Abstract
Background
In Arabidopsis thaliana (A. thaliana) the WD40 protein TRANSPARENT TESTA GLABRA1 (TTG1) controls five traits relevant for the adaptation of plants to environmental changes including the production of proanthocyanidin, anthocyanidin, seed coat mucilage, trichomes and root hairs. The analysis of different Brassicaceae species suggests that the function of TTG1 is conserved within the family.
Results
In this work, we studied the function of TTG1 in Arabis alpina (A. alpina). A comparison of wild type and two Aattg1 alleles revealed that AaTTG1 is involved in the regulation of all five traits. A detailed analysis of the five traits showed striking phenotypic differences between A. alpina and A. thaliana such that trichome formation occurs also at later stages of leaf development and that root hairs form at non-root hair positions.
Conclusions
The evolutionary conservation of the regulation of the five traits by TTG1 on the one hand and the striking phenotypic differences make A. alpina a very interesting genetic model system to study the evolution of TTG1-dependent gene regulatory networks at a functional level.
Keywords: Arabis alpina, TTG1, Trichomes, Root hairs, Pro-anthocyanidin, Anthocyanidin, Seed coat mucilage
Background
One approach towards a mechanistic understanding of phenotypic changes is evolutionary developmental biology (also called Evo-Devo) [1]. As most of our knowledge is based on a few well-characterized model systems that are separated by large evolutionary distances, evolutionary comparisons are often descriptive and have little functional depth. Typically Evo-Devo approaches aim to characterize the key players or pathways known to be relevant for a given process in one model organism in an evolutionarily distantly related species. As outlined by Sommer [2] this often leads to an almost descriptive list of the molecular inventories rather than a functional understanding. For a functional evolutionary comparison of developmental processes it is necessary to study clearly homologous processes in closely related species. This enables the understanding of changes in the regulatory network at a mechanistic level.
We focused on the TTG1-dependent gene regulatory network that is well-described in A. thaliana. Here it controls five traits that all have an adaptive value for the plant and are likely to be variable on the one hand, but also interdependent as they are controlled by the same regulatory genes [3]. TTG1 encodes a WD40 protein [4]. In A. thaliana TTG1 acts together with R2R3-MYB and bHLH proteins (called MBW complex) to regulate different aspects of epidermal cell differentiation including the production of proanthocyanidin, anthocyanidin, seed coat mucilage, trichomes and root hairs [5-12]. The bHLH factor is represented by three homologous, partially redundant acting genes. TT8 regulates seed coat mucilage production, seed coat pigment production and anthocyanin biosynthesis. EGL3 controls seed coat pigmentation, anthocyanin biosynthesis, trichome and root hair development and GL3 is involved in anthocyanin biosynthesis, trichome and root hair development. High trait specificity is found for the R2R3-MYB factors such that one specific R2R3-MYB gene regulates each trait [3]. GL1 regulates trichome initiation [13], WER the non-root hair development [14], PAP1 and PAP2 anthocyanidin production [15,16], TT2 pro-anthocyanidin production and MYB61 regulates seed coat mucilage production [17]. During trichome and root hair development additional R3 single repeat MYBs are important as negative regulators mediating cellular interactions during pattern formation [18-24].
The function of the MBW complex in epidermal cell differentiation is evolutionary conserved in plants, though their regulation of anthocyanin and proanthocyanidin production seems to be the most ancient function. This is suggested by the finding that the MBW complex in maize is only involved in anthocyanin production [25,26], in petunia in anthocyanidin and proanthocyanidin production [27-29] and in A. thaliana in all five traits [6,7]. Based on the phylogenetic tree of the MYB proteins, Serna and Martin suggested that the additional role of the MBW complex in trichome formation has been adopted after the Asterid-Rosid division [5]. This view is supported by the findings that GL1 (A. thaliana) or C1 (Zea mays) overexpression in tobacco has no effect on trichome formation [30]. Conversely, overexpression of TTG1 homologs from various species has been successfully used to complement the corresponding A. thaliana mutant phenotypes. These include AN11 from Petunia hybrida[27], PAC1 from maize [31], GhTTG1 and GhTTG3 from Gossypium hirsutum ([32], InWDR1/Ca from Ipomoea nil[33], MtWD40-1 from Medicago truncatula ([34], MdTTG1 from Malus domestica[35], VvTTG1 from Vitis vinifera L [36], and PgWD40 from Punica granatum L. [37]. This indicates that the biochemical function of TTG1 is functionally conserved over a large evolutionary distance.
Apart from A. thaliana, genetic data are available for two other species within the Brassicaceae family. In Brassica rapa it was shown that two traits, glabrous and yellow seeds, strictly co-segregated and that these two traits map to the BrTTG1 locus [38]. In addition a yellow seed mutation was mapped to the BrTT8 locus suggesting that also the function of the corresponding bHLH factor is conserved. In Matthiola incana it was shown that a line displaying white flowers, yellow seeds, seed mucilage defects and a glabrous phenotype exhibits a relevant point mutation in the MiTTG1 gene [39]. Together these data indicate that a function of TTG1 in the regulation of trichome, seed coat differentiation, anthocyanin and proanthocyanin pathways is conserved within the Brassicaceae.
As a complex gene regulatory network governs the regulation of the TTG1-dependent five traits it seems very attractive to study network evolution in this family. Towards this end it is desirable to systematically establish a second genetic model system enabling the functional characterisation by mutant analysis. We chose A. alpina for several reasons: On the one hand, A. alpina is sufficiently closely related to enable the identification of clear ortholog genes by sequence similarity and synteny in the fully sequenced genome. On the other hand the evolutionary distance of about 26 million years [40] to 40 million [41] between A. thaliana and A. alpina promised phenotypic variations for these traits and variations in the underlying gene regulatory networks. Finally, A. alpina can be transformed by Agrobacterium mediated gene transfer [42].
In this work we studied the function of TTG1 in Arabis alpina (A. alpina). We demonstrate that all five traits are affected in two Aattg1 alleles in A. alpina. As considerable phenotypic variation was already observed for some of these traits between different members of the Brassicaceae family [43-45] we did a detailed phenotypic description of the five traits to provide a reference for future studies. Our analysis revealed striking differences in the case of trichome and root hair patterning in A. alpina as compared to A. thaliana.
Results
Identification of Aattg1 mutants in A. alpina
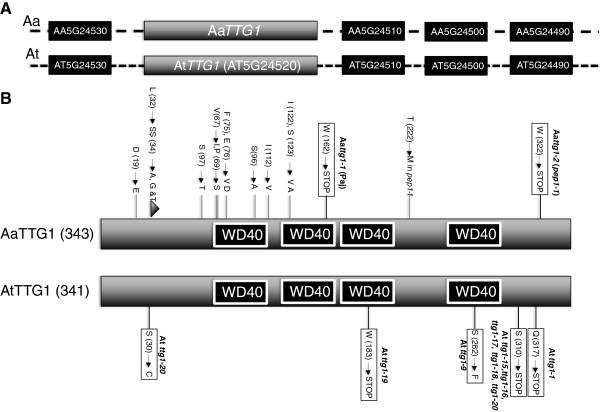
To initiate a genetic dissection of TTG1-dependent regulation cascades in A. alpina, we identified the putative A. alpina TTG1 gene by homology and synteny comparisons. The A. alpina TTG1 gene (AaTTG1) has a putative coding sequence of 1032 base pairs representing a putative protein of 343 amino acids. The AaTTG1 gene shows 90% sequence identity when compared to the A. thaliana AtTTG1 gene. The two flanking genes of AaTTG1 are homologs of those two flanking AtTTG1: AT5G24530 and AT5G24510 (Figure 1A). This indicates that AaTTG1 and AtTTG1 are orthologs. A comparison of the amino acid sequence revealed differences in 12 amino acids and two additional amino acids (Figure 1B, Additional file 1: Table S1).
Figure 1.
Schematic presentation of AaTTG1 and AtTTG1. A) Genomic regions of the TTG1 loci including the neighboring genes in A. alpina and A. thaliana. The black boxes represent the homologous flanking genes. The gene names used for A. alpina are derived from the corresponding A. thaliana genes by exchanging the “At” for A. thaliana to “Aa” for Arabis alpina. As the distance between genes differs in the two species they are shown with different types of dashed lines. B) Schematic presentation of the protein structures. The position of the WD40 domains was determined using the ELM [46]. Differences of amino acids between A. alpina and A. thaliana are indicated. The amino acid position changed in mutant alleles in A. alpina (this work) and A. thaliana[4] have been marked by boxes.
We identified two mutants in EMS mutagenized M2 populations of A. alpina showing a phenotype similar to ttg1: glabrous trichomes and yellow seeds. One mutant was recovered from the EMS treated wild-type background A. alpina Pajares (Paj) [42]. The other mutant was found in a mutagenized A. alpina (Paj) background carrying the pep1-1 mutation [42,47,48]. In the A. alpina (pep1-1) background the TTG1 gene contained a mutation that would lead to a T to M exchange at position 222 of the TTG1 protein, which, however, does not affect the function of TTG1 as this background shows no effect on any assumed TTG1-dependent phenotype. When sequencing the AaTTG1 gene in the ttg1 mutant isolated from the Paj wild-type background we found a mutation leading to a stop codon after 161 amino acids in the second WD40 domain and called this allele Aattg1-1. The putative ttg1 allele induced in the pep1-1 background carried a mutation leading to a stop codon after 321 amino acids (called Aattg1-2). As a premature STOP codon two amino acids C-terminal to this position leads to strong ttg1 phenotypes in A. thaliana[4], it is conceivable that also the A. alpina Aattg1-2 allele shows the observed strong phenotypes (Figure 1B). Together these data indicate that the two mutants identified by the trichome and seed color phenotypes are two A. alpina Aattg1 alleles. We tested this by crossing Aattg1-1 and Aattg1-2 plants. F1 plants were glabrous confirming the allelism (Additional file 2: Figure S1A-E). To test whether AaTTG1 protein can rescue the Arabidopsis ttg1 mutant phenotype we expressed the Arabis alpina coding sequence under the promoter of the Arabidopsis TTG1 gene [49]. Towards this end we used the wild type coding sequence from A. alpina Pajares and pep1-1. We recovered 7 and four lines, respectively, displaying partial rescue of the trichome phenotype (Additional file 2: Figure S1F-H).
(Pro-) anthocyanidin production in A. alpina wild type and Aattg1 mutants
Brown colour of Arabidopsis seeds is caused by oxidized proanthocyanidins [50]. Screening for differences in seed pigmentation revealed a group of mutants with transparent testa - including ttg1 - that was impaired in flavonoid accumulation [51,52]. Most of these genes were subsequently identified either as structural enzymes or regulators (e.g. TTG1) of the proanthocyanidin pathway [50].
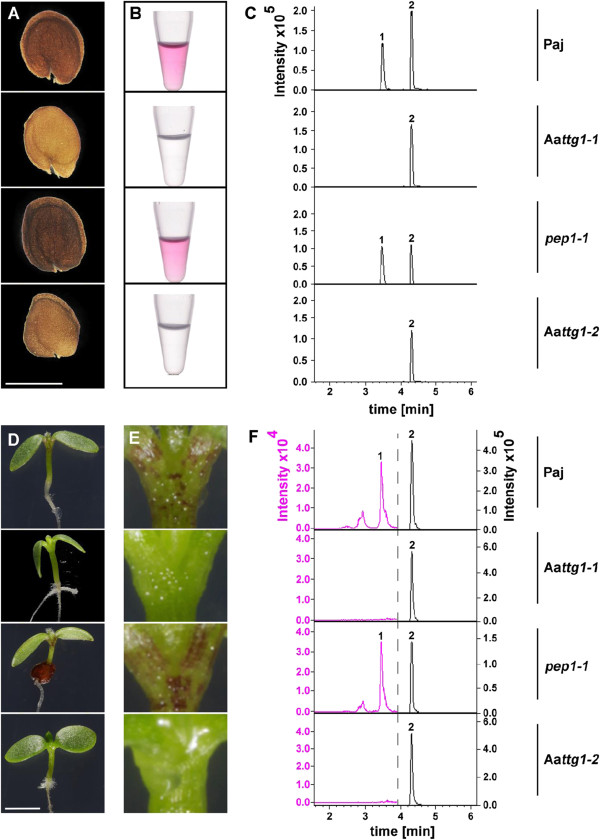
Compared to the corresponding backgrounds, the seed colour of both Aattg1 mutants is yellowish, sharing the transparent testa phenotype with the mutants in A. thaliana (Figure 2A). This finding points to a lack of proanthocyanidins. The majority of procyanidin is found in the insoluble fraction of flavonoid extracts and can be analyzed by acidic hydrolysis to cyanidin [53,54]. We hydrolyzed the insoluble substances upon extraction for visual inspection (Figure 2B). A pink staining that is characteristic for cyanidin was obtained for the backgrounds but not for the Aattg1 mutants. In subsequent HPLC-MS analysis of extracted soluble, hydrolyzed flavonoids no cyanidin was detectable, proofing its absence in Aattg1 mutants (Figure 2C, Additional file 3: Figure S2). In contrast, kaempferol, was detected in the mutants and backgrounds, serving as a control for the successful extraction of flavonoids. This indicates that AaTTG1 either regulates the activity of the AaLDOX (leucoanthocyanidin dioxygenase) enzyme that catalyses the last step of cyanidin biosynthesis and/or earlier enzymes of the pathway.
Figure 2.
TTG1 is needed for the accumulation of anthocyanidins in A. alpina. Seeds (A-C) and seedlings (D-F) of Aattg1-1 and Aattg1-2 are devoid of cyanidin - a late component of the (pro-) anthocyanidin biosynthesis pathway - but contain kaempferol – an early component of this pathway. The genotype for each row in A-C and D-F is given on the right. A) stereo-microscopy of seeds. Aattg1 mutants have a yellowish seed colour as compared to the respective backgrounds indicating a lack of proanthocyanidin; B) extracted and hydrolyzed insoluble components of the seeds‘ (pro-) anthocyanidin pathway, presence (backgrounds) or absence (Aattg1 mutants) of pink colour relates to the presence or absence of (pro-) anthocyanidin in seeds; C), F) HPLC-MS analysis of extracted soluble, hydrolyzed components of the anthocyanidin biosynthesis pathway. Shown are extracted ion chromatograms for m/z = 287.055 +/- 0.005. Note that cyanidin and kaempferol have the same m/z value. Different scales were chosen to highlight the absence of cyanidin in F). Full chromatograms are provided in Additional file 3: Figure S2. D) photography of 5 day-old seedlings grown on MS medium with 1% sucrose at constant light. E) zoom in on the petiole- and SAM-region of the seedlings shown on the left in D). Aattg1 mutants do not accumulate anthocyanidins in the seedling‘s hypocotyl. All pictures within one subfigure were taken at the same light settings. Bar in A: 1 mm; bar in C: 2 mm; 1: cyanidin (late biosynthesis compound); 2: kaempferol (early biosynthesis compound).
The so called late genes of the anthocyanidin biosynthesis have been classified by their regulation through the TTG1 containing MBW complexes [3,55-59]. Therefore, our result identifies kaempferol as an early and cyanidin as a late component of the proanthocyanidin biosynthesis pathway in A. alpina.
The anthocyanidin biosynthesis pathway is part of the proanthocyanidin biosynthesis pathway. Red colour of A. thaliana hypocotyls and young leaves is the result of accumulating UV-protective anthocyanidins and their derivatives upon exposure to light or other stresses [60-62].
In A. thaliana, TTG1 as a general regulator also promotes visible accumulation of purple anthocyanins in the seedlings’ hypocotyls [3]. Our finding that Aattg1 confers a transparent testa phenotype to A. alpina seeds suggests an absence of anthocyanins also in the hypocotyl. It is, however, possible that a redundant regulator exists for anthocyanin accumulation.
To address this, we grew seedlings of the Aattg1 alleles and their respective backgrounds under constant light on plates supplemented with sucrose. We found that in the hypocotyls of Aattg1 mutant seedlings no visible pink anthocyanin was accumulated in contrast to Paj and pep1-1 (Figure 2D-E). Moreover, HPLC-MS analysis for both Aattg1 mutants showed that AaTTG1 is also essential for the accumulation of cyanidin in seedlings and its function is not taken over by any other gene (Figure 2F, Additional file 3: Figure S2).
Seed coat differentiation in A. alpina wild type and Aattg1 mutants
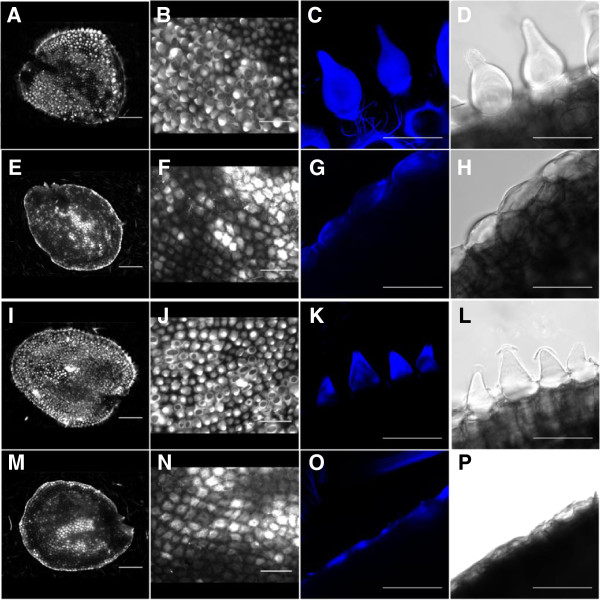
In A. thaliana, seed epidermal cells of the outer integument differentiate into highly specialized seed coat cells. These are characterized by the formation of a central column surrounded by a secondary cell wall (the columella) and the accumulation of polysaccharide mucilage between the plasma membrane and the primary cell wall that is released in the presence of water [63,64]. In A. thaliana, ttg1 mutants neither form a columella nor produce mucilage [51]. We compared columella formation in the two ttg1 mutants with the respective genetic backgrounds Paj and pep1-1 by staining with calcofluor white (Figure 3), Ruthenium Red (Additional file 4: Figure S3) and by Scanning Electron Microscopy analysis (Additional file 5: Figure S4). Wild type and pep1-1 seeds have an irregular but smooth surface with domes representing the columella (Figure 3A-D, I-L). In the two corresponding Aattg1 mutants, the columella is completely missing indicating that epidermal differentiation of the seed coat is affected similar as observed in Atttg1 mutants [51,63,64] (Figure 3E-H, M-P).
Figure 3.
Calcofluor white stained wild type and Aattg1 mutant seeds. Fluorescence microscope (A,B,E,F,I,J,M,N) and CLSM (fluorescence channel: C,G,K,O, transmission channel: D,H,L,P) pictures of the surface of A. alpina seeds stained with calcofluor white. A to D) wild type Paj. I to L) pep1-1 mutant. Note, that in wild type and pep1-1 mutants the columella is seen as large domes. E to H) Aattg1-1 mutant induced in the wild type Paj background. M to P) The Aattg1-2 mutant induced in the pep1-1 background. In both mutants only the rim of the epidermal cells is left and columellas are absent. Scale bar: A,E,I,M = 300 μm, B,F,G,N = 100 μm, C,D,G,H,K,L,O,P = 50 μm.
Trichome patterning in A. alpina wild type and Aattg1 mutants
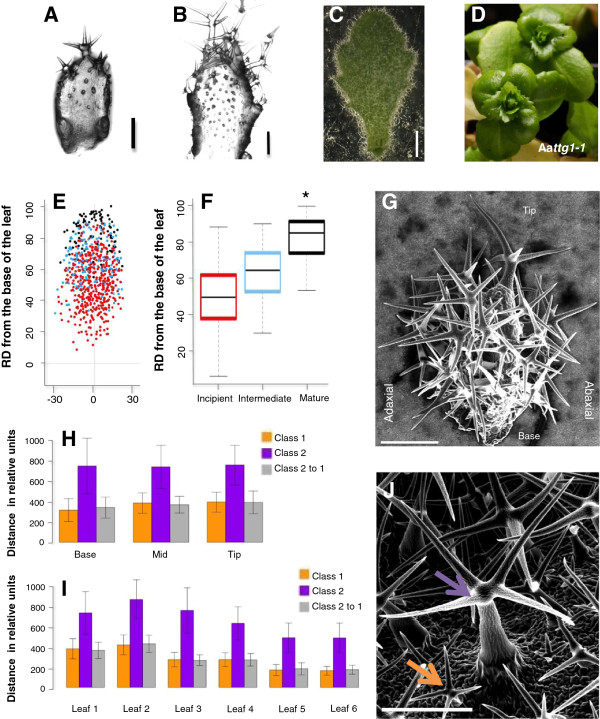
In A. thaliana, trichomes are initiated on young leaves and become separated by division and expansion of the epidermal cells lying in between [65]. At the first glance, trichome initiation on A. alpina leaves is very similar to the situation in A. thaliana (Figure 4A-C): At the base of young leaves, we found incipient trichomes. These comprise trichome stages preceding branch initiation (Figure 4A). Mature trichomes with several branches were found at the leaf tip and intermediate stages were observed in the mid region (Figure 4B). Adult leaves are covered densely with trichomes (Figure 4C). However, we noted one striking difference: on mature leaves we found two classes of mature trichomes that differed in size and height (Figure 4J). Our visual impression was that the larger trichomes (class 2) are arranged in a regular pattern with the smaller trichomes (class 1) being scattered between them (Figure 4J). To test this, we systematically measured the distances between the two classes of trichomes. We analyzed three regions of the third leaf: a region at the base, in the middle and the tip. In all three regions, on average the larger trichomes showed about twice the distance from each other as compared to the distances in between smaller trichomes and between smaller trichomes and the larger trichomes (Figure 4H). The same pattern was also found on the second to the sixths leaves indicating that it is a general feature (Figure 4I). These distance patterns suggested to us two superimposed trichome patterns. We therefore studied young leaf stages in more detail using the analysis tool TrichEratops [66]. A meta leaf was generated, in which the position of three developmental trichome stages are shown at their relative positions on the leaf with respect to the basal-distal axis (Figure 4E). As described in A. thaliana, we found a general gradient of trichome developmental stages with mature trichomes at the tip and young trichomes at the base of the leaf. In contrast to A. thaliana, we observed early developmental stages of trichome development between already mature trichomes (Figure 4B, F). These patterns may either be explained by the formation of new trichomes between more mature trichomes or by spatial differences in the growth characteristics such that the smaller trichomes are initiated normally, but grow slower. During the course of our experiments we noted that all A. alpina leaves are densely covered with trichomes on both the adaxial and the abaxial sides (Figure 4G). This is in contrast to A. thaliana where only late leaves produce abaxial trichomes as a consequence of the phase change from vegetative to generative growth [67]. To analyse the role of A. alpina TTG1 in trichome formation we analysed the two ttg1 mutants Aattg1-1 and Aattg1-2. As known from strong ttg1 mutants in A. thaliana both alleles lacked trichomes completely (Figure 4D).
Figure 4.
Trichome patterning in A. alpina. A-C) Three developmental stages of an A. alpina leaf. D) Aattg1-1 leaves. E) Meta leaf generated using TrichEratops integrating the relative position of trichomes from 22 young leaves [66]. Red dots are incipient trichomes, blue dots intermediate developmental stages and black dots represent mature trichomes. Note that incipient trichomes are found in a region in which already mature trichomes have developed. F) Boxplot depicting the relative position (0 is the base of the leaf and 100 the tip of the leaf) of three developmental classes of trichome development. Note that the average position of the three developmental classes is clearly distinct. “*” indicate that all three classes are significantly different from each other according to Student’s t-test (P < 0.001). G) SEM picture of a leaf 3 from the side showing that trichomes are found on the abaxial and adaxial side. H) Average minimum distance to the nearest neighboring trichome on different regions of leaf 2. Distances between smaller trichomes (class 1) are shown in orange, distances between larger trichomes (class 2) in pink and distances between the two classes in grey. I) Average minimum distance to the nearest neighboring trichome on leaves 1 to 6. J) SEM of an adult A. alpina leaf showing the large (pink arrow) and the small classes of trichomes (orange arrow). Scale bar = 100 μm in A,B. Scale bar = 500 μm in C. Scale bar = 1 mm in D,E.
Root hair patterning in A. alpina wild type and Aattg1 mutants
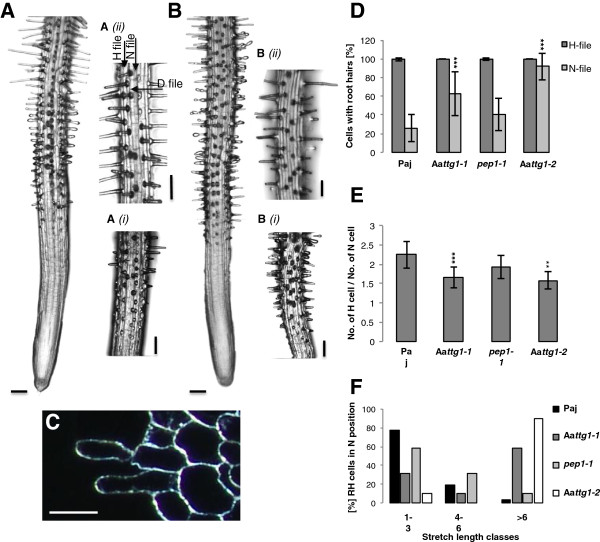
In A. thaliana, epidermal root cells are arranged in files. Cells overlaying the cleft between two underlying cortex cells are short and differentiate into hair cells (H–file) [68]. Cells in all other files are long and do not develop root hairs (called N-files). To understand root hair patterning in A. alpina, we analysed file-specific root hair production in two regions of the differentiation zone to determine the temporal spatial development of the root hair pattern (Figure 5A). Both, A. alpina Paj and pep1-1 showed a similar frequency of root hair cells in the lower (Figure 5Ai) and in the upper region (Figure 5Aii) indicating that the final root hair pattern is determined already in the lower region (data not shown). Similar to A. thaliana almost all cells in the H–position develop into root hairs. However, in A. alpina cells in N-file positions also frequently form root hairs (Figure 5A, C, D). We found between 30% (Paj) and 40% (pep1-1) of the N-position cells to develop into root hairs (Figure 5D). This finding raised the question, whether the cells in the H– and N-positions also differ in other characteristics. One difference reported in A. thaliana is the length of the individual cells such that cells in N-files are about twice as long as H–file cells [69]. As the cell length continuously changes along the root axis due to cell growth, we did not measure the actual cell lengths, but rather determined the H- to N-file cell number ratios. In A. alpina Paj we found a ratio of 2.3, in pep1-1 the ratio was 2.0, indicating that the N-file cells are approximately twice as long as H-file cells (Figure 5E). During the course of experiments we noticed that the cells carrying a root hair in N position were not evenly distributed along the file, but were arranged in continuous stretches of N-file cells differentiating into root hairs. As depicted in Figure 5F, more than 50% of all stretches consisted of 1 to 3 cells, indicating a higher probability for short (1–3) than for long (>6) stretches. Typically, we found root hairs in N-position always on only one side of an H-file. Out of 542 Paj H-file cells, just one was flanked symmetrically by two N-file cells carrying a root hair (pep1-1: 20 out of 511). To study the function of A. alpina TTG1 in root hair patterning we studied root hair patterning in Aattg1-1 and Aattg1-2. We found ectopic root hairs at N-positions similar as described in A. thaliana (Figure 5B, D).
Figure 5.
Root hair patterning in A. alpina. A) Wild type A. alpina root; A(i), Higher magnification of the lower root hair differentiation zone; A(ii), Higher magnification of upper root hair differentiation zone. Root hair file (H), Non-root hair file (N) and non-root hair file with root hair stretches (D-file) is indicated by arrows. B) Aattg1-1 root; B(i), Higher magnification of the lower root hair differentiation zone; B(ii), Higher magnification of the upper root hair differentiation zone. C) Wild type A. alpina root cross section depicting one root hair at the H-position over the cleft of two underlying cortex cells and a neighboring root hair in an N-position. D) Percentage of root hairs in root hair files (H) and non-root hair files (N). E) Relative number of cells in H and N files. A ratio of about two reflects that the non-hair cells are twice as long as the H-cells. F) Number of cells in continuous stretches of root hair cells in the N files. Scale bar: 100 μm in A,B; Scale bar: 40 μm in C. ***: P < 0.001, **: P < 0.01.
Discussion
TTG1 containing MBW complexes regulate five different traits in A. thaliana including proanthocyanidin, anthocyanidin and mucilage production as well as trichome and root hair patterning. Taking together the data on single or multiple TTG1-dependent traits studied in different Brassicaceae species indicates that TTG1 has the same range of functions as described in A. thaliana throughout this family. For Brassica rapa a function in trichome and seed color was reported [38]. In Matthiola incana a role in (pro)-anthocyanin, mucilage and trichome formation was shown [39]. In support of these findings, we show that these four traits and in addition root hair patterning are affected in two independent Aattg1 alleles in A. alpina.
These five traits have an adaptive value for the plant. Therefore, on the one hand, variability between species might be expected. On the other hand, these traits are regulated by differential TTG1 complexes encoded by members of the TTG1 gene regulatory network. Due to this genetic interdependence variability might be limited. In support of these considerations we found no apparent differences between A. alpina and A. thaliana for some but not all traits.
According to Serna and Martin, the TTG1-dependent regulation of (pro-) anthocyanidin production is a more ancient trait than trichome formation [5]. In agreement with this, the TTG1-dependent definition of early and late substances between kaempferol and cyanidin is maintained in A. alpina as compared to A. thaliana[3,55-59]. Similarly, AaTTG1 is needed for columella formation as described for A. thaliana[51].
While the phenotypes of these traits are generally very similar to those in A. thaliana, we noted some striking deviations for trichome and root hair patterning that are not trivial to explain in the light of the known regulation schemes. The observed two superimposed trichome patterns can be explained in two ways. One possibility is that small and large trichomes are initiated around the same time and that the subsequent cell differentiation differs. In this scenario, genes regulating cell differentiation and/or morphogenesis would be differentially expressed in the two types of trichomes. This could occur at different levels including genes regulating differentiation such as MYB5 and MYB23 [70], GL2 [71,72], TTG2 [73] or genes controlling endoreduplication such as for example the group of KAK genes [74]. The second possibility is that trichome initiation occurs in developmentally advanced stages of leaf development. In A. thaliana this phenotype was found in lines overexpressing the GL3/EGL3 homolog from maize, the R-gene [75]. By analogy, the intercalation phenotype in A. alpina could be explained by changes in the spatial/temporal expression of A. alpina GL3. The formation of root hairs in N-files is also not found in wild type A. thaliana under normal conditions but is reminiscent of Atttg1 and Atwer mutants [14,76]. Thus, one possibility to derive the wild-type pattern in A. alpina roots is a reduction of the AaTTG1 or AaWER activity. Given that mutations reducing trichome number lead to the production of additional root hairs because the MBW complex serves to activate trichomes and non-root hair fates the production of additional root hairs and extra trichome formation between mature trichomes cannot easily be explained by the A. thaliana network.
Conclusions
Taken together, our results demonstrate that the phenotypes of the five TTG1-dependent traits studied here are in general very similar in A. thaliana and A. alpina. The various phenotypic differences make A. alpina a very interesting genetic model system to study the evolution of gene regulatory networks at a functional level.
Methods
Plant Material and growth conditions
The Aattg1 alleles were isolated from EMS mutagenized A. alpina Pajares [42] and pep1-1 populations. For root hair file analysis, seeds were surface sterilized with 70% (v/v) ethanol (5 min) and 2% sodium hypochlorite (w/v, 8 min). Sterilized seeds were sown on 1× Murashige-Skoog plates lacking sucrose and stratified at 4°C for 5 days. Plants were grown on vertically positioned plates for 7 days under long day (LD) conditions (16 h light, 8 h darkness) at 21°C. For inter-species rescue experiments we used the Arabidopsis ttg1-1 mutant (Ler background, [77]).
For soil-grown A. alpina, seeds were stratified in darkness at 4°C for four days and then placed in growth chambers under LD conditions at 21°C.
Sequence and synteny analysis
Extracted AaTTG1 (Paj and pep1-1) sequences were analysed on CLC DNA Workbench 5.6.1 (CLC bio, Aarhus, Denmark) by comparison with CDS of AT5G24520 downloaded from TAIR (http://www.arabidopsis.org). NCBI Blastn 2.2.28 was used to confirm the synteny of the neighbouring genes [78]. Primers rev_ttg1_arabis_out (5‘-GCAATCAAGAATCTCTAGAACCAAG-3‘) and fwd_ttg1_arabis_out (5’-CAAATGTATGGACCGAATTATCAAG-3‘) were designed outside the CDS of AaTTG1 to sequence it from the wild types and the mutants.
Trichome analysis
The first true leaf of soil-grown A. alpina was labeled as leaf 1 and the following ones accordingly. When leaf 6 had reached a size of approximately 2 cm in length, leaves 1–6 were used for trichome analysis. For this, all leaves of one developmental stage were photographed at a magnification that enabled the distinction of different trichome classes. The pictures where analyzed using the TrichEratops software by marking the two different classes of trichomes [66]. The distance between trichomes of one class and trichomes of different classes were calculated with R (http://www.r-project.org/). Young leaves were essentially analyzed as described previously [66].
Root hair file analysis
H-files of 7-day-old plate-grown seedlings were microscopically identified by the position over cortical cell boundaries. Following 10 to 15 H-file cells per root and zone, the number of cells and root hairs in the flanking N-files was determined. Further, the length of continuous stretches of N-file cells carrying a root hair was determined. For cross-sections, all root tissues were fixed and embedded as previously described [79]. Agarose was used for mechanical fixation. 100 μm sections were made using a Leica EM UC7 ultra microtome (Leica Microsystems, Wetzlar, Germany) with glass knives.
Flavonoid analysis
For extraction of flavonoids, seeds were homogenized in 100 μl MeOH/water/formic acid (50:49:1, v/v) with glass beads in a tissue lyser (Qiagen, Hilden, Germany) at 30 Hz for 180 sec. Following centrifugation, the pellet was extracted with 200 μl MeOH/water/formic acid (50:49:1, v/v) over night at 4°C. 60 μl of centrifuged pooled supernatants were mixed with 440 μl MeOH:HClconc. (95:5, v/v), hydrolyzed for 90 min and diluted 1:1 with MeOH prior to LC-MS analysis. For photography, pellets were treated in the same way with 200 μl MeOH:HClconc. (95:5, v/v). For seedlings, 200 μl of MeOH:HClconc. (95:5, v/v) were used for homogenization and over night extraction followed by direct hydrolysis.
Mass analysis was done with a Dionex 3000 UPLC (Thermo Scientific, Dreiech, Germany) - maXis 4G (Bruker Daltonics, Bremen, Germany) LC-MS system equipped with an Apollo II ESI source (Bruker Daltonics, Bremen, Germany). 5 μl of samples were separated with a Poroshell 120, EC-C18, (3×50 mm, 2.7 μm) C18 column (Agilent, Waldbronn, Germany) and mix of solvent A water (0.1% formic acid) and solvent B MeOH (0.1% formic acid) with gradient profile (starting with 95:5, v/v, for 0.5 min; linear gradient up to 0:100, A/B, over 3.9 min and maintained for 2 min and re-equalibration to 95:5, A/B with a total runtime of 8.6 min) at a flow rate of 0.3 ml/min. LC-MS analysis, data processing and annotation of kaempferol and cyanidin were carried out with Compass DataAnalysis Version 4.0 SP5 (Bruker Daltonics, Bremen, Germany). Metabolites were identified by comparison to kaempferol and cyanidin (Sigma, Germany).
Constructs and transformation
The binary vector proAtTTG1pAMPAT-GW [49] was used to create proAtTTG1:AaTTG1(Paj) and proAtTTG1:AaTTG1(pep1-1) using the GatewayR system (Invitrogen) using the following primers for the cloning of AaTTG1: Fwd: GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGGATAACTCAGCTCCAGA.
Rev: GGGGACCACTTTGTACAAGAAAGCTGGGTTTCAAACTCTAAGGAGCTGCA. The constructs were introduced in the A. thaliana ttg1-1 mutant (Ler background, [77]) by Agrobacterium-mediated (strain GV3101-pMP90RK) transformation using the floral dip method described previously [80]. Transformants were selected in the T1 generation on soil by screening for trichomes on leaf number 3 or 4.
Photography and microscopy
Whole leaves were captured using a Canon EOS 5D Mark (Canon, Krefeld, Germany). Dry A. alpina seeds were mounted on a conductive carbon tab covered SEM stub and analyzed using a FEI Quanta FEG 250 Scanning Electron Microscope (SEM; FEI, Eindhoven, The Netherlands) at an accelerating voltage of 15 kV. A. alpina seeds were stained with 0.01% (w/v) aqueous solution of calcofluor white (Fluroscent Brightner, Sigma-Aldrich, Germany) overnight and analysed by creating manual z-stacks with Leica DM5000B microscope fitted with a LEICA DFC 360 FX camera and a Leica LAS AF software (Leica Microsystems, Wetzlar). Optical sections of the calcofluor white stained seeds were obtained by confocal laser scanning (CLSM) microscopy using the Leica TCS SPE CLSM, Leica LCS software (Leica Microsystems, Wetzlar). 0.05%(w/v). Aqueous ruthenium red (Sigma-Aldrich, Germany) stained seeds (overnight) were analysed with a conventional light microscope, Leica DMRE microscope, Leica LCS software (Leica Microsystems, Wetzlar). Root hair files were analyzed by creating z-stacks by conventional light microscopy using a Leica DM5000B fluorescence microscope (Leica Microsystems, Wetzlar). Images were processed using ImageJ (Rasband, W.S., ImageJ, U.S. National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/, 1997–2012) and Photoshop 7.0.1, Adobe. Pictures of seeds (Figure 2) and of the adult leaf (Figure 4C) were acquired using a Leica stereomicroscope (MZ FLIII) with the MultiFocus and Montage option of the Leica Application Suite V3 (Leica Microsystems, Wetzlar, Germany).
Statistical analysis and software
Statistical analysis was done as described before [81]. We used the Tricharatops [66] and R software (http://www.r-project.org/) to create the meta leaf, box plots and graphs for the analysis of the minimum distance to the nearest neighbouring trichome. Microsoft Excel (Microsoft, Redmond, USA) was used for the diagrams analyzing the root hair pattern.
Authors’ contributions
DC, HW, JS, SS, MCA AS performed the experiments, GC, AS and MH supervised the project, AS and MH wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Comparison of amino acid (aa) sequences between AtTTG1 and AaTTG1.
Complementation test between the Aattg1-1 and Aattg1-2 mutant alleles and rescue experiments in A. thaliana. First true leaves of A) wild type Paj, B) Aattg1-1, C) pep1-1, D) Aattg1-2, E) First true leaf of a F1 plant from the cross between Aattg1-1 and Aattg1-2. Leaves are glabrous indicating allelism. Scale bar = 1 mm. F) A. thaliana ttg1-1 rosette leaves. Plants are completely devoid of trichomes in this allele. G) A. thaliana Atttg1-1ProAtTTG1:AaTTG1Paj plant showing partial rescue of the trichome phenotype. H) Atttg1-1ProAtTTG1:AaTTG1pep1-1 plant showing partial trichome rescue. Scale bar = 1 mm.
HPLC-MS analysis of cyanidin and kaempferol in seeds and seedlings of A. alpina – full chromatograms. Shown are total ion chromatogram (TIC, left) and extracted ion chromatogram (EIC, right) for 750 nM cyanidin (MeOH), 750 nM kaempferol (MeOH) and for all samples shown in Figure 2C and Figure 2F. The m/z value for cyanidin ([M]+) and kaempferol ([M + H]+) is 287.055. EICs for m/z = 287.055 +/− 0.005 were generated based on the corresponding TICs using the Compass DataAnalysis software Version 4.0 SP5 (Bruker Daltonics, Bremen, Germany). In the TIC the retention time and in the EIC the peak of cyanidin and kaempferol is marked. 1: cyanidin; 2: kaempferol. Dashed lines mark the begin and end of each sample. Before the first dashed line the mass calibration for each run can be seen in the TIC.
Ruthenium red stained seeds of wild type and Aattg1 mutants. Light microscopy image of the surface of A. alpina seeds. The dome shaped columella is stained with ruthenium red labeling the seed coat mucilage. A) Wild type Paj. B) pep1-1 mutant. C) Aattg1-1 mutant induced in the wild type Paj background. D) Aattg1-2 mutant induced in the pep1-1 background. Note, the absence of ruthenium red stained columellas in both mutants. Scale bar = 500 μm, inset = 50 μm.
SEM pictures of wild type and Aattg1 mutant seeds. Scanning Electron Micrographs of the surface of A. alpina seeds. A, B) wild type Paj and pep1-1 mutant, respectively. Note, that the surface is irregularly but smooth and that the columella is seen as small domes. C, D) Aattg1-1 mutant induced in the wild type Paj background and the Aattg1-2 mutant induced in the pep1-1 background. Only the rim of the epidermal cells is left. Columellas are absent. Scale bar: 100 μm.
Contributor Information
Divykriti Chopra, Email: div.mailme@gmail.com.
Heike Wolff, Email: hwolff@smail.uni-koeln.de.
Johannes Span, Email: jspan0@smail.uni-koeln.de.
Swen Schellmann, Email: swen.schellmann@uni-koeln.de.
George Coupland, Email: coupland@mpipz.mpg.de.
Maria C Albani, Email: albani@mpipz.mpg.de.
Andrea Schrader, Email: Andrea.Schrader@uni-koeln.de.
Martin Hülskamp, Email: martin.huelskamp@uni-koeln.de.
Acknowledgements
We thank Bastian Welter for excellent technical assistance, Louai Rishmawi for help and discussions on root hair patterning, Benjamin Jaegle for his support in statistical analysis using R, Britta Mueller for her help in embedding, Sebastian Hess for his guidance in microtoming, Hans-Peter Bollhagen and Frank Nitsche for SEM assistance. Henrik Failmetzger and Achim Tresch provided help with the use of the TrichEratops software. We thank Eva-Maria Willing, Geo Velikkakam James and the Arabis alpina genome consortium for access to genomic information prior to publication. This work was supported by SFB680 and a DFG grant to M.H., an IMPRS fellowship to D.C and by the SPP1530 to MCA. The authors acknowledge the significant contributions by the Biocenter-MS facilities, University of Cologne.
References
- Rudel D, Sommer RJ. The evolution of developmental mechanisms. Dev Biol. 2003;264(1):15–37. doi: 10.1016/S0012-1606(03)00353-1. [DOI] [PubMed] [Google Scholar]
- Sommer R. The future of evo-devo:model systems and evolutionary theory. Nat Rev Genet. 2009;10:416–422. doi: 10.1038/nrg2567. [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne T, Lloyd A. A network of redundant bHLH proteins function in all TTG1 dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–1349. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna L, Martin C. Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci. 2006;11(6):274–280. doi: 10.1016/j.tplants.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10(5):236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10(2):63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66(1):94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- Balkunde R, Pesch M, Hulskamp M. Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr Top Dev Biol. 2010;91:299–321. doi: 10.1016/S0070-2153(10)91010-7. [DOI] [PubMed] [Google Scholar]
- Broun P. Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol. 2005;8(3):272–279. doi: 10.1016/j.pbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Grotewold E. The genetics and biochemistry of floral pigments. Annu Rev Plant Biol. 2006;57:761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R, Ishida T, Wada T. New insights into the mechanism of development of Arabidopsis root hairs and trichomes. Int Rev Cell Mol Biol. 2011;286:67–106. doi: 10.1016/B978-0-12-385859-7.00002-1. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991;67:483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/S0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. lant Cell. 2000;12(12):2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanin accumulation in developing seed. Plant Cell. 2001;13(9):2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell. 2001;13(12):2777–2791. doi: 10.1105/tpc.010265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science. 1997;277(5329):1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jurgens G, Hulskamp M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. Embo J. 2002;21(19):5036–5046. doi: 10.1093/emboj/cdf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Simon M, Hulskamp M, Schiefelbein J. The ENHANCER OF TRY AND CPC1 (ETC1) gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol. 2004;268:506–513. doi: 10.1016/j.ydbio.2003.12.037. [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Wester K, Schiefelbein J, Hulskamp M. ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol Biol. 2004;55:389–398. doi: 10.1007/s11103-004-0893-8. [DOI] [PubMed] [Google Scholar]
- Wester K, Digiuni S, Geier F, Timmer J, Fleck C, Hulskamp M. Functional diversity of R3 single-repeat genes in trichome development. Development. 2009;136(9):1487–1496. doi: 10.1242/dev.021733. [DOI] [PubMed] [Google Scholar]
- Gan L, Xia K, Chen JG, Wang S. Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biol. 2011;11:176. doi: 10.1186/1471-2229-11-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Hubbard L, Chang Y, Guo J, Schiefelbein J, Chen JG. Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 2008;8:81. doi: 10.1186/1471-2229-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci USA. 1989;86(18):7092–7096. doi: 10.1073/pnas.86.18.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. Embo J. 1987;6(12):3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetten N, Quattrocchio F, Mol J, Koes R. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 1997;11:1422–1434. doi: 10.1101/gad.11.11.1422. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, van der Woude K, Mol JN, Koes R. Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 1998;13(4):475–488. doi: 10.1046/j.1365-313X.1998.00046.x. [DOI] [PubMed] [Google Scholar]
- Spelt C, Quattrocchio F, Mol JN, Koes R. anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell. 2000;12(9):1619–1632. doi: 10.1105/tpc.12.9.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne T, Clement J, Arnold D, Lloyd A. Heterologous myb genes distinct from GL1 enhance trichome production when overexpressed in Nicotiana tabacum. Development. 1999;126(4):671–682. doi: 10.1242/dev.126.4.671. [DOI] [PubMed] [Google Scholar]
- Carey CC, Strahle JT, Selinger DA, Chandler VL. Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell. 2004;16(2):450–464. doi: 10.1105/tpc.018796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JA, Walker AR, Timmis JN, Orford SJ. Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol Biol. 2005;57(1):67–81. doi: 10.1007/s11103-004-6768-1. [DOI] [PubMed] [Google Scholar]
- Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S. Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol. 2006;47(4):457–470. doi: 10.1093/pcp/pcj012. [DOI] [PubMed] [Google Scholar]
- Pang Y, Wenger JP, Saathoff K, Peel GJ, Wen J, Huhman D, Allen SN, Tang Y, Cheng X, Tadege M. et al. A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development. Plant Physiol. 2009;151(3):1114–1129. doi: 10.1104/pp.109.144022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggemann J, Weisshaar B, Sagasser M. A WD40-repeat gene from Malus x domestica is a functional homologue of Arabidopsis thaliana TRANSPARENT TESTA GLABRA1. Plant Cell Rep. 2010;29(3):285–294. doi: 10.1007/s00299-010-0821-0. [DOI] [PubMed] [Google Scholar]
- Matus JT, Poupin MJ, Canon P, Bordeu E, Alcalde JA, Arce-Johnson P. Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera L.) Plant Mol Biol. 2010;72(6):607–620. doi: 10.1007/s11103-010-9597-4. [DOI] [PubMed] [Google Scholar]
- Ben-Simhon Z, Judeinstein S, Nadler-Hassar T, Trainin T, Bar-Ya'akov I, Borochov-Neori H, Holland D. A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta. 2011;234(5):865–881. doi: 10.1007/s00425-011-1438-4. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lu Y, Yuan Y, Zhang X, Geng J, Chen Y, Cloutier S, McVetty PB, Li G. Map-based cloning and characterization of a gene controlling hairiness and seed coat color traits in Brassica rapa. Plant Mol Biol. 2009;69(5):553–563. doi: 10.1007/s11103-008-9437-y. [DOI] [PubMed] [Google Scholar]
- Dressel A, Hemleben V. Transparent Testa Glabra 1 (TTG1) and TTG1-like genes in Matthiola incana R. Br. and related Brassicaceae and mutation in the WD-40 motif. Plant Biol (Stuttg) 2009;11(2):204–212. doi: 10.1111/j.1438-8677.2008.00099.x. [DOI] [PubMed] [Google Scholar]
- Koch MA, Kiefer C, Ehrich D, Vogel J, Brochmann C, Mummenhoff K. Three times out of Asia Minor: the phylogeography of Arabis alpina L. (Brassicaceae) Mol Ecol. 2006;15(3):825–839. doi: 10.1111/j.1365-294X.2005.02848.x. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2010;107(43):18724–18728. doi: 10.1073/pnas.0909766107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Farrona S, Vincent C, Joecker A, Schoof H, Turck F, Alonso-Blanco C, Coupland G, Albani MC. PEP1 regulates perennial flowering in Arabis alpina. Nature. 2009;459(7245):423–427. doi: 10.1038/nature07988. [DOI] [PubMed] [Google Scholar]
- Beilstein M, Al-Shehbaz IA, Kellogg EA. Brassicaceae phylogeny and trichome evolution. Am J Bot. 2006;93(4):607–619. doi: 10.3732/ajb.93.4.607. [DOI] [PubMed] [Google Scholar]
- Vancaeseele L, Mills JT, Sumner M, Gillespie R. Cytology of mucilage production in the seed coat of candle canola (brassica-campestris) Can J Bot. 1981;59(3):292–300. doi: 10.1139/b81-042. [DOI] [Google Scholar]
- Beilstein MA, Al-Shehbaz IA, Mathews S, Kellogg EA. Brassicaceae phylogeny inferred from phytochrome A and ndhF sequence data: tribes and trichomes revisited. Am J Bot. 2008;95(10):1307–1327. doi: 10.3732/ajb.0800065. [DOI] [PubMed] [Google Scholar]
- Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, Altenberg B, Toedt G, Uyar B, Seiler M, Budd A. et al. ELM–the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40(Database issue):D242–251. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albani MC, Castaings L, Wotzel S, Mateos JL, Wunder J, Wang R, Reymond M, Coupland G. PEP1 of Arabis alpina is encoded by two overlapping genes that contribute to natural genetic variation in perennial flowering. PLoS Genet. 2012;8(12):e1003130. doi: 10.1371/journal.pgen.1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonzi S, Albani MC, Themmat PEVL, Nordström KJV, Wang R, Schneeberger K, Moerland PD, Coupland G. Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science. 2013. [DOI] [PubMed]
- Bouyer D, Geier F, Kragler F, Schnittger A, Pesch M, Wester K, Balkunde R, Timmer J, Fleck C, Hulskamp M. Two-dimensional patterning by a trapping/depletion mechanism: the role of TTG1 and GL3 in Arabidopsis trichome formation. PLoS Biol. 2008;6(6):e141. doi: 10.1371/journal.pbio.0060141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M. Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol. 2006;57:405–430. doi: 10.1146/annurev.arplant.57.032905.105252. [DOI] [PubMed] [Google Scholar]
- Koornneef M. The complex syndrome of ttg mutants. Arabidopsis Inf Serv. 1981;18:45–51. [Google Scholar]
- Koornneef M. Mutations affecting the testa colour in Arabidopsis. Arabidopsis Inf Serv. 1990;27:1–4. [Google Scholar]
- Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, Einhorn J, Lepiniec L. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta. 2006;224(1):96–107. doi: 10.1007/s00425-005-0197-5. [DOI] [PubMed] [Google Scholar]
- Porter LJ, Hrstich LN, Chan BG. The conversion of procyanidins and prodelphinidins to cyanidin and delphinidin. Phytochemistry. 1985;25(1):223–230. doi: 10.1016/S0031-9422(00)94533-3. [DOI] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53(5):814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- Martin C, Prescott A, Mackay S, Bartlett J, Vrijlandt E. Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J. 1991;1(1):37–49. doi: 10.1111/j.1365-313X.1991.00037.x. [DOI] [PubMed] [Google Scholar]
- Pelletier MK, Murrell JR, Shirley BW. Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis. Further evidence for differential regulation of "early" and "late" genes. Plant Physiol. 1997;113(4):1437–1445. doi: 10.1104/pp.113.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier MK, Shirley BW. Analysis of flavanone 3-hydroxylase in Arabidopsis seedlings. Coordinate regulation with chalcone synthase and chalcone isomerase. Plant Physiol. 1996;111(1):339–345. doi: 10.1104/pp.111.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen H, Mol J, Koes RE. Regulatory genes controlling anthocyanin pigmentation Are functionally conserved among plant species and have distinct sets of target genes. Plant Cell. 1993;5(11):1497–1512. doi: 10.1105/tpc.5.11.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker-Scott L. Environmental significance of anthocyanins in plant stress responses. Photochem Photobiol. 1999;70(1):1–9. doi: 10.1111/j.1751-1097.1999.tb01944.x. [DOI] [Google Scholar]
- Misyura M, Colasanti J, Rothstein SJ. Physiological and genetic analysis of Arabidopsis thaliana anthocyanin biosynthesis mutants under chronic adverse environmental conditions. J Exp Bot. 2013;64(1):229–240. doi: 10.1093/jxb/ers328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubasek WL, Shirley BW, McKillop A, Goodman HM, Briggs W, Ausubel FM. Regulation of flavonoid biosynthetic genes in germinating arabidopsis seedlings. Plant Cell. 1992;4(10):1229–1236. doi: 10.1105/tpc.4.10.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Skinner DJ, Haughn GW. Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 2000;122(2):345–355. doi: 10.1104/pp.122.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windsor JB, Symonds VV, Mendenhall J, Lloyd AM. Arabidopsis seed coat development: morphological differentiation of the outer integument. Plant J. 2000;22(6):483–493. doi: 10.1046/j.1365-313x.2000.00756.x. [DOI] [PubMed] [Google Scholar]
- Hulskamp M, Misera S, Jürgens G. Genetic dissection of trichome cell development in Arabidopsis. Cell. 1994;76:555–566. doi: 10.1016/0092-8674(94)90118-X. [DOI] [PubMed] [Google Scholar]
- Failmezger H, Jaegle B, Schrader A, Hulskamp M, Tresch A. Semi-automated 3D leaf reconstruction and analysis of trichome patterning from light microscopic images. PLoS Comput Biol. 2013;9(4):e1003029. doi: 10.1371/journal.pcbi.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development. 1997;124:645–654. doi: 10.1242/dev.124.3.645. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Berger F, Hung C, Dolan L, Schiefelbein J. Control of cell division in the root epidermis of Arabidopsis thaliana. Dev Biol. 1998;194:235–245. doi: 10.1006/dbio.1997.8813. [DOI] [PubMed] [Google Scholar]
- Li SF, Milliken ON, Pham H, Seyit R, Napoli R, Preston J, Koltunow AM, Parish RW. The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell. 2009;21(1):72–89. doi: 10.1105/tpc.108.063503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD. The glabra 2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 1994;8:1388–1399. doi: 10.1101/gad.8.12.1388. [DOI] [PubMed] [Google Scholar]
- DiCristina M, Sessa G, Dolan L, Linstead P, Ruberti S, Morelli G. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996;10:393–402. doi: 10.1046/j.1365-313X.1996.10030393.x. [DOI] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14(6):1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Herzog M, Hulskamp M, Brown S, Dorne A, Bonneville J. Trichome cell growth in Arabidopsis thaliana can be depressed by mutations in at least five genes. Genetics. 1999;152(1):461–476. doi: 10.1093/genetics/152.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AM, Walbot V, Davis RW. Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science. 1992;258:1773–1775. doi: 10.1126/science.1465611. [DOI] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol. 1994;166:740–754. doi: 10.1006/dbio.1994.1352. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Dellaert LWM, VanderVeen JH. EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana. Mutat Res. 1982;93:109–123. doi: 10.1016/0027-5107(82)90129-4. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr AP. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/S0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Clough S, Bent A. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Schrader A, Welter B, Hulskamp M, Hoecker U, Uhrig JF. MIDGET connects COP1- dependent development with endoreduplication in Arabidopsis thaliana. Plant J. 2013;75(1):67–79. doi: 10.1111/tpj.12199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of amino acid (aa) sequences between AtTTG1 and AaTTG1.
Complementation test between the Aattg1-1 and Aattg1-2 mutant alleles and rescue experiments in A. thaliana. First true leaves of A) wild type Paj, B) Aattg1-1, C) pep1-1, D) Aattg1-2, E) First true leaf of a F1 plant from the cross between Aattg1-1 and Aattg1-2. Leaves are glabrous indicating allelism. Scale bar = 1 mm. F) A. thaliana ttg1-1 rosette leaves. Plants are completely devoid of trichomes in this allele. G) A. thaliana Atttg1-1ProAtTTG1:AaTTG1Paj plant showing partial rescue of the trichome phenotype. H) Atttg1-1ProAtTTG1:AaTTG1pep1-1 plant showing partial trichome rescue. Scale bar = 1 mm.
HPLC-MS analysis of cyanidin and kaempferol in seeds and seedlings of A. alpina – full chromatograms. Shown are total ion chromatogram (TIC, left) and extracted ion chromatogram (EIC, right) for 750 nM cyanidin (MeOH), 750 nM kaempferol (MeOH) and for all samples shown in Figure 2C and Figure 2F. The m/z value for cyanidin ([M]+) and kaempferol ([M + H]+) is 287.055. EICs for m/z = 287.055 +/− 0.005 were generated based on the corresponding TICs using the Compass DataAnalysis software Version 4.0 SP5 (Bruker Daltonics, Bremen, Germany). In the TIC the retention time and in the EIC the peak of cyanidin and kaempferol is marked. 1: cyanidin; 2: kaempferol. Dashed lines mark the begin and end of each sample. Before the first dashed line the mass calibration for each run can be seen in the TIC.
Ruthenium red stained seeds of wild type and Aattg1 mutants. Light microscopy image of the surface of A. alpina seeds. The dome shaped columella is stained with ruthenium red labeling the seed coat mucilage. A) Wild type Paj. B) pep1-1 mutant. C) Aattg1-1 mutant induced in the wild type Paj background. D) Aattg1-2 mutant induced in the pep1-1 background. Note, the absence of ruthenium red stained columellas in both mutants. Scale bar = 500 μm, inset = 50 μm.
SEM pictures of wild type and Aattg1 mutant seeds. Scanning Electron Micrographs of the surface of A. alpina seeds. A, B) wild type Paj and pep1-1 mutant, respectively. Note, that the surface is irregularly but smooth and that the columella is seen as small domes. C, D) Aattg1-1 mutant induced in the wild type Paj background and the Aattg1-2 mutant induced in the pep1-1 background. Only the rim of the epidermal cells is left. Columellas are absent. Scale bar: 100 μm.