Abstract
There is a great demand for rapid tests that can be used on-site for the detection of small analytes, such as pesticides, persistent organic pollutants, explosives, toxins, medicinal and abused drugs, hormones, etc. Dipsticks and lateral flow devices, which are simple and provide a visual readout, may be the answer, but the available technology for these compounds requires a competitive format that loses sensitivity and produces readings inversely proportional to the analyte concentration, which is counterintuitive and may lead to potential misinterpretation of the result. In this work, protein-multipeptide constructs composed of anti-immunocomplex peptides selected from phage libraries and streptavidin/avidin as core protein, were used for direct detection of small compounds in a non-competitive two-site immunoassay format that performs with increased sensitivity and positive readout. These constructs that we termed “Nanopeptamers” allow the development of rapid point-of-use tests with a positive visual endpoint of easy interpretation. As proof of concept, lateral flow assays for the herbicides molinate and clomazone were developed and their performance was characterized with field samples.
Keywords: Immunoassay, carbon black, noncompetitive
Immunoassays are analytical tests that exploit the high capacity of antibodies for recognizing target analytes with high affinity and specificity. Due to their simplicity, sensitivity, cost-effectiveness and matrix tolerance, immunoassays have been used in countless applications in pharmacology, food safety, environment, homeland security, narcotics, etc.1–3 In these methods, the antigen-antibody reaction is generally coupled to a signal generating molecule (tracer) which translates the interaction into a quantitative readout. If the analyte is a macromolecule, typically the antigen is first captured by a primary antibody and then detected with a secondary antibody coupled to the tracer. This two-site format allows the use of excess concentrations of reacting antibodies, promoting the formation of the trivalent immunocomplex, even in the presence of trace amounts of the analyte (high assay sensitivity). Unfortunately, this is not possible for the important group of small analytes (drugs, metabolites, toxins, additives, pesticides, explosives, etc.), because they cannot bind two antibodies simultaneously. The measurement of such small molecules (immunochemically classified as haptens) is typically accomplished by competitive reactions between the analyte and a labeled variant of the analyte in the presence of limiting amounts of the anti-hapten antibody. This competitive format performs with inferior sensitivity, precision, kinetics, and working range than the two-site noncompetitive format,4 and produces readouts inversely proportional to the analyte concentration, making more difficult its adaptation into microarrays or microfluidic devices, or into rapid “on-site” visual assays such as lateral flow tests.
Several attempts have been made to implement small-molecule noncompetitive assays, but most of them are limited to particular chemical structures or require analyte labeling.5–7 By focusing the recognition of the analyte-antibody immunocomplex (IC) into changes of the hapten binding pocket upon analyte binding, we were able to isolate short-peptide loops from phage display libraries that specifically react with the analyte-capture antibody IC.8 Using M13 phage particles displaying these peptides, we developed two-site phage anti-IC assays (PHAIA) (figure 1A) for the herbicides molinate, atrazine and clomazone,8,9 the drugs digoxin and cyclosporine,8 the flame-retardant brominated diphenyl ether,10 and the pyrethroid metabolite phenoxybenzoic acid.11 Recently, other groups have also isolated anti-IC peptides for gibberellins12 and a 1,3-diketone hapten derivative13 further showing the general applicability of the concept. The PHAIA method provides higher sensitivity than the conventional format (one or more orders of magnitude) and the two-site recognition also accounts for a higher specificity.9 The phage particles proved to be robust and versatile assay components, 14 but they are “unconventional” reagents for the immunoassay industry and their biological nature (an E. coli infecting phage) could be a matter of concern in some laboratories. For this reason we sought to develop PHAIA into a phage-free detection method by using synthetic peptides designed from the anti-immunocomplex peptide sequences isolated from phage display libraries attached to a protein scaffold. These constructs that we termed “Nanopeptamers” allow sensitive detection of ICs and could be developed into lateral flow tests, a feat that turned up to be particularly difficult with phage particles due to their filamentous nature.
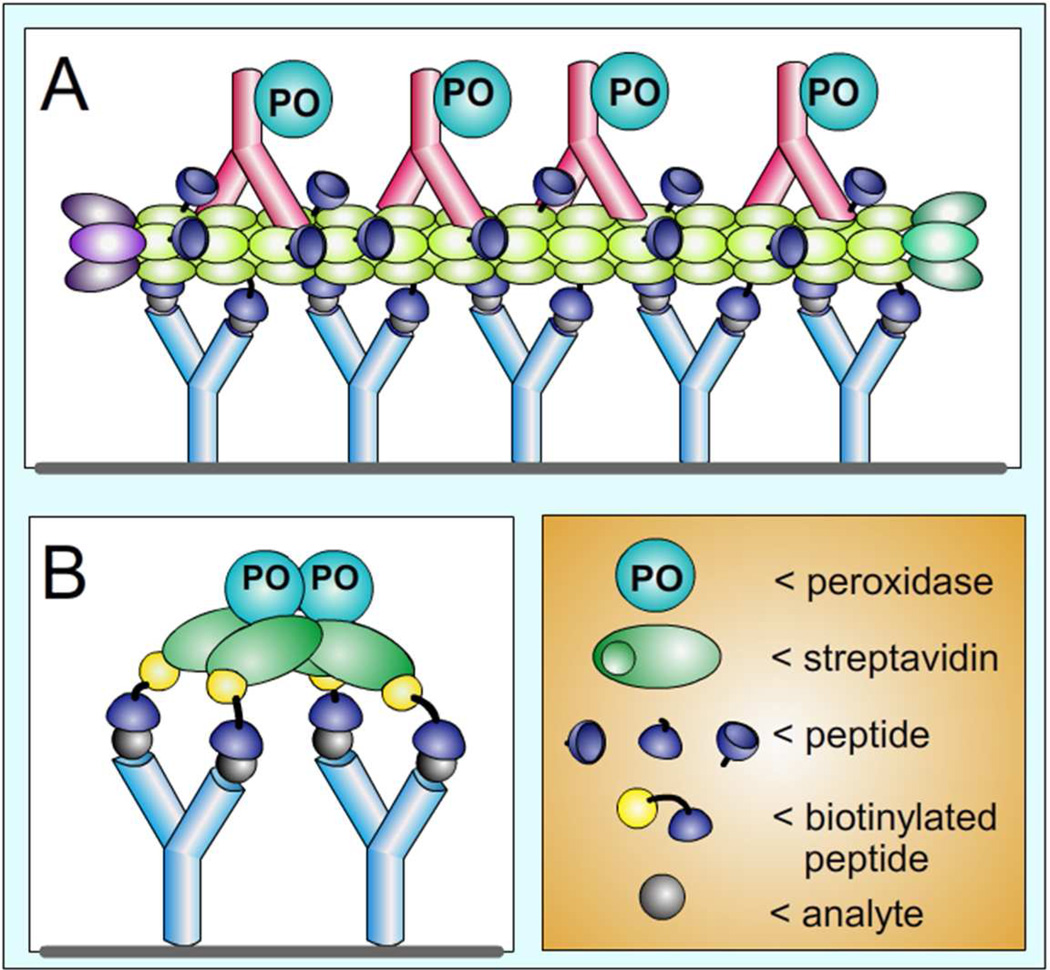
Figure 1.
Scheme of PHAIA (phage anti-immunocomplex assay) and the Nanopeptamer noncompetitive assays. Panel A shows the scheme of PHAIA that typically uses filamentous M13 phage (light green) expressing disulfide constrained peptides (~100–200 copies along the ~2700 copies of the coat protein pVIII). Binding of the phage to the analyte-antibody (light blue) immunocomplex is detected with an anti-M13 antibody (pink) coupled to horse radish peroxidase (PO). Panel B schematizes the substitution of the phage particle by a Nanopeptamer (a streptavidin conjugate, combined with four molecules of the biotinylated peptide)
EXPERIMENTAL SECTION
Materials
Molinate and the thiocarbamate standards were gifts from Stauffer Chemical Co. Thiobencarb was a gift from Chevron Chemical Co. Development of the monoclonal anti-molinate antibody (MoAb 14D7) and anti-clomazone (MoAb 5.6) has been described in detail previously1,2. Clomazone was purchased from Riedel-de Haen (Seelze, Germany), high-sensitivity Streptavidin-Peroxidase (SPO) from Pierce (Rockford, IL), and Bovine Serum Albumin (BSA), Tween 20, and 3,3′,5,5′-tetramethylbenzidine (TMB) and avidin from Sigma (St.Louis, MO). ELISA and dilution microtiter polystyrene plates were purchased from Greiner (Solingen, Germany). Hi-Flow Plus 120 nitrocellulose membrane cards and cellulose absorbent pads were purchased from Millipore (Bedford, MA). Carbon black nanoparticles were from Degussa AG (Frankfurt, Germany).
Formation of anti-molinate and anti-clomazone Nanopeptamers
Two anti-IC peptide sequences previously isolated for molinate 3 and one for clomazone 4 were produced by a commercial manufacturer (Peptron Co., Daejeon, Korea). These peptides were synthetized to eighty percent of purity by HPLC, with intramolecular disulfides bonds between cysteines, an N-terminal biotin molecule and amidated C-terminus. The synthesized peptides were: p1M, biotin-SGSGCSTWDTTGWC; pA, biotin-SGSGCSLWDTTGWC; and pX11, biotin-SGSGCLEAPNIEGC; for molinate, molinate and clomazone, respectively, (the anti-IC sequence is in black and the SGSG was used as spacer). Two hundred and fifty picomoles of SPO or avidin in 100µL phosphate-buffered saline (PBS), 1% BSA were incubated with 50, 6.5, 1.5 and 0.6 fold molar excess of biotinylated peptides for 15 minutes on ice. The Nanopeptamers were initially separated from the excess of biotinylated peptides by gel filtration, but this step proved to be unnecessary.
Nanopeptamer ELISA
Microtiter plates were coated with 100 µL of the anti-herbicide monoclonal antibody in PBS at different concentrations (10 to 2.5 µg/mL). After incubation for 1 hour at 37 °C and blocking with PBS BSA 1% 1 hour at 37 °C, the microtiter plates were washed three times with PBS 0.05% Tween 20 (PBST). Nanopeptamer (prepared with SPO and the corresponding anti-IC biotinylated peptide) were diluted with PBST to the appropriate dilution, and 50 µL were mixed with the 50 µL of the herbicide standard or water sample in non-treated polystyrene plates (low binding capacity), transferred to the reaction microtiter plate and incubated for 1h at room temperature. After thoroughly washing, 100 µL of the peroxidase substrate (0.4 ml of a 6 mg/ml DMSO solution of 3,3´,5,5´-tetramethylbenzidine, 0.1 mL of 1% H2O2 in water in a total of 25 mL of 0.1 M citrate acetate buffer, pH 5.5) were dispensed into each well. The enzyme reaction was stopped after 20 min by addition of 50 µL of 2 N H2SO4, and the absorbance was read at 450/650 nm in a microtiter plate reader (Multiskan MS, Thermo Labsystems, Waltham, MA). Absolute or normalized values were fitted to a four-parameter logistic equation using Genesis Lite 3.03 (Life Sciences, London) package software.
Molinate Nanopeptamer assay cross-reactivity
The specificity of the assay was characterized with related S-thiocarbamate pesticides. Cross-reactant concentrations in the 0–10000 ng/mL range were used in the noncompetitive ELISAs. Standard curves were normalized by expressing experimental absorbance values (B), as (B/B0) ×100, where B0 is the absorbance value at zero analyte concentration, and the molar concentration corresponding to the midpoint of the curve, SC50), was used to express the cross-reactivity of the assay according to the equation % cross-reactivity = 100 × [SC50 (analyte) / SC50 (cross-reacting compound)]
Carbon black avidin labeling
A 5% solution of carbon black in MilliQ water was homogenized in sonication water bath, and was diluted 25 fold with 5 mM boric buffer, pH 8.8. One hundred µL of avidin (4 mg/mL) in 5 mM boric buffer, pH 8.8 were combined with 900 µL of 0.2% carbon black and incubated for 3 h at room temperature. The suspension was then centrifuged a 14,000 rpm for 15 min and the pellet resuspended in 1 mL of 100 mM boric buffer, pH 8.8, 0.02% Tween 20 by sonication. After repeating this step three more times, the carbon-labeled avidin was resuspended in 1 mL of 100 mM boric buffer, pH 8.8, 0.02% Tween 20, and used to prepare the Nanopeptamers as described below.
Lateral Flow Immunocromatography
MoAb 14D7 or MoAb 5.6 lines were printed on Hi-Flow Plus 120 nitrocellulose membrane cards at 0.92 µg/cm using a BioDot AD 1500 Liquid Dispenser. Avidin was labeled with carbon black nanoparticles as described. Approximately 40 µg (100 µL) of carbon black labeled avidin was preincubated with 15 µg of pA or pX11 (6 µL, 2.5 mg/mL in DMSO) in a final volume of PBS containing 0.025 % Tween-20. After incubation for 15 min on ice, 2.5 µL of this carbon black labeled Nanopeptamers mixture were transferred to microtiter plate wells containing 100 µL of PBS, 0.025% Tween previously spiked with known amounts of molinate and clomazone standards. Hi-Flow Plus 120 nitrocellulose membrane cards (printed with MoAbs 14D7 or 5.6) assembled with an absorbent cellulose pad were dipped into the wells and let stand for 10 minutes. After that, the strips were read for the formation of a visible reaction line by the naked eye by four independent observers in three different repetitions of the tests. Molinate assays were validated by performing spiking in runoff water samples of agricultural areas of Uruguay (95 µL water sample + 5 µL PBS × 10, 0.025% Tween 20), and measuring them with strips as described above.
RESULTS AND DISCUSION
Anti-IC peptides can react specifically with ICs when displayed in a multivalent manner
As starting model system we used the thiocarbamate herbicide molinate, the anti-molinate monoclonal antibody 14D715 and the anti-IC synthetic peptides p1M (biotin-SGSGCSTWDTTGWC) and pA (biotin-SGSGCSLWDTTGWC). Both peptides were previously isolated from phage libraries panned against the molinate-14D7 IC. Peptide pA, which differs only by having Leu in position 7th, was isolated from a mutagenesis library derived from the p1M sequence, and while performing with similar sensitivity in PHAIA tests, showed less residual cross-reactivity with the unliganded antibody 8. Initial ELISA experiments aimed at detecting binding of the biotynilated peptides to the molinate-MoAb14D7 IC failed. This was assumed to be caused by the loss of the avidity effect inherent to the display of the peptide on pVIII. This prompted us to express the peptides as N-terminal fusion with the E. coli alkaline phosphatase that forms dimers and therefore a ows bivalent display of the peptides, but only marginal reactivity with the IC was observed (not shown). In order to further increase the avidity of the interaction, we used the tetrameric structure of streptavidin as scaffold for tetravalent display of the biotinylated synthetic peptides. These streptavidin-peptide complexes, that we termed ‘Nanopeptamers’, have the additional advantage that several ready-to-use streptavidin commercial conjugates exist, which can be used as versatile signal-generation elements (figure 1).
Preliminary experiments with different commercial streptavidin-peroxidase conjugates showed the best results when streptavidin conjugated to a polymeric form of horse radish peroxidase conjugated to streptavidin (SPO, high-sensitivity Streptavidin-Peroxidase, Pierce (Rockford, IL)) was used. The Nanopeptamers were initially formed by using an estimated 50-fold molar excess of biotinylated peptide and their reactivity was assayed in microtiter plates coated with MoAb 14D7, in the presence or absence of molinate. Similarly to what we observed with anti-IC phage, the Nanopeptamers reacted specifically with the IC but showed some residual reactivity with the uncombined antibody. This cross-reactivity was more evident at high concentrations of Nanopeptamers and high density of coating antibody (figure S-1, Supporting Information). Since these conditions (excess of reagents) are expected to provide the best assay sensitivity, a trade-off selection of these parameters needs to be done by checkerboard experiments. This is exemplified in figure S2 (Supporting Information) where increasing concentrations of the Nanopeptamer (SPO-pA) improve the assay sensitivity but it also shows that the background signal at zero concentration of analyte deteriorates after a certain point due to the residual cross-reactivity with the uncombined antibody. The influence of the peptide to SPO ratio on assay performance was also studied. The formation of the streptavidin-biotin complex is fast, and due to the exceptionally high affinity of the interaction,16 it is expected that rapidly after mixing all the biotinylated peptide is complexed. However, since the exact stoichiometry of the SPO commercial polymeric conjugate is unknown, the amount of saturating biotynilated peptide was determined empirically (figure S-3, Supporting information).
PHAIA and Nanopeptamer ELISAs perform in a similar way
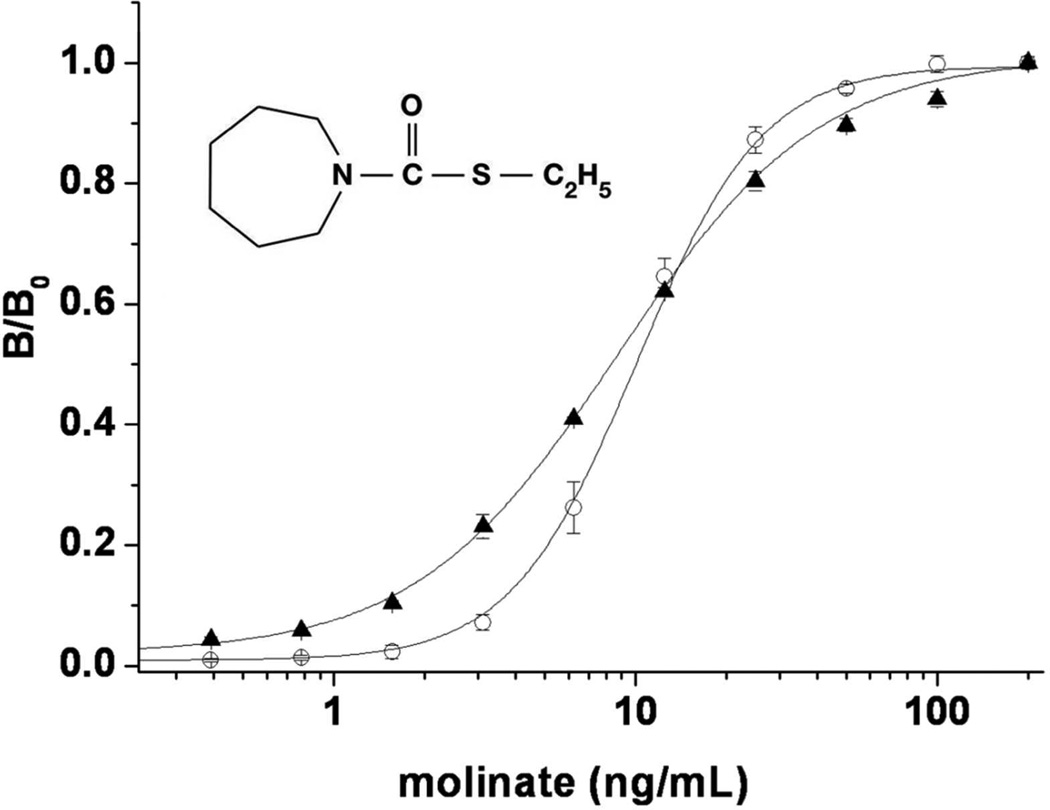
After optimization of these parameters, both Nanopeptamers (SPO-pA and SPO-p1M) were used to develop noncompetitive assays for molinate, figure 2. The dose response curves had a typical sigmoid shape with signal saturation at high concentration of molinate. The midpoint corresponding to the concentration of analyte giving 50% of signal saturation (SC50) were 8.3 ± 0.2 and 10.0 ± 0.3 ng/ mL, and the limit of detection (LOD = analyte concentration giving a 10% increase over the zero signal) were 1.2 and 3.2 ng/mL for Nanopeptamers pA and p1M, respectively. The LOD attained with the Nanopeptamers were up to 18 fold better than that of the competitive ELISA set up with the same antibody (LOD 22 ng/mL, IC50 69 ± 0.5 ng/mL).15
Figure 2.
Noncompetitive Nanopeptamer ELISA for molinate using peptides pA and p1M. MoAb 14D7 (10 µg/mL) was used for coating and SPO (0.75 µg/mL) complexed with a 50 fold molar excess of pA (triangles) or p1M (circles) was used for detection.
The SC50 obtained with Nanopeptamer p1M was not as good as the SC50 obtained in the PHAIA format (SC50 = 5.0 ± 0.4 ng/mL), but this was not the case for the pA complex that performed with a SC50 value equal to that obtained with the phage borne peptide (8.5 ± 0.5 ng/mL).8 Based on these results, Nanopeptamer pA was then used to characterize the cross-reactivity, matrix interference and for adaptation of the Nanopeptamer assay to other formats. Cross-reactivity was tested using a panel composed of common agrochemicals utilized in rice culture (quinclorac, glyphosate, molinate, bispiribac, propanil, and atrazine), as well as other S-thiocarbamate pesticides, table 1. Only minor cross-reactivity with closely related thiocarbamate compounds was observed, which was similar to that obtained with the PHAIA assay. Likewise, no matrix effect was observed when standard molinate curves were performed with undiluted agricultural run off-water samples from different areas of Uruguay (figure S-4, Supporting Information).
Table 1.
Cross reactivity (%) of Nanopeptamer pA and PHAIA assays with related thiocarbamate pesticides.
| Compound | Structure | SPO-pA | PHAIA |
|---|---|---|---|
| Molinate | 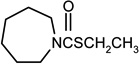 |
100 | 100 |
| Thiobencarb | 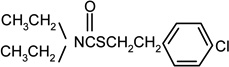 |
0 | 0 |
| Butylate | 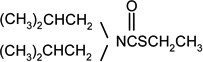 |
1 | 0 |
| EPTC | 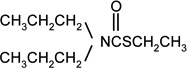 |
2 | 5 |
| Cycloate | 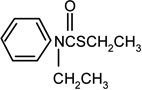 |
10 | 9 |
| Pebulate | 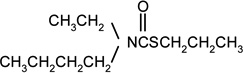 |
6 | 7 |
| Vernolate | 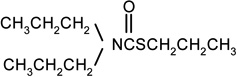 |
5 | 4 |
All data are the mean of two independent experiments. A value of 0 means that there was no observable cross-reactivity at the highest concentration tested 104 ng/mL
Nanopeptamers allow the development of noncompetitive lateral-flow tests
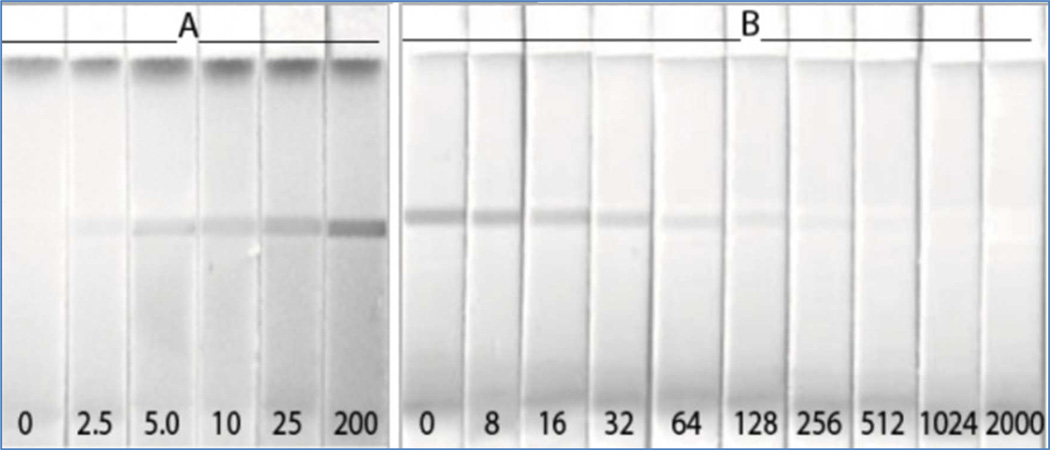
A major advantage of noncompetitive assays is that they can be developed into simple formats with a positive visual endpoint. Our initial attempts to adapt PHAIA into lateral-flow assays were not successful, most probably due to the filamentous nature of the phage that promoted the formation of aggregates with the colloidal labels. Fortunately, this limitation was overcome using Nanopeptamers. Figure 3A shows the results of a lateral-flow assay set up using MoAb 14D7 as capture reagent immobilized on polyester-backed nitrocellulose membranes and colloidal-carbon labeled avidin complexed to pA for detection (see supporting information). For the sake of comparison, we also developed the molinate assay in a lateral-flow competitive format using MoAb 14D7 as capture antibody and the molinate derivative 7b (S-2-(p-aminophenyl)-ethyl-hexahydroazepine-1-carbothioate) coupled to conalbumin, which was labeled with colloidal carbon for detection, figure 3B. A molinate concentration of 2.5 ng/mL caused a visible test line in the noncompetitive assay, while 32 ng/mL produce a weaker test line than the negative control in the competitive assay; as was agreed upon by four independent observers in three different repetitions of the test. This is in agreement, with the densitometry analysis of the strips, table S-1A (Supporting Information). In addition to the positive reading, the noncompetitive test performed with a 10 fold improved sensitivity. The assay was then tested for matrix interference using ten runoff water samples from agricultural areas of Uruguay spiked with 0, 1.0, 1.5, 2.5, 5, and 20 ng/mL of molinate. The strips were read by four independent observers, all of whom detected a visible reaction line with concentrations of 2.5 ng/mL or higher, figure S-5 (Supporting information).
Figure 3.
Noncompetitive Nanopeptamer pA-based (A) and competitive (B) lateral-flow assays for molinate. The nitrocellulose strips were tested with buffer containing various concentrations of molinate (ng/mL) as denoted in the figure.
Nanopeptamer based ELISA and lateral-flow test for clomazone
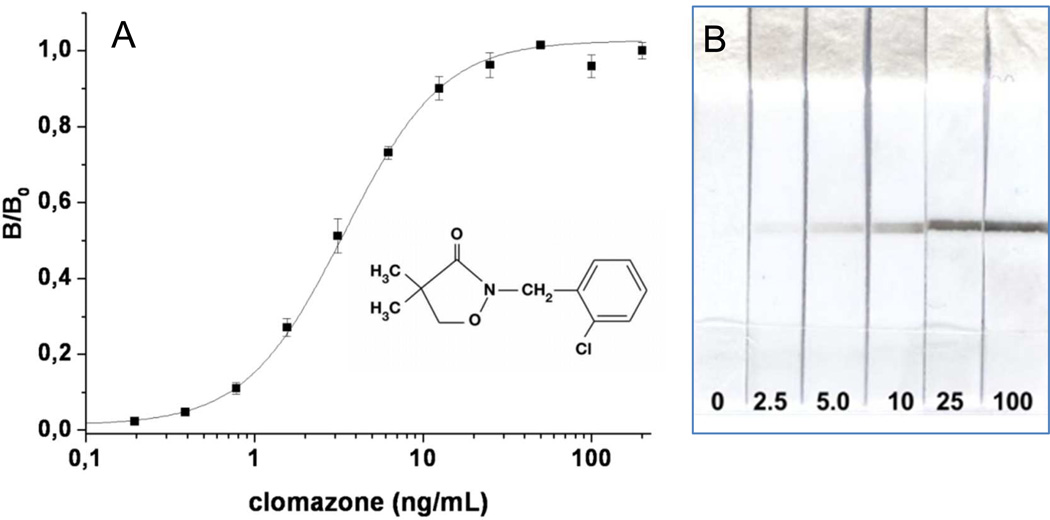
To explore the general utility of the method, we also developed Nanopeptamer-based assays for the herbicide clomazone. To this end we used the anti-clomazone MoAb 5.6 17 and the anti-IC synthetic peptide X11 (biotin-SGSGCLEAPNIEGC) complexed with SPO or avidin. This peptide has been previously isolated from phage libraries panned against the clomazone-MoAb 5.6 IC.9 For ELISA, the assay conditions were optimized essentially as described for the molinate test, which allowed to obtain a LOD = 1.2 ng/mL and SC50 = 3.4 ± 0.2 ng/mL using SPO-pX11 (figure 4A). This represents an improvement of 3.3 and 8.3-fold regarding the assay set up with the same antibody in a competitive format (LOD = 4 ng/ml and IC50 = 28 ± 1.1 ng/mL, respectively).17 The assay was also adapted into a lateral-flow format using avidin-pX11 labeled with carbon black, which allowed to detect up to 2.5 ng/mL of clomazone (figure 4B); the densitometry data is shown in table S-1B.
Figure 4.
Noncompetitive Nanopeptamer ELISA and lateral-flow test for clomazone using peptide pX11. A) MoAb 5.6 (10 µg/mL) was used for coating and SPO (0.75 µg/mL) complexed with a 50 fold-molar peptide excess for detection. B) Nitrocellulose strips were tested with buffer containing various concentrations of clomazone (ng/mL) as denoted in the lower part of the figure.
Conclusions
Our results show that in spite of big structural differences, the streptavidin/avidin-based Nanopeptamer properly reproduces the binding characteristics of the phage particles. Indeed, the dihedral D2 molecular symmetry of the streptavidin homo-tetramer positions two pairs of biotin binding sites on opposite faces of the protein, and thus the distance between the biotin carboxylated oxygens that anchor the peptides are ~2 nm on the same face, and ~3 nm on opposite faces of the complex.18 On the other hand, M13 has a ~6 × 1000 nm rod-like structure, covered by ~2700 copies of the minor protein pVIII arranged in a fish-scale pattern, and the use a phagemid system for the generation of the virons yields phage particles with approximately 200 copies of pVIII expressing the peptide, meaning an average distance between peptides close 10 nm.19 In addition, the peptide is tethered in a different way in both systems. For the sake of simplicity, the peptides were synthesized with a biotin residue added to their N-terminus, while they are expressed as pVIII N-terminal fusions on the phage. Considering the differences in valences and display features of both systems, it seems that the specific recognition of the IC by the peptide can be transferred out of the ‘phage context’ with a considerable degree of flexibility.
As demonstrated here, the availability of phage-free reagents for noncompetitive assays could have a major impact in the development of rapid two-site tests for small molecules with a positive visual endpoint, a feat not possible with the available technology. Since the strategy for the selection of anti-IC phage borne peptides is well established, and considering the varied offer commercially available streptavidin conjugates, Nanopeptamers may be the long-sought detection reagent for the development of fast noncompetitive assays for small analytes. In that sense a major proof of concept contributed by our work is their use in the development of lateral flow test for small molecules with a positive reading improving the sensitivity and interpretability of these tests.
Supplementary Material
ACKNOWLEDGMENT
We are in debt to Patricia Noguera, Department of Chemistry, Polytechnic University of Valencia, for her help with the initial lateral-flow experiments with phage.
Funding Sources
This work was supported with funds provided by grants FMV 3138 ANII (Agencia Nacional de Investigación e Innovación, Uruguay) and TW05718 Fogarty Center NHI.
ABBREVIATIONS
- PHAIA
phage anti-immunocomplex assay
- SPO
high-sensitivity Streptavidin-Peroxidase
- MoAb
monoclonal antibody
Footnotes
ASSOCIATED CONTENT
Supporting Information. Five figures and one table. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- 1.Hage DS. Anal. Chem. 1995;67:455R. doi: 10.1021/ac00108a030. [DOI] [PubMed] [Google Scholar]
- 2.Ng AH, Uddayasankar U, Wheeler AR. Anal. and Bioana. Chem. 2010;397:991. doi: 10.1007/s00216-010-3678-8. [DOI] [PubMed] [Google Scholar]
- 3.Schneider RJ. Anal. and Bioana. Chem. 2003;375:44. doi: 10.1007/s00216-002-1659-2. [DOI] [PubMed] [Google Scholar]
- 4.Jackson TM, Ekins RP. J. Immunol. Methods. 1986;87:13. doi: 10.1016/0022-1759(86)90338-8. [DOI] [PubMed] [Google Scholar]
- 5.Piran U, Riordan WJ, Livshin LA. Clin. Chem. 1995;41:986. [PubMed] [Google Scholar]
- 6.Pradelles P, Grassi J, Creminon C, Boutten B, Mamas S. Anal. Chem. 1994;66:16. doi: 10.1021/ac00073a005. [DOI] [PubMed] [Google Scholar]
- 7.Ueda H. J. Bioscience and Bioeng. 2002;94:614. [PubMed] [Google Scholar]
- 8.Gonzalez-Techera A, Vanrell L, Last JA, Hammock BD, Gonzalez-Sapienza G. Anal. Chem. 2007;79:7799. doi: 10.1021/ac071323h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossotti MA, Carlomagno M, Gonzalez-Techera A, Hammock BD, Last J, Gonzalez-Sapienza G. Anal. Chem. 2010 doi: 10.1021/ac101476f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HJ, Rossotti MA, Ahn KC, Gonzalez-Sapienza GG, Gee SJ, Musker R, Hammock BD. Anal. Chem. 2010;401:38. doi: 10.1016/j.ab.2010.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Techera A, Kim HJ, Gee SJ, Last JA, Hammock BD, Gonzalez-Sapienza G. Anal. Chem. 2007;79:9191. doi: 10.1021/ac7016713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inaba J, Nakamura S, Shimizu K, Asami T, Suzuki Y. Anal. Biochem. 2009;388:63. doi: 10.1016/j.ab.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka F, Hu Y, Sutton J, Asawapornmongkol L, Fuller R, Olson AJ, Barbas CF, 3rd, Lerner RA. Bioorganic Med. Chem. 2008;16:5926. doi: 10.1016/j.bmc.2008.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HJ, McCoy M, Gee SJ, Gonzalez-Sapienza GG, Hammock BD. Anal. Chem. 2011;83:246. doi: 10.1021/ac102353z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rufo C, Hammock BD, Gee SJ, Last JA, Gonzalez-Sapienza G. J. Agric. Food Chem. 2004;52:182. doi: 10.1021/jf034710a. [DOI] [PubMed] [Google Scholar]
- 16.Srisa-Art M, Dyson EC, deMello AJ, Edel JB. Anal. Chem. 2008;80:7063. doi: 10.1021/ac801199k. [DOI] [PubMed] [Google Scholar]
- 17.Carlomagno M, Matho C, Cantou G, Sanborn JR, Last JA, Hammock BD, Roel A, Gonzalez D, Gonzalez-Sapienza G. J. Agric. Food. Chem. 2010;58:4367. doi: 10.1021/jf9043259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrickson WA, Pahler A, Smith JL, Satow Y, Merritt EA, Phizackerley RP. Proc. Natl. Acad. Sci. 1989;86:2190. doi: 10.1073/pnas.86.7.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martens CL, Cwirla SE, Lee RY, Whitehorn E, Chen EY, Bakker A, Martin EL, Wagstrom C, Gopalan P, Smith CW, et al. J. Biol. Chem. 1995;270:21129. doi: 10.1074/jbc.270.36.21129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.