Abstract
We previously developed cell-based kinetics assays of chloride channel modulators utilizing genetically encoded yellow fluorescent proteins. Fluorescence platereader-based high-throughput screens yielded small-molecule activators and inhibitors of the cAMP-activated chloride channel CFTR and calcium-activated chloride channels, including TMEM16A. Here, we report a microfluidics platform for single-shot determination of concentration-activity relations in which a 1.5 × 1.5 mm square area of adherent cultured cells is exposed for 5–10 min to a pseudo-logarithmic gradient of test compound generated by iterative, two-component channel mixing. Cell fluorescence is imaged following perfusion with an iodide-containing solution to give iodide influx rate at each location in the image field, thus quantifying modulator effects over a wide range of concentrations in a single measurement. IC50 determined for CFTR and TMEM16A activators and inhibitors by single-shot microfluidics were in agreement with conventional plate reader measurements. The microfluidics approach developed here may accelerate the discovery and characterization of chloride channel-targeted drugs.
Keywords: drug discovery, CFTR, calcium-activated chloride channel, microfluidic channel
Introduction
Chloride channels are an important class of targets for drug discovery.1 The cAMP chloride channel CFTR (Cystic Fibrosis Transmembrane conductance Regulator) is expressed in the airways, pancreas, intestine, testes and other tissues.2 Excessive activation of CFTR in intestinal enterocytes by enterotoxins causes secretory diarrheas, and CFTR-mediated fluid secretion is involved in cyst growth in polycystic kidney disease for cyst growth. Loss-of-function CFTR mutations cause the genetic disease cystic fibrosis.3,4 Calcium-activated chloride channels (CaCCs), such as TMEM16A, are widely expressed in various tissues, including salivary gland, airways, smooth muscle and nociceptive neurons, in which they facilitate epithelial fluid secretion, smooth muscle contraction and neural signal transduction.5,6 Modulators of CFTR and CaCC chloride channel function have potential therapeutic efficacy in a wide range of diseases, including cystic fibrosis, polycystic kidney disease, secretory diarrheas, pain and dry eye/mouth syndromes.
Our lab developed cell-based functional assays for discovery of small-molecule modulators of chloride channel activity. The assays utilize mutated yellow fluorescent proteins, such as YFP-H148Q/I152L,7–9 whose fluorescence is strongly quenched by iodide or chloride. In one adaptation for plate reader assays, cells expressing the fluorescent sensor, together with a chloride channel, are subject to an inwardly directed iodide gradient. Intracellular iodide accumulation in response to channel-mediated influx causes fluorescence quenching, whose kinetics provides a quantitative, linear measure of channel activity. Rates of iodide influx (JI, in mM/s) are computed from initial slope of the fluorescence curve (dF/dt)t=0 and the relationship between YFP fluorescence and iodide concentration: JI = (KIFo)−1(dF/dt)t=0, where Fo is initial fluorescence in the absence of iodide, and KI is the Stern-Volmer quenching constant (in M−l).10 Yellow fluorescent protein-based high-throughput screens have yielded activators and inhibitors of CFTR,9,11,12 mutant CFTRs that cause cystic fibrosis,13–16 and CaCCs.5,17–19
An important step in the characterization of active compounds emerging from high-throughput screens is the determination of concentration-activity relationships. Here, we report a simple microfluidics platform for single-shot determination of concentration-dependent chloride channel activity over a wide range of activator/inhibitor concentrations. As diagrammed in Fig. 1, a concentration gradient was generated using a multiplex microchannel design, based on that reported by Jeon et al. and Stephan et al.20,21 Cells cultured on a coverglass in a microfluidics device are exposed for several minutes to a wide range of concentrations of a chloride channel modulator. Chloride channel activity is then determined from the kinetics of cell fluorescence following infusion of an iodide-containing solution, in which quantitative imaging of the area containing cultured cells gives influx data at each location, corresponding to different concentrations of modulator compound. The method was applied to dose-response analysis of small-molecule activators and inhibitors of CFTR and TMEM16A chloride channels.
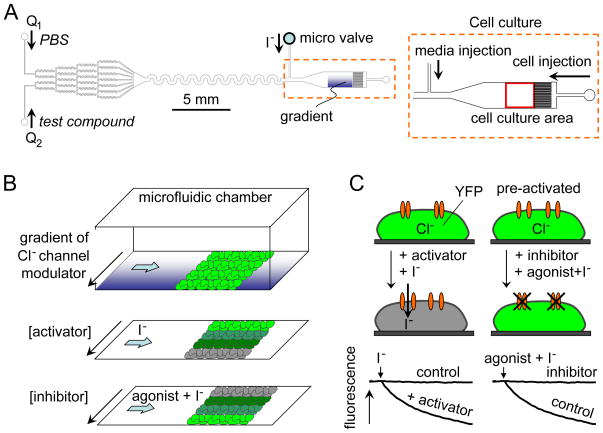
Figure 1.
Cell-based microfluidics fluorescence assay of chloride channel activity. A. Microfluidic chamber design showing gradient-generating channels at the left and cell culture area at the right. B. Assay method showing perfusion with a concentration gradient of chloride channel modulator followed by rapid exchange (top) to generate an inwardly directed iodide gradient of activator (middle) or inhibitor (bottom). C. Schematic showing fluorescence quenching of a cytoplasmic YFP iodide sensor following addition of iodide with an activator (left) and inhibitor (right).
Experimental
Microfluidics chamber design and fabrication
As diagrammed in Fig. 1A, the microfluidics chamber was designed with three major parts, including a: (a) gradient generator to give a continuous and stable pseudo-logarithmic profile of compound concentration by repeatedly splitting, mixing and recombining using a symmetrical channel network, (b) smoothing component to generate a smooth concentration gradient profile in a 300-μm wide channel, and (c) 1.5 × 1.5 mm square observation area for viewing using a 4× magnification lens in an epifluorecence microscope.
The design of the gradient-generating component was based on prior work by Jeon et al. and Dertinger et al.20,21 As the fluid streams travel through the microfluidic network they were repeatedly split, mixed, and recombined. After several generations of branching, all branches are recombined into a single channel, established a concentration gradient across the channel.
The microfluidics chamber was fabricated using conventional soft lithography22,23 with film mask printing, SU-8 master fabrication, and PDMS base and curing agent (Dow Corning, Sylgard 184), with molding at 10:1 w/w ratio and baking for 2 h at 80 °C. After thermal curing the PDMS structure layer was peeled from the master, and inlet and outlet holes were created using a cylindrical punch. The structured PDMS layer and cover glass were treated with air plasma (Harrick Plasma, PlasmaFlo PDC-FMG and Plasma cleaner PDC-32G) at 700 mTorr for 50 s and thermally bonded for 5 min, which produced strong bonding with hydrophilic surface characteristics.
Cell culture
Fisher rat thyroid (FRT) cells stably co-expressing human CFTR and the halide sensor YFP-H148Q were generated as described.11 Briefly, full-length wild-type human CFTR cDNA was subcloned into pcDNA3.1/Zeo (Invitrogen) and co-transfected into FRT cells with the YFP-H148Q expression plasmid (pcDNA3.1/Neo-YFP). Cells were maintained with F-12 modified Coon’s medium (Sigma) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 2 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 100 μg/ml zeocin and 500 μg/ml G418. To establish FRT cells stably co-expressing human TMEM16A (abc isoform) and halide sensor YFP-F46L/H148Q/I152L, human TMEM16A cDNA was subcloned into pcDNA3.1/Neo (Invitrogen) and co-transfected into FRT cells with the YFP-F46L/H148Q/I152L expression plasmid (pcDNA3.1/Hygro-YFP). Cells were maintained with selection agents (100 μg/ml hygromycin B and 500 μg/ml G418) as described.18
Prior to cell injection the microfluidic chamber was filled with 100 μg/ml bovine serum fibronectin (Sigma Aldrich) and place in a humidified incubator at 37 °C for 1 h to improve cell attachment. The fibronectin solution was washed out with PBS and the chamber filled with media. Cells were then slowly infused by gravity into the cell culture area through a 100-μL pipette tip (~2×105 cells/mL, ~2×103 cells/mm2 in the culture area). The inlet and the outlet holes were filled with media to prevent evaporation and cell movement during 2 h incubation to allow attachment. Culture was then continued on the rectangular observation area by continuous perfusion using a syringe pump or by gravity with medium for 8–24 h in a 37 °C cell culture incubator.
Measurement protocol
The microfluidics chamber and syringe pump (KD Scientific, Gemini 88 Dual Syringe pump) containing 250-μl Luer-lock glass syringes (Hamilton, 1700 series) were connected using Teflon tubing and connectors (Upchurch, P-659 and F-331Nx), and mounted on the stage of an inverted epifluorescence microscope (Nikon, ECLIPSE TE2000-U). Chloride channel activators or inhibitors (Table 1) were infused through two inlets using a syringe pump. A stable concentration gradient was generated in less than 30 seconds. To minimize effects of compound exposure during the pre-steady-state, PBS was injected initially into prior to infusion of test compound. Cells were incubated with compounds by continuous perfusion for 5–15 min. During compound incubation the iodide (or ATP/iodide) inlet port remained closed using a micro-mechanical valve (Upchurch, P-732 shut-off valve) to generate a stable concentration gradient. YFP fluorescence from a 1.5 × 1.5 mm rectangular observation area was imaged at 2 Hz using a 4x dry objective lens (Nikon, Plan Fluor, working distance 17.1 mm, numerical aperture 0.13), YFP fluorescence filter cube (Nikon, ET-YFP 49003) and CCD camera (Hamamatsu, ORCA-ER). Cellular iodide influx was initiated by rapid perfusion via the side port to displace the compound-containing solution (Fig. 1B). The two inlets were switched off during perfusion with the iodide-containing solution (Q1=Q2=0), which filled the full observation area. The solution exchange time was < 100 ms. The kinetics of YFP fluorescence provides a quantitative measure of chloride channel activity. Fig. 1C schematically shows quenching of YFP fluorescence following chloride channel activator (left), which is prevented by chloride channel inhibitor (right).
Table 1.
Compositions of solutions containing chloride channel activators and inhibitors.
| CFTR activator study | CFTR inhibitor Study | TMEM16A inhibitor study | |
|---|---|---|---|
|
|
|
||
| Inlet 1 | PBS | 5 μM forskolin | PBS |
| Inlet 2 | 30 μM forskolin | 5 μM CFTRinh-172 +5 μM forskolin | 30 μM TMinh-A01 |
| I− flux | 140 mM I− | 140 mM I− | 70 mM I−+ 50 μM ATP |
Data analysis
Microfluidics data were analyzed using Matlab (The Mathworks, version 7.1). For analysis, the 1.5 × 1.5 mm square observation area was arbitrarily divided into rectangular regions of 300 μm × 100 μm, giving a 5 by 15 array. The YFP fluorescence signal was summed in each rectangular region, with the 5-array used for standard error determination and 15-array for the pseudo-logarithmic concentration profile. Different array sizes can be used for different resolutions. Following threshold analysis to eliminate bright spots from damaged cells (< 1 % cells) integrated fluorescence intensities over 300 μm-wide rectangles were averaged and standard errors computed. Initial iodide influx rates were determined from the initial slope of fluorescence decrease, by exponential regression, following infusion of iodide or ATP/iodide. Dose-response curves were computed by the Hill equation: % activation or inhibition = 100/[1+10(log IC50 − log C)×nH)], where C is drug concentration and nH is the Hill coefficient.
Plate reader assays
Plate reader assays were done as described.11,19 CFTR or TMEM16A-expressing FRT cells were plated in 96-well black-walled microplates (Corning). After growth to confluence, each well was washed 3 times with 200 μl PBS, leaving 50 μl PBS. Activators and/or inhibitors were added. After 10 min incubation, fluorescence was measured using a plate reader (Tecan, Infinite F500) for 1 s (baseline), then 50 μl of a 280 mM iodide-containing solution was injected over 1 s. The initial rate of iodide influx was computed by nonlinear regression.
Results and discussion
Gradient generation and cell culture
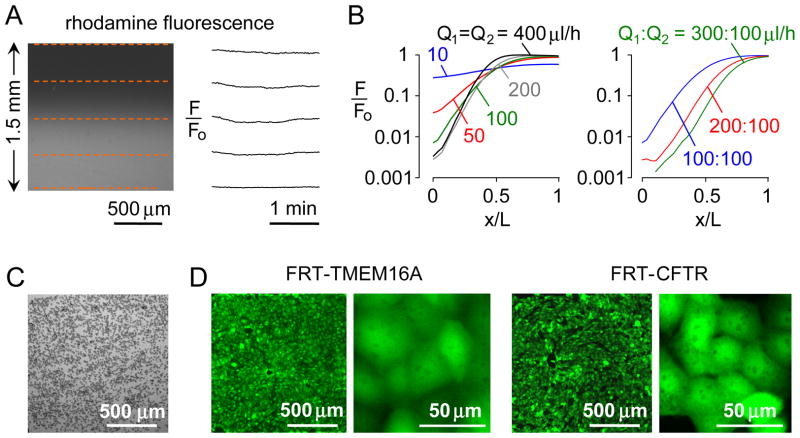
The concentration gradient generated in the microfluidics chamber was quantified and optimized. The chamber was perfused with water in one inlet and a dilute solution of a polar rhodamine-based fluorescent dye in the other inlet. Linearity between rhodamine fluorescence and concentration was verified using the detection system here (Fig. S1). Fig. 2A shows that the gradient is stable over time. Following perfusion of the chamber with the same fluorescent solution in both inlets, we found minimal variation in fluorescence over the square measurement area, with a standard deviation of < 1 % of fluorescence (Fig. S2). Fig. 2B shows a pseudo-logarithmic concentration profile, as deduced from fluorescence intensities, which depended on inlet flow rates. A near-logarithmic concentration profile was found for inlet flow rates of 300 and 100 μl/h, establishing the conditions for further measurements, below.
Figure 2.
Gradient generation and cell culture. A. The microfluidic chamber was perfused with water in one inlet and 5 μM rhodamine fluorescent dye in the other inlet, each at 200 μl/h. Fluorescence micrograph of 1.5 × 1.5 mm observation area (left), showing time-invariant gradient generation (F/Fo, relative fluorescence) at locations indicated by red dashed lines (right). B. Relative fluorescence across the observation area for same flow rate (Q1 = Q2) (left), and different flow rates (right). C. Brightfield micrograph just after injection of TMEM16A-expressing FRT cells (2×105 cells/mL, 2×103 cells/mm2) in the culture area. D. Fluorescence micrographs at 1 day after cultures of TMEM16A- and CFTR-expressing FRT cells shown at two magnifications.
Several methods were tested to culture cells uniformly in the observation area. As explained under Experimental, a cell suspension (~2×105 cells/mL) was infused into the 1.5 mm × 1.5 mm × 50 μm (or 100 μm) rectangular observation volume and allowed to adhere to the fibronectin-coated coverglass (Fig. 2C). Cells adhered tightly within 2 h and a confluent monolayer was seen after 8–12 h culture in a 37 °C incubator in which culture medium was gravity-perfused at a rate of ~20 μl/h (Fig. 2D).
Dose-response analysis of a CFTR activator and inhibitor
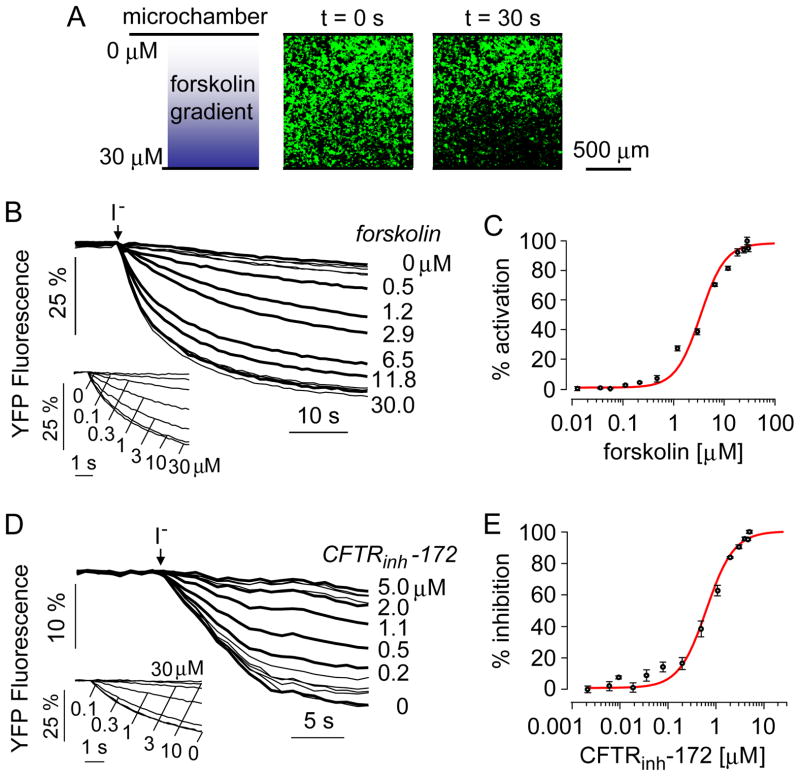
A dose-response measurement was first done of a CFTR activator, the cAMP agonist forskolin. FRT cells expressing human wildtype CFTR were exposed to a concentration gradient of forskolin by infusion of PBS through one inlet of the microfluidic chamber and PBS containing 30 μM forskolin in the other inlet. After 10 min incubation the solution was exchanged rapidly with a buffer containing 140 mM iodide. Iodide was used to quantify channel transport because CFTR and TMEM16A transport iodide efficiently and because of the high sensitivity of YFP fluorescence to iodide. Fig. 3A shows cell fluorescence in the square observation area at t = 0 and 30 s. The reduced fluorescence in the lower part of the panel at 30 s indicates greater CFTR activation, which increases iodide influx and YFP fluorescence quenching. Fig. 3B shows the time course of fluorescence in rectangular areas across the observation area, with deduced forskolin concentrations shown. The single-shot measurements were comparable to data obtained using a conventional plate reader assay in which measurements at single forskolin concentrations were made in individual wells of 96-well plates (Fig. 3B, inset). The signal-to-noise ratio, as computed from the ratio of the signal amplitude and the noise amplitude, was ~1000 and 2100, respectively, for the microfluidics and plate reader measurements. Increasing forskolin resulted in more rapid fluorescence quenching, from which I− influx rates were computed. Fig. 3C shows the deduced dose-response relation from the single-shot microfluidics study, giving an IC50 of 3.2 ± 0.2 (n=5) μM, which was in good agreement with that of 2.9 ± 0.4 μM (n=4) measured in plate reader assays.
Figure 3.
Dose-response analysis of a CFTR activator (forskolin) and inhibitor (CFTRinh-172) measured in CFTR-expressing FRT cells. A. The microfluidic chamber was perfused for 10 min with PBS containing 0 or 30 μM forskolin at 600 μl/h and 200 μl/h flow rates, respectively. Gradient shown on the left and fluorescence micrographs of indicated areas on the right, measured just before and 30 s after perfusion with I− buffer. B. Time course of cellular fluorescence in different rectangular areas corresponding to indicated forskolin concentrations. Inset shows dose-response study done using a conventional platereader assay. C. Deduced single-shot dose-response with fitted IC50 of 3.2 μM and nH of 1.8. D. The microfluidic chamber was perfused for 10 min with PBS containing 0 or 5 μM CFTRinh-172, each together with 5 μM forskolin. Inset shows dose-response analysis done by platereader assay. E. Deduced single-shot dose-response with fitted IC50 of 0.65 μM and nH of 1.4.
A dose-response study was also done for inhibition of CFTR by the thiazolidinone CFTRinh-172.17 CFTR was activated by 5 μM forskolin and exposed for 10 min to a concentration gradient of CFTRinh-172, from 0 to 5 μM. Fig. 3D shows fluorescence time course data, with platereader data shown in the inset. Fig. 3E shows the deduced dose-response relation from the single-shot microfluidics study, giving an IC50 of 0.65 ± 0.2 (n=5) μM, which was in good agreement with that of 1.0 ± 0.2 (n=3) measured in plate reader assays.
Dose-response analysis of a TMEM16A inhibitor
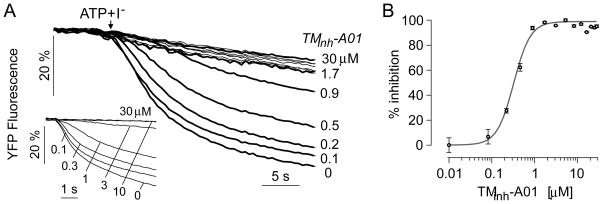
A dose-response study was also done of an inhibitor of the calcium-activated chloride channel TMEM16A, TMinh-A01.25 FRT cells expressing human TMEM16A were exposed to a concentration gradient of TMinh-A01 by infusion of PBS through one inlet of the microfluidic chamber and PBS containing 30 μM TMinh-A01 in the other inlet. After 10 min incubation the solution was exchanged rapidly with a buffer containing 50 μM ATP and 70 mM iodide, which rapidly activates TMEM16A. Fig. 4A shows the time course of fluorescence in rectangular areas across the observation area, with deduced TMinh-A01 concentrations shown. Increasing TMinh-A01 resulted in slowed fluorescence quenching, from which iodide influx rates were computed. The results of a plate reader assay are shown in the figure inset. Fig. 4B shows the deduced dose-response relation from the single-shot microfluidics study, giving an IC50 of 0.35 ± 0.1 (n=5) μM, which was in good agreement with that of 0.4 ± 0.1 (n=5) μM measured in parallel plate reader assays.
Figure 4.
Dose-response analysis of a TMEM16A inhibitor (TMinh-A01) measured in TMEM16A-expressing FRT cells. A. The microfluidic chamber was perfused for 10 min with PBS containing 0 or 30 μM TMinh-A01. Time course of cellular fluorescence in different rectangular areas corresponding to indicated TMinh-A01 concentrations. Inset shows dose-response analysis done by platereader assay. B. Deduced single-shot dose-response with fitted IC50 of 0.35 μM and nH of 2.2.
The microfluidics approach reported here affords rapid, single-shot determination of dose-response relationships for chloride channel activators and inhibitors over a large ~4 orders-of-magnitude concentration range. Image analysis of a square field of adherent cultured cells allowed parallel kinetic analysis of halide flux rates at all points in the image field, providing quantitative information about chloride channel activity as a function of concentration of test activator or inhibitor. Though the approach here was applied to dose-response analysis of chloride channel modulators, it is generally applicable to measurement of cell membrane transport of a wide variety of ions and solutes for which a suitable cellular fluorescent indicator exists.
The technical challenges in our study included development of methodologies for cell culture in the microfluidics chamber, the generation of a stable compound concentration gradient, and the rapid solution exchange to establish a halide gradient. Cell culture methods were optimized for uniform, confluent growth of cells on the square observation surface, which involved fibronectin coating, slow cell injection and adhesion, and continuous infusion with media in a cell culture incubator.
In prior work on cell-based drug screening, concentration-dependence studies were done using multiple, separate cell culture chambers, in which concentration resolution is limited by the number of chambers.24–27 The experimental set-up here gave a continuous and stable concentration profile of test compound in single cell culture chamber with high resolution of compound concentration. Our approach involving iterative mixing required a narrow range of inlet flow rates, which were determined empirically, to produce a stable, pseudo-logarithmic concentration profile and good uniformity along the direction of flow in the observation area. A stable concentration profile in the cell culture chamber was established using simple micro-mechanical valve without requiring a pneumatic valve and vaccum pump. Rapid solution exchange to establish the ion concentration gradient was accomplished using relatively high flow rates, such that solution exchange time was much faster than the kinetics of the transport process.
The microfluidics device reported here was developed specifically for cell-based functional assays for discovery of small-molecule modulators of chloride channel activity. This work built on our prior development of chloride- and iodide-sensing yellow fluorescent proteins and their application to chloride channel drug discovery. In general, the microfluidics approach developed here should be widely applicable for single-shot dose-response analysis of transport activators and inhibitors for which a kinetic, cell-based assay is possible. A wide variety of fluorescent dyes are available to measure pH, cations (sodium, potassium, calcium), membrane potential, and other cellular parameters. Because test compounds are washed out at the time of assay, a requirement of the approach here is that compound washout time should be much greater than assay time. For the chloride channel modulators studied here, washout, which takes tens of minutes is much slower than the iodide influx assay, which takes seconds. Another limitation of our approach is the need for accurate knowledge of concentration gradient, which is best accomplished by mixing of a fluorescent dye-containing solution with a non-fluorescent solution, as done here.
Conclusion
The microfluidics platform developed and validated here allowed for single-shot dose-response analysis of chloride channel modulators with excellent signal-to-noise ratio. The platform, which involves fluorescence imaging of a field of adherent cells, allows for reuse of single chambers and for fabrication of chambers for parallel, medium-throughput measurements.
Supplementary Material
Acknowledgments
This work was supported by NIH grants DK72517, EB00415, HL73856, DK35124 and EY13574, and a Research Development Program grant from the Cystic Fibrosis Foundation.
References
- 1.Verkman AS, Galietta LJ. Nature Rev Drug Disc. 2009;8:153–71. doi: 10.1038/nrd2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang W, Fujii N, Naren AP. Future Med Chem. 2012;4(3):329–45. doi: 10.4155/fmc.12.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verkman AS, Synder D, Tradtrantip L, Thiagarajah JR, Anderson MO. Curr Pharm Des. 2013 doi: 10.2174/13816128113199990321. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiagarajah JR, Verkman AS. Clin Pharmacol Ther. 2012;92(3):287–90. doi: 10.1038/clpt.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Namkung W, Yao Z, Finkbeiner WE, Verkman AS. The FASEB Journal. 2011;25:1–15. doi: 10.1096/fj.11-191627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho H, Yang YD, Lee J, Lee B, Kim T, Jang Y, Back SK, Na HS, Harfe BD, Wang F, Raouf R, Wood JN, Oh U. Nature Neurosci. 2012;15:1015–21. doi: 10.1038/nn.3111. [DOI] [PubMed] [Google Scholar]
- 7.Wachter RM, Remington SJ. Curr Biol. 1999;9(17):R628–R629. doi: 10.1016/s0960-9822(99)80408-4. [DOI] [PubMed] [Google Scholar]
- 8.Jayaraman S, Haggie P, Wachter RM, Remington SJ, Verkman AS. J Biol Chem. 2000;275(9):6047–50. doi: 10.1074/jbc.275.9.6047. [DOI] [PubMed] [Google Scholar]
- 9.Galietta LJ, Haggie PM, Verkman AS. FEBS Lett. 2001;499(3):220–4. doi: 10.1016/s0014-5793(01)02561-3. [DOI] [PubMed] [Google Scholar]
- 10.Verkman AS. Am J Physiol Cell Physiol. 1990;259:C375–88. doi: 10.1152/ajpcell.1990.259.3.C375. [DOI] [PubMed] [Google Scholar]
- 11.Galietta LJ, Springsteel MF, Eda M, Niedzinski EJ, By K, Haddadin MJ, Kurth MJ, Nantz MH, Verkman AS. J Biol Chem. 2001;276(23):19723–8. doi: 10.1074/jbc.M101892200. [DOI] [PubMed] [Google Scholar]
- 12.Ma T, Thiagarajah JR, Yang H, Sonawane ND, Folli C, Galietta LJV, Verkman AS. J Clin Invest. 2002;110(11):1651–58. doi: 10.1172/JCI16112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Shelat AA, Guy RK, Gopinath VS, Ma T, Du K, Lukacs GL, Taddei A, Folli C, Pedemonte N, Galietta LJ, Verkman AS. J Biol Chem. 2003;278(37):35079–85. doi: 10.1074/jbc.M303098200. [DOI] [PubMed] [Google Scholar]
- 14.Pedemonte N, Lukacs GL, Du K, Caci E, Zegarra-Moran O, Galietta LJ, Verkman AS. J Clin Invest. 2005;115(9):2564–71. doi: 10.1172/JCI24898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pedemonte N, Sonawane ND, Taddei A, Hu J, Zegarra-Moran O, Suen YF, Robins LI, Dicus CW, Willenbring D, Nantz MH, Kurth MJ, Galietta LJ, Verkman AS. Mol Pharmacol. 2005;67(5):1797–807. doi: 10.1124/mol.105.010959. [DOI] [PubMed] [Google Scholar]
- 16.Phuan PW, Yang B, Knapp J, Wood A, Lukacs GL, Kurth MJ, Verkman AS. Mol Pharmacol. 2011;80:683–93. doi: 10.1124/mol.111.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De La Fuente R, Namkung W, Mills A, Verkman AS. Mol Pharmacol. 2008;73(3):758–768. doi: 10.1124/mol.107.043208. [DOI] [PubMed] [Google Scholar]
- 18.Namkung W, Thiagarajah JR, Phuan PW, Verkman AS. FASEB J. 2010;24(11):4178–86. doi: 10.1096/fj.10-160648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Namkung W, Phuan PW, Verkman AS. J Biol Chem. 2011;286(3):2365–74. doi: 10.1074/jbc.M110.175109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeon NL, Dertinger SKW, Chiu DT, Choi IS, Stroock AD, Whitesides GM. Langmuir. 2000;16:8311–6. [Google Scholar]
- 21.Dertinger SKW, Chiu DT, Jeon NL, Whitesides GM. Anal Chem. 2001;73:1240–6. [Google Scholar]
- 22.Xia Y, Whitesides GM. Annu Rev Mater Sci. 1998;28:153–84. [Google Scholar]
- 23.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJA, Whitesides GM. Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 24.Hung PJ, Lee PJ, Sabounchi P, Lin R, Lee LP. Biotech Bioengin. 2005;89:1–8. doi: 10.1002/bit.20289. [DOI] [PubMed] [Google Scholar]
- 25.Chen CY, Andrew MW, Jong DS. Lab Chip. 2012;12:794–801. doi: 10.1039/c1lc20548j. [DOI] [PubMed] [Google Scholar]
- 26.Lee K, Kim C, Kim Y, Ahn B, Bang J, Kim J, Panchapakesan R, Yoon YK, Kang JY, Oh KW. Microfluid Nanofluid. 2011;11:75–86. [Google Scholar]
- 27.Ye N, Qin J, Shi W, Liu X, Lin B. Lab Chip. 2007;7:1696–704. doi: 10.1039/b711513j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.