miRNA expression in several cancer cell lines39-41,61,63. Other modifications such as 2'-O- methoxyethyl groups (2'-MOE) increase affinity and specificity to RNA compared to the 2' -O- methyl analogs87.
As a proof of principle, Krutzfeldt et al. developed chemically modified (2-OMe-modified nucleotides, with a phosphorothioate linkage), cholesterol-conjugated single-stranded RNA analogues (named ‘antagomirs’), complementary to miR-122, which is abundant in the liver These antagomirs were injected into the tail vein of mice, and specific targeting of miR-122 in the liver was observed after 24 hours89. The silencing of endogenous miRNAs by this novel method was specific, efficient and long lasting, and the effects were still observed 23 days after injection89. Gene expression and bioinformatics analysis of the whole transcriptome (mRNA) from antagomir-treated animals revealed that the 3' UTRs of up-regulated transcripts were strongly enriched in miR-122 recognition motifs, whereas down-regulated genes were depleted of these motifs. Using an antagomir against the ubiquitously expressed miR-16, the authors also investigated the bioavailability and silencing activity of antagomirs in different tissues. In mice treated with this antagomir, miR-16 was efficiently silenced in all tissues tested except brain (Figure 4)89.
Locked nucleic acid (LNA) antimiR constructs
LNA nucleosides are a class of nucleic acid analogues in which the ribose ring is “locked” by a methylene bridge connecting the 2’-O atom and the 4’-C atom (Figure 4). By “locking” the molecule with the methylene bridge, LNA oligonucleotides display unprecedented hybridization affinity toward complementary single-stranded RNA and complementary single- or double-stranded DNA90. In addition, they display excellent mismatch discrimination and high aqueous solubility. LNA antimiR have been used successfully in several in vitro studies to knock down specific miRNA expression41, 64-65.
Studies in mice using LNA antimiR have shown the feasibility and high efficiency of this approach. Recently, Elmen and colleagues examined whether combining LNA antimiR with phosphorothioate modifications could improve delivery of the compounds and silence miR-122 in mice without requiring additional chemical modifications91. The authors chose to target miR-122 based on previous data that indicate that miR-122 binds to the hepatitis C virus and stimulates its replication92. In a mouse model, intravenous injections of about 1–25 mg/kg of such LNA antimiR showed markedly improved efficiency in antagonizing miR-122 compared to cholesterol-conjugated antagomir-12291. A previous report indicated that three doses of 80 mg/kg of a cholesterol-conjugated oligonucleotide were needed to silence miR-122 in mice 89. This data suggest that LNA antimiRs are able to effectively silence their targets at much lower doses than cholesterol-based oligonucleotides.
The simple systemic delivery of an unconjugated LNA-antimiR-122 (SPC3649). has also been shown to effectively antagonize liver-expressed miR-122 in nonhuman primates93. Using three intravenous doses of 10 mg/kg in African green monkeys, the authors observed an effective depletion of miR-122 in the liver without any evidence of LNA-associated toxicities or histopathological changes in the animals. LNA-mediated antagonism of miR-122 in primates was effective and long-lasting93. The same group recently investigated the potential of miR-122 antagonism by LNA antimiR-122 (SPC3649) as a new anti-HCV therapy in a chimpanzee model system of chronic infection94. The animals were treated at two dose levels (5 mg kg−1 and 1 mg kg−1) by i.v. injections of SPC3649 on a weekly basis for 12 weeks followed by a treatment-free period of 17 weeks. While a significant decline of HCV RNA was observed in the serum after 3 weeks of treatment with the higher dose of SPC3649, high variability was observed at the lower dose levels94. Measurements of miR-122 expression revealed substantial and durable silencing of miR-122 expression in the liver. Furthermore, while there was no evidence of viral resistance or side effects in the treated animals, transcriptome and histological analyses of liver biopsies demonstrated de-repression of target mRNAs with miR-122 seed sites, down-regulation of interferon-regulated genes, and improvement of HCV-induced liver pathology94.
Overall, these studies not only support the rationale for targeting miR-122 as a novel treatment of chronic hepatitis C virus (HCV) infection to prevent cirrhosis and associated liver cancer, but also provide proof of principle for antagomir and LNA-antimiR therapies in inflammatory, degenerative or neoplastic human diseases characterized by aberrant upregulation of a specific miRNA family. Indeed, based on these encouraging results, Santaris Pharma is conducting early clinical studies using LNA-antimiR-122 (PSC-3649) in healthy human subjects in Denmark. A clinical Phase 1 trial, which is currently ongoing, will provide valuable information about pharmacokinetics and safety profiles.
MiRNAs Sponges
In addition to antisense oligonucleotides against miRNAs, other strategies have been developed to block miRNA function. Ebert and colleagues recently reported the use of competitive miRNA inhibitors known as miRNA sponges in mammalian cells95. MiRNA sponges are transcripts that contain multiple, tandem binding sites to a miRNA of interest that are transcribed from mammalian expression vectors (Figure 4). The authors reasoned that miRNA target sequences expressed at high levels could compete with bona fide targets within a cell for miRNA binding. To increase the affinity of these decoy transcripts, the authors introduced not only multiple miRNA binding sites, but also a bulge at the position normally cleaved by Argonaute 2, therefore enabling stable association of miRNA sponges with ribonucleoproteins complexes loaded with the corresponding miRNA. Using these constructs, a de-repression of miRNA targets was observed and indicated effective in vitro silencing of miRNAs96. Theses effects were comparable to that of obtained with 2’O-ME modified or LNA antisense oligonucleotides. Furthermore, sponges that contained only the heptameric seed were shown to effectively repress an entire miRNA family that shares by definition the same seed sequence96. In a recent study, Loya and colleagues applied miRNA sponges to inhibit miRNA activity in transgenic Drosophila in vivo97.
The art of disguise: miR-Mask
MiRNA-Masking Antisense oligonucleotides Technology (miR-Mask) is another decoy based mechanism developed by Xiao et al (Figure 4)98. In contrast to miRNA sponges, miR-MASKs consist of single-stranded 2′-O-methyl-modified antisense oligonucleotides that are fully complementary to predicted miRNA binding sites in the 3' UTR of the target mRNA98. In this way, the miR-Mask covers up the miRNA binding site to de-repress its target gene (mRNA), thereby its effects are gene-specific. This technology has been applied successfully to prevent repressive actions of miR-430 in TGF-β signaling pathways in zebrafish model98. While unwanted effects or off targets effects can be dramatically reduced with this approach, this may be a disadvantage for cancer therapy where the targeting of multiple pathways might be desirable.
Small- molecule inhibitors
Several drugs may have the ability to modulate the expression of miRNAs by targeting signaling pathways that ultimately converge on the activation of transcription factors that regulate miRNA encoding genes. Furthermore, it is possible to modulate the machinery that contributes to miRNA maturation and degradation processes. The identification of these compounds, however, is not straightforward and requires efficient screening of chemical libraries. Recently, Gumireddy and colleagues identified a method to screen for small molecule inhibitors of miRNAs (Figure 4)99. As a proof-of-principle for this approach, the authors selected miR-21, since this miRNA is frequently up-regulated in cancer (Table 1). Complementary sequences of miR-21 were cloned into a luciferase reporter gene, which was then used as sensor to detect the presence of specific mature miRNA molecules. The construct was transfected into HeLa cells, which express high miR-21 levels, resulting in low luciferase activity. Subsequently, a primary screen of more than 1000 small molecule compounds was conducted and an initial hit compound, diazobenzene 1, produced a 250% increase in the intensity of the luciferase signal relative to the untreated cells99. Further characterization revealed that this compound affects the transcription of miR-2199. This strategy could be applied to the screening of small molecules as inhibitors for other distinct oncogenic miRNAs. These could be utilized with conventional cancer therapeutics to develop novel combinatorial approaches for cancer treatment.
Restoring tumor suppressor miRNA expression
The loss or down-regulation of a tumor suppressor miRNA could be overcome by introducing synthetic oligonucleotides that are identical to the selected miRNA, known as miRNA mimics (Figure 4). Ectopic expression of synthetic miRNAs mimics with tumor suppressor function in cancer cells have been shown to induce cell death and block proliferation in several studies 40-42,44,49,63. For example, restoring miR-15-a and miR-29 in prostate and AML cell lines respectively, induced apoptosis44,63 . These miRNA mimics are small, usually double stranded and chemically modified (2’O-Me with phosphorothioate modifications). Some of these include longer sequences such as the miRNA precursor (for example, a pre-miRNA developed by Ambion). It has been reported that intratumoral injection of miR-29 mimics are effective in decreasing tumorigenicity in human rhabdomyosarcoma, liver and AML xenograft murine models 40,42,63. However, there is no in vivo data using miRNA mimics delivered by intravenous injection.
Another strategy to increase the expression of a tumor suppressor miRNA in cancer utilizes adenovirus associated vectors (AAV)(Figure 4). These vectors do not integrate into the genome and are eliminated efficiently with minimal toxicity, as shown in phase I and phase II clinical trials in about 200 patients 100-101. Another advantage of AAV vectors is the efficient transduction of target cells100. The development of self-complementary genome and nonhuman primate AAV serotypes allow more than 90% transduction efficiency of hepatocytes and long-term gene expression without toxicity following a single systemic administration of recombinant virus100. Kota and colleagues recently showed that miR-26 expression is lost in human liver cancers, while it was expressed at high levels in normal tissue102. Ectopic expression of this miRNA in liver cancer cell lines was shown to induce cell cycle arrest. The authors further cloned miR-26 into an AAV vector and viral particles were tested in an established MYC-dependent liver cancer mouse model102. Intravenous injection of this miRNA resulted in suppression of tumorigenicity by inducing tumor apoptosis and repressing cell growth, without signs of toxicity. Interestingly, significant anticancer effects were shown even when proteins other than the initiating oncogene (MYC) were targeted. This work represents the first evidence that restoring the expression of a tumor suppressor miRNA blocks cancer progression in vivo102. This strategy could be viable for the treatment of liver cancer, since it is easily targeted by both viral and non viral gene and small molecule delivery systems101. However, the efficacy of this system for other types of tumors and in different locations is unknown. Since there are multiple AAV serotypes available that allow efficient targeting of many tissues of interest, it is possible to target cancers that arise from different tissues. For example, muscle targeting could be desirable for the treatment of soft tissue sarcoma.
Reprogramming Cancer Cells: Turning around a bad network
So far, all the strategies to modulate miRNA expression are designed to modify only one miRNA or a family of miRNAs. Since it is likely that miRNAs act coordinately in cancer pathogenesis, and the phenotypic effects results from multiple interactions between miRNAs and the transcriptome, it is reasonable to search for strategies that aim to reprogram aberrant miRNA networks in cancer. Reprogramming could be achieved by modulating several of the key miRNAs within a network using antisense oligonucleotides or mimics. However, targeting multiple miRNAs using antisense oligonucleotides or mimics may be technically challenging.
Another strategy to rewire miRNA expression is the use of chemotherapeutic drugs (Figure 4). Several groups reported miRNA expression changes upon drug treatment in vitro and in vivo53-54, 103-104, suggesting that such changes may be responsible at least in part for the anti cancer effects. In acute promyelocytic leukemia (APL), a subtype of AML characterized by maturation arrest at the promyelocytic stage and caused by the fusion protein PML/RARa, pharmacological doses of all-trans-retinoic acid (ATRA) have been shown to reverse the dominant-negative effect of PML-RARa fusion and induce granulocytic differentiation of the AML blasts and apoptosis103. Our group reported that the apoptotic effect observed after ATRA treatment of APL cells and patients is partially explained by ATRA-induced activation of miR-15a-miR-16-1 cluster expression, which is known to target the antiapoptotic gene BCL-2103.
Decitabine and 5-azacytidine are two well known hypomethylating agents currently approved for the treatment of myelodysplastic syndrome (MDS), although they have shown activity in many other malignancies including AML105. It has long been known that these drugs work through DNA methyltransferase inhibition, resulting in tumor suppressor gene re-expression mediated by promoter hypomethylation105. More recently, miRNAs have been shown to be actively re-expressed after treatment with these drugs, and to play important roles for the therapeutic effects of these compounds. It is tempting to hypothesize that many of the biological effects of decitabine and 5-azacytidine may be mediated by re-expression of non coding RNAs53-55.
Once a cancer miRNA network is identified, one can envision the use of drugs or other agents to modify the expression of such miRNAs and thereby restore normal patterns of miRNA expression. In a few years, we may able to generate cell-specific miRNA expression profiles after drug treatment that may enable the discovery of functional connections between drugs, genes and diseases, similar to that of the connectivity map (a collection of genome-wide transcriptional expression data from cultured human cells treated with bioactive small molecules)106. Since miRNAs are fewer in number compared to mRNA, it could be assumed that there will be less noise and background in high-throughput based experiments such as microarrays for miRNAs than the ones performed using mRNA106. These miRNA-drug maps could be then used to discover novel drug applications and establish drug combination treatments.
Challenges of miRNA-based therapies
The challenges for developing miRNA-based therapeutics are the same as for siRNA therapeutics and comprise delivery, potential off-target effects and safety (Box 1 and Table 2).. One of the major problems for the use of miRNA therapeutics in vivo relates to tissue-specific delivery and cellular up-take of sufficient amounts of synthetic oligonucleotides to achieve sustained target inhibition101,107. The first obstacle to overcome is the biological instability of these compounds in biological fluids or tissues, since unmodified ‘naked’ oligonucleotides are rapidly degraded by cellular and serum nucleases101,107. The second obstacle is the poor cellular uptake of oligonucleotides due to their size and negative charge, which could prevent them from crossing through cell membranes101,107.
Box 1. Delivering synthetic miRNAs.
To overcome small RNA oligonucelotides deliveries hurdles, non-viral and viral strategies have been developed101,107. The non-viral strategies include: oligonucleotides with chemical modifications, liposomes, polymers, hydrogels and nanoparticles101,107,109-113. The most widely studied oligonucleotide modification is the replacement of each non-bridging oxygen in the backbone with a sulfur atom, thereby forming a phosphorothioate (PS) linkage101,107 (Figure 4). PS oligonucleotides exhibit a dramatically improved in vivo half-life compared with naked oligonucleotides101,107. However, these therapies have generally been administered by continuous intravenous injection and have been associated with several toxicities101,107. The efficacy data from Phase 2 studies show tumor regression, however major responses are rare101,107.
Liposomes are composed of a phospholipid bilayer with an enclosed aqueous compartment. They interact with oligonucleotides to form complexes stabilized by electrostatic interactions107,109. Cationic liposomes protect oligonucleotides from degradation by nucleases, and increase circulating half time and cellular up-take107,109. However, they are toxic to the cell and elicit hypersensitive reactions in vivo109-110. Several efforts are underway to make liposomes safer, such as improving their formulation by adding chemical additives to reduce cell toxicity101,107. Another limitation of liposomes is that tend to accumulate preferentially in the reticuloendothelial system, leading to a short life in the serum and reduced access to other tissues101,107.
Since chemically modified oligonucleotides alone or in combination with liposomes exhibit a short half life and require either continuous infusion or frequent administration, a possible approach to overcome this problem was to develop sustained-release polymer formulations112. Polymers are biodegradable compounds that protect RNA from degradation and facilitate sustained delivery to the tissues112. There are many different types of polymers that vary in size, chemistry and pharmacological properties101,112. In vitro and in vivo studies have shown that biodegradable polymer-antisense oligonucleotide combinations achieve sustained delivery and improved tissue biodistribution112. More research is still needed to guide the polymer architecture and chemical structure that are most suited for oligonucleotide delivery and cell and tissue targeting.
Nanoparticles, microspheres and hydrogels have also been developed as gene delivery vehicles. These strategies are promising since they provide improved oligonucleotide delivery and stability with minimal toxicity in animal models107,113.
Target specific delivery could also be achieved by direct injection of the synthetic oligonucleotides into solid tumors. This may be a feasible strategy for mesothelioma (intrapleural injections), ocular tumors, brain tumors or sarcomas, and should reduce or eliminate off target effects. This also could be achieved by tagging nanoparticle-miRNA oligonucleotide complexes with antibodies that bind the desired target cell101,107. For example, one could envision the development of miR-15a/miR-16-1 oligonucleotide-nanoparticles coated with anti-CD20 antibodies to treat CLL. This could be a potential strategy to overcome off targets effect in hematological cancers.
Table 2.
Limitations and advantages of direct miRNA-based therapeutic approaches
| Strategy | Limitations | Advantages | Experimental data | Solutions/ Future directions |
|---|---|---|---|---|
| 2-O-Me PS oligos | Delivery: Short serum half-life Poor cellular up-take Off target effects Limited biological effects |
Safe Improved stability Nuclease resistance Increased binding affinity |
In vitro and in vivo data Animal models, Phase I II and III clinical trials |
Improve delivery |
|
2-O-Me PS oligos
cholesterol backbone |
Toxicity Requires high doses |
Good bioavailability | In vitro and in vivo (animals) | Improve safety profile |
| LNA | Off targets effects Potential dose toxicity effects |
Safe Good biodistribution Effective |
In vitro and in vivo (mice and chimpances) Human trials ongoing |
Detailed PK, PD and toxicity studies in humans Develop Tissue-specific delivery |
|
Liposomes-Oligos
complexes |
Toxicity Hypersensitivity Potential dose toxicity effects |
Improved stability and delivery |
In vitro and in vivo (animals) | Develop better formulations |
|
Polymers-Nanoparticles
Oligos complexes |
Off target effects Potential dose toxicity effects |
Improved stability and delivery, minimal toxicity |
In vitro and in vivo (animals) | Develop Tissue-specific delivery (antibody tagging) |
| miR-mask | Limited scope (one target) Delivery |
Effects are gene-specific No off target effects |
In vitro studies | Achieve delivery in vivo Asses activity in vivo |
| miR-sponge | Delivery Off targets effects |
Able to silence family of miRNAs |
In vitro studies | Achieve delivery in vivo Asses activity in vivo |
| AAV vectors | Potential dose toxicity effects Off target effects |
Safe Efficient transduction Long term expression |
In vitro and in vivo (animals) Human trials for siRNA Phase I, II and III trials |
More extensive animal data is needed (in particular with other tumors) |
To overcome these delivery hurdles, viral and non-viral strategies have been developed (Box 1). A variety of chemical modifications in oligonucleotides have been investigated, such as morpholinos, peptide nucleic acids, cholesterol conjugation (see antisense section) and phosphorothioate backbone modification (see antisense section, Figure 3 and Box 1)108-110. Although chemical modifications have improved the delivery of oligonucleotides to tissues, this is often associated with impaired biological activity and increased toxicity, in particular when cholesterol-conjugated oligonucleotides are used (Table 2)108-111. Other strategies such as the use of cationic lipids, polymers and nanoparticles have recently become popular, in an attempt to enhance the cellular uptake and pharmacological effectiveness of antisense oligonucleotides in vivo108,111-113. While cationic lipids are too toxic to the cell and elicit hypersensitive reactions in vivo, polymers and nanoparticle strategies are promising since they have improved antisense delivery and stability with minimal in vivo toxicity (Box 1 and Table 2)108, 111-113.
Finally, since miRNAs regulate many genes, the potential off target effects of miRNA therapeutics is a major concern, since it may cause toxic phenotypes 101,108. As discussed above, in vitro and in vivo data for several types of cancer support the use of miR-29 oligonucleotide mimics as anti-cancer drugs40-43,63. While miR-29 targets several oncogenic pathways like apoptosis (MCL-1), proliferation (CDK6), methylation (DNMT1, DNMT3a and 3b), it also modulates other processes like bone development114, immune function (Th1 responses)115 and granulocytic differentiation63. Systemic over-expression of miR-29 using a synthetic mimic could target genes in particular in non cancerous tissues, and cause unwanted side effects such as autoimmunity or myeloid hyperproliferation. These problems could be solved by engineering effective systems that deliver the synthetic miRNA oligonucleotides specifically to the diseased tissue and cancer cells (Box 1).
Early clinical trials using DNA antisense technologies showed that severe side effects such as cytokine-release syndrome, hematological toxicity (thrombocytopenia) and liver damage may occur108,111. In some cases, these side effects were mainly due to problems with formulation, for example liposomes being directly toxic or inducing hypersensitive reactions (Table 2)111. In other circumstances they could be related to non-specific immunologic activities triggered by certain CpG motifs in the oligonucleotides that activated mechanisms of innate immunity mediated by toll-like cell receptors (TLR) and other inflammasome effectors, leading to interferon and other cytokine responses (Box 1)108,116. However, no data hitherto suggest that exogenous miRNAs may elicit cytokine responses in mammalian organisms. The use of LNA antimiR has been proven safe so far when tested in non-human primates published to date 93-94. The ongoing Phase1 clinical trial in humans using antimiR-122 LNA will be critical to assess the safety of this approach (Table 2).
More concerning is a recent report suggesting that toxicity is closely linked to the small RNA concentration. Grim and colleagues elegantly showed that sustained high expression of short hairpin RNAs by AAV vectors induced severe dose-dependent liver injury due to interference with endogenous miRNA processing in the liver, resulting in liver-specific miRNA down-regulation and injury117. Since both shRNAs and miRNAs used the same processing pathways7, these effects could be explained by the saturation of the processing machinery by exogenous shRNA, leading to loss of miRNA function. This work underscores the challenge for using vector based therapies to over-express miRNAs. Similar problems could arise using synthetic mature oligonucleotides, since they may also saturate RISC complexes and compete and displace other endogenous miRNAs.
Potential Applications for MicroRNA-Based Treatments in Cancer
What are the potential tumors that could be amenable for miRNA-therapy? Certain miRNAs, such as miR-155, miR-21, miR-17-92 and miR-29 are consistently deregulated in many cancers25,27-32 (Table 1), therefore, developing anticancer treatments targeting these miRNAs may be applicable to multiple malignancies. Silencing miR-155 and miR-21 expression in cancer cells would unblock the expression of critical tumor suppressor targets such as the phosphatases SHIP-1 and PTEN respectively, restoring normal patterns of cell differentiation and proliferation and inducing cancer cell death73,67. Since miRNAs, in particular the miR-29 family, have been shown to down-regulate DNA methyltransferases (DNMT1, DNMT3a and DNMT3b) and induce global DNA hypomethylation and tumor suppression gene re-expression118-119, restoring miR-29 expression could be used as an epigenetic hypomethylating strategy in malignancies. For example, certain subsets of AML have been shown to have low miR-29 expression63 and aberrant epigenetics play a critical role105. In addition, due to their hypomethylating effects, miR-29s are also negative regulators of apoptosis and cell proliferation by targeting the pro-apoptotic MCL-1 and the cell cycle regulator CDK6, respectively63. Thus, therapeutic modulation of miR-29 would impact on three pathways deregulated in cancer; epigenetics, apoptosis and cell proliferation/cell cycle (Figure 3).
MiRNAs could also be targeted for therapeutic applications besides cancer.. For example, since miR-155 is not only involved in cancer25,28, but also in inflammation and immunity120-121, therapies targeting miR-155 could be potentially applicable to a variety of autoimmune and inflammatory disorders.
Future Directions
As the miRNA field continues to evolve, a better understanding of miRNA biogenesis and function will certainly impact on the development of miRNA-based therapies. MiRNA effects are currently largely interpreted as the result of miRNA:mRNA 3' UTR interactions that cause target postranslational inhibition or degradation; However, focusing on this mechanism to design miRNA therapeutics is likely to prove too simplistic, due to the emerging miRNA mechanisms, which include decoy activity, and 5' UTR and direct DNA regulatory activities 17-21.
Research efforts should focus on maximizing the benefit of target diversity, while preventing off target effects. Improving the chemical design of antisense and miRNA mimics as well as developing novel delivery methods will be critical to achieve this goal. Detailed pharmacokinetic (PK) and pharmacodynamic (PD) studies will be needed to assure that the desired miRNA concentrations are achieved in tissues and the targets are down-regulated. These PK and biological PD effects need to be be correlated with clinical outcome, including treatment responses.
As more miRNA profiling studies are performed after drug treatment in cell lines and patients, distinctive drug specific miRNA maps could be obtained. Based on these profiles, it might be possible to use drugs alone or in combination, to reprogram the miRNome of cancer patients. We also envision that miRNA targeted therapies could be used to enhance or prevent resistance to standard chemotherapies agents or other biological agents. For example, miR-128 has been shown to modulate steroid refractoriness in acute lymphoblastic leukemia (ALL), therefore one strategy could be to use a synthetic miRNA in combination with chemotherapy to overcome this problem in ALL122.
However, outstanding challenges intrinsic to the oligonucleotide-based approaches remain to be overcome, including low bioavailability and poor cellular uptake resulting in suboptimal delivery, as well as off target effects and long term safety concerns in humans. Novel miRNA-formulations including nanoparticles and polymers as well as virus-based approaches could be employed to overcome these problems. Overall, targeting miRNAs to reprogram miRNA networks in cancer constitutes a reasonable and evidence based strategy with strong potential and chance for success. The enthusiasm for miRNA-based treatments is high and is reflected by the large number of pharmaceutical companies pursuing this strategy.
Definitions
Antisense: The term ‘antisense’ is generally used for nucleic acid based approaches that interferes, in a sequence selective way, with the processing of RNA from its transcription via mRNA to protein or with the effects of other forms of functional RNA
Phosphorothioate (PS): an oligonucleotide in which the oxygen atom normally linking two consecutive nucleotides has been replaced with sulfur and which resists degradation by cellular enzymes. The phosphorothioate backbone, while reducing affinity to target RNA, confers significant stability to nuclease degradation, and is essential for in vivo delivery of AMOs to tissues, as the phosphorothioate promotes protein binding and delays plasma clearance.
Functional definition of miRNA: miRNAs are evolutionarily conserved gene-regulatory molecules that can carry out integrated biological functions by regulating gene networks at multiple levels.
MiRNA sponges: are transcripts that contain multiple, tandem binding sites to a miRNA of interest that are transcribed from mammalian expression vectors
miR-MASK is a single-stranded 2′-O-methyl-modified oligonucleotide, which is antisense to the sequences of predicted miRNA binding sites in the protein-coding mRNA.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee Y, Ahn C, Han J, Choi H, Kim J. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 5.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammond S, Bernstein E, Beach D, Hannon G. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–229. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 8.Thimmaiah P, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutvanger G, Zamore PD. A microRNA in a multiple-turnover RNAi enxyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis B, Shih I, Jones-Rhoades M, Bartel D, Burge C. Prediction of Mammalian MicroRNA Targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 12.Krek D, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 13.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanovska I, Cleary MA. Combinatorial microRNAs: Working together to make a difference. Cell Cycle. 2008;20:3137–3142. doi: 10.4161/cc.7.20.6923. [DOI] [PubMed] [Google Scholar]
- 16.Stark A, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ørom UA, Nielsen FN, Lund AH. MicroRNA-10a Binds the 5' UTR of Ribosomal Protein mRNAs and Enhances Their Translation. Mol. Cell. 2008;4:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Eiring A, et al. A decoy binding activity of miR-328 controls hnRNP-E2 translation regulatory function in leukemic blasts. Cell. 2010;140:652–655. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beitzinger M, Meister G. MicroRNAs: From Decay to Decoy. Cell. 2010;140:612–614. doi: 10.1016/j.cell.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 20.Khraiwesh B, et al. Transcriptional Control of Gene Expression by MicroRNAs. Cell. 2010;140:112–122. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez S, Pisano DG, Serrano M. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle. 2008;7:2601–2608. doi: 10.4161/cc.7.16.6541. [DOI] [PubMed] [Google Scholar]
- 22.Kim DH, Saetrom P, Snøve O, Jr., Rossi JJ. MicroRNA directed transcriptional gene silencing in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calin GA, et al. Frequent deletions and downregulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 25.Volinia S, et al. A microRNA expression signature in human solid tumors defines cancer targets. Proc. Natl Acad. Sci. USA. 2005;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ota A, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 27.Petrocca F, et al. E2F1-regulated microRNAs impair TGFb-dependent cell cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 29.Diosdado B, et al. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer. 2009;101:707–714. doi: 10.1038/sj.bjc.6605037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calin GA, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl. J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 31.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 32.Garzon R, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schetter AJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciafre SA, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 35.Bloomston M, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 36.Pichiorri F, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc. Natl Acad. Sci. USA. 2008;105:12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metzler M, et al. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 38.Kluiver J, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 39.Garzon R. Distinctive miRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc. Natl Acad. Sci. USA. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–81. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mott JL, Kobayashi S, Bronk SF, Gores GJ. Mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiong Y, et al. Effects of MicroRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010 doi: 10.1002/hep.23380. (EPUB) [DOI] [PubMed] [Google Scholar]
- 43.Zhao JJ, et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010 doi: 10.1182/blood-2009-09-243147. (EPUB) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonci D, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 45.Bottoni A, et al. miR-15a and miR-16-1 down-regulation in pituitary adenomas. J Cell Physiol. 2005;204:280–285. doi: 10.1002/jcp.20282. [DOI] [PubMed] [Google Scholar]
- 46.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Takamizawa J, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 48.Yu F, et al. let-7 Regulates Self Renewal and Tumorigenicity of Breast Cancer Cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 49.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–6. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 50.Yang N, et al. MicroRNA Microarray Identifies Let-7i as a Novel Biomarker and Therapeutic Target in Human Epithelial Ovarian Cancer. Cancer Res. 2008;68:10307–10314. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Motoyama K, et al. Clinical Significance of High Mobility Group A2 in Human Gastric Cancer and Its Relationship to let-7 MicroRNA Family. Clin. Cancer Res. 2008;14:2334–2340. doi: 10.1158/1078-0432.CCR-07-4667. [DOI] [PubMed] [Google Scholar]
- 52.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saito Y, et al. Specific activation of microRNAs-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 54.Lujambio A, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 55.Hackanson B, et al. Epigenetic modification of CCAAT/enhancer binding protein alpha expression in acute myeloid leukemia. Cancer Res. 2008;68:3142–51. doi: 10.1158/0008-5472.CAN-08-0483. [DOI] [PubMed] [Google Scholar]
- 56.Kumar MS, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merritt WA, et al. Dicer, Drosha, and Outcomes in Patients with Ovarian Cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang TC, et al. Transactivation of miR-34a by p53 Broadly Influences Gene Expression and Promotes Apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang TS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nature Genetics. 2007;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cimmino A, et al. 2006;102:13944–13949. [Google Scholar]
- 62.Klein U, et al. The DLEU2/miR-15a/16-1 Cluster Controls B Cell Proliferation and Its Deletion Leads to Chronic Lymphocytic Leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 63.Garzon R, et al. MicroRNA-29 functions in AML. Blood. 1142009:5331–41. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sampson VB, et al. MicroRNA let-7a Down-regulates MYC and Reverts MYC-Induced Growth in Burkitt Lymphoma Cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 65.Lujambio A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl Acad. Sci. USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 67.Meng F, et al. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frankel LB, et al. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 69.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with elevated miR-17-92 expression in lymphocytes. Nat. Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mu P, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–11. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olive V, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–49. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl Acad. Sci. USA. 2006;103:7024–9. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Connell RM, et al. Sustained expression of micro-RNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp. Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O'Connell RM, Chaudhuri A,A, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl Acad. Sci. USA. 2009;106:7113–8. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Costinean S, et al. Ship and C/ebpβ are targeted by miR-155 in B cells of Eμ-miR-155 transgenic mice. Blood. 2009;114:1374–1382. doi: 10.1182/blood-2009-05-220814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Q, et al. The inositol polyphosphate 5-phosphatase ship is a crucial negative regulator of B cell antigen receptor signaling. J Exp Med. 1998;188:1333–42. doi: 10.1084/jem.188.7.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 80.Ma L, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341–7. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. 2008:z47–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Linsley PS, et al. Transcripts targeted by the microRNA-16 family cooperatively regulates cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Calin GA, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc. Natl Acad. Sci. USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li QJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 85.Croce CM. Oncogenes and Cancer. N Engl. J Med. 2008;358:502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 86.Boutla A, Delidakis C, Tabler M. Developmental defects by antisense-mediated inactivation of micro-RNAs 2 and 13 in Drosophila and the identification of putative targets. Nucleic Acid Res. 2003;31:4973–4980. doi: 10.1093/nar/gkg707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hutzvanger G, Simard MJ, Mello CC, Zamore PD. Sequence specific inhibition of small RNA function. Plos Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs); amunition to target miRNAs implicated in human disease? Gene Therapy. 2006;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 89.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 90.Vester B, Wengel J. LNA (locked nucleic acid): high affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 91.Elmén J, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to upregulation of a large set of predicted target RNAs in the liver. Nucleic Acids Res. 2008;36:1153–62. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 93.Elmén J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 94.Lanford RE, et al. Therapeutic Silencing of MicroRNA-122 in Primates with Chronic Hepatitis C Virus Infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiao J, et al. Novel approaches for gene-specific interference via manipulating actions of micro-RNAs: examination on the pacemaker channel genes HCN2 and HCN4. J Cell Physiol. 2007;212:285–92. doi: 10.1002/jcp.21062. [DOI] [PubMed] [Google Scholar]
- 97.Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–4. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 99.Gumireddy K, et al. Small-molecule inhibitors of microrna miR-21 function. Angew Chem. Int. Ed Engl. 2008;47:7482–4. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Michelfelder S, Trepel M. Adeno-associated viral vectors and their redirection to cell-type specific receptors. Adv Genet. 2009;67:29–60. doi: 10.1016/S0065-2660(09)67002-4. [DOI] [PubMed] [Google Scholar]
- 101.Aagaard L, Rossi JJ. RNAi therapeutics: Principles, prospects and challenges. Adv. Drug Deliv. Rev. 2007;59:75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kota J, et al. Therapeutic microRNA Delivery Suppresses Tumorigenesis in a Murine Liver Cancer Model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Garzon R, et al. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26:4148–4157. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 104.Roccaro A, et al. microRNA expression in the biology, prognosis, and therapy of Waldenström macroglobulinemia. Blood. 2009;113:4391–4402. doi: 10.1182/blood-2008-09-178228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Galm O., Herman, J.G., Baylin SB. The fundamental role of epigenetics in hematopoietic malignancies. Blood Rev. 2006;20:1–13. doi: 10.1016/j.blre.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 106.Lamb J, et al. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 107.Zhao X, et al. Controlled delivery of antisense oligonucleotides: a brief review of current strategies. Exp. Opin. Drug Deliv. 2009;6:673–686. doi: 10.1517/17425240902992894. [DOI] [PubMed] [Google Scholar]
- 108.Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol. cancer Therap. 2002;1:347–355. [PubMed] [Google Scholar]
- 109.Malik R, Roy L. Design and development of antisense drugs. Exp. Opin. Drug Discov. 2008;3:1189–207. doi: 10.1517/17460441.3.10.1189. [DOI] [PubMed] [Google Scholar]
- 110.Chiarantini L, et al. Comparison of a novel delivery systems for antisense peptide nucleic acids. J Control Release. 2005;109:24–36. doi: 10.1016/j.jconrel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 111.Zuhorn IS, Engberts JBF, Hoeckstra D. Gene delivery by cationic lipid vectors: overcoming cellular barriers. Eur Biophys J. 2007;36:349–62. doi: 10.1007/s00249-006-0092-4. [DOI] [PubMed] [Google Scholar]
- 112.Chirila TV, et al. The use of synthethic polymers for delivery of therapeutic antisense oligodeoxynucleotides. Biomaterials. 2002;23:321–42. doi: 10.1016/S0142-9612(01)00125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rosi NL, et al. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–30. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 114.Li Z, et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–84. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu H, Cheung IY, Guo HF, Cheung NK. MicroRNA miR-29 modulates expression of immunoinhibitory molecule B7-H3: potential implications for immune based therapy of human solid tumors. Cancer Res. 2009;69:6275–81. doi: 10.1158/0008-5472.CAN-08-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hornung V, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 117.Grimm D, et al. Fatality in mice due to oversaturation of cellular MicroRNA/short hairpin RNA pathways. Nature. 2006;441:537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 118.Garzon R, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–8. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl Acad. Sci. U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 121.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kotani Ai., et al. miR-128b is a potent glucocorticoid sensitizer in MLL-AF4 acute lymphocytic leukemia cells and exerts cooperative effects with miR-221. Blood. 2009;114:4169–4178. doi: 10.1182/blood-2008-12-191619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Trang P, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–7. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes & Develop. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pekarsky Y, et al. TCL1 expression in CLL is regulated by miR-29 and miR-181. Cancer Res. 2006;66:1590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 126.Lodygin D, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 127.Toyota M, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 128.Sun F, et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 129.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl Acad. Sci. USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pigazzi M, Manara E, Baron E, Basso G. miR-34b targets cyclic AMP-responsive element binding protein in acute myeloid leukemia. Cancer Res. 2009;69:2471–2478. doi: 10.1158/0008-5472.CAN-08-3404. [DOI] [PubMed] [Google Scholar]
- 131.Cole KA, et al. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol. Cancer Res. 2008;6:735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li Z, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc. Natl Acad. Sci. USA. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.O’Donnell KA, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 134.Velu CS, Baktula AM, Grimes HL. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. 2009;113:4720–8. doi: 10.1182/blood-2008-11-190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Löffler D, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 136.Zhu S, Si. M-L, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. 2007. [DOI] [PubMed] [Google Scholar]
- 137.Voorhoeve PM, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2007;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]