Abstract
All cancers are believed to arise by dynamic, stochastic somatic genomic evolution with genome instability, generation of diversity and selection of genomic alterations that underlie multi-stage progression to cancer. Advanced esophageal adenocarcinomas (EAs) have high levels of somatic copy number alterations. Barrett’s esophagus (BE) is a risk factor for developing EA, and somatic chromosomal alterations (SCAs) are known to occur in BE. The vast majority (~95%) of individuals with BE do not progress to EA during their lifetimes, but a small subset develop EA, many of which arise rapidly even in carefully monitored patients without visible endoscopic abnormalities at the index endoscopy. Using a well-designed, longitudinal case-cohort study, we characterized SCA as assessed by SNP arrays over space and time in 79 “progressors” with BE as they approach the diagnosis of cancer and 169 “nonprogressors” with BE who did not progress to EA over 20,425 person-months of follow-up. The genomes of nonprogressors typically had small localized deletions involving fragile sites and 9p loss/copy neutral LOH that generate little genetic diversity and remained relatively stable over prolonged follow-up. As progressors approach the diagnosis of cancer, their genomes developed chromosome instability with initial gains and losses, genomic diversity, and selection of SCAs followed by catastrophic genome doublings. Our results support a model of differential disease dynamics in which nonprogressor genomes largely remain stable over prolonged periods whereas progressor genomes evolve significantly increased SCA and diversity within four years of EA diagnosis, suggesting a window of opportunity for early detection.
Keywords: Somatic genomic evolution, Barrett’s esophagus, overdiagnosis, genome doubling in cancer, temporal and spatial neoplastic evolution
Introduction
The incidence and mortality of esophageal adenocarcinoma (EA) have been increasing at an alarming rate over the past four decades (1). EA typically presents as a symptomatic, highly aggressive, lethal cancer with a low cure rate (2). Even 15% of small superficial submucosal T1b tumors are lethal, which has been attributed to early lymphatic invasion and metastasis (3–5). This dismal prognosis has led to attempts to detect intramucosal EA, a stage that is nearly 100% curable, by screening and surveillance of BE, the only known precursor of EA (6). However, advanced EAs have been reported to arise even in carefully monitored patients with BE without visible endoscopic abnormalities at the index endoscopy (7–9), and most EAs are detected within three years of the index endoscopy as reported in multiple population and clinical cohort studies (9–13). In the surveillance study with the highest number of EA outcomes, 96% of EAs were detected at a highly curable intramucosal stage, but such early detection required a mean of 163 four-quadrant biopsies obtained every one centimeter throughout the BE segment without visible evidence of cancer during periodic surveillance (7). The sudden appearance of advanced EAs, many with lymphatic invasion, after diagnosis of BE in cohort studies despite careful surveillance, combined with evidence that the great majority (90–95%) of patients with BE die of causes unrelated to the esophagus, has led experts to question the benefit of screening and surveillance for BE (12, 14, 15).
Cancer is a disease of dynamic, stochastic somatic genomic evolution (16, 17). All cancers are believed to arise as a result of (i) somatic genomic instability, (ii) generation of somatic genomic diversity and (iii) natural selection of variants that underlie progression to cancer (16). Based largely on studies of advanced cancers at a single point in time, it has been inferred that different types of genomic instability have different rates of progression. For example, colorectal and some other cancers are believed to develop through gradual accumulation of point mutations at a normal mutation rate over decades (18, 19), which would provide a prolonged time window of opportunity for early detection. However, many cancers show manifestations of chromosome instability, which has been defined as an increased rate of gains or losses of whole chromosomes or large regions of chromosomes (17, 20, 21). Recent studies of advanced cancers at single points in time have inferred that somatic chromosome evolution may occur suddenly, perhaps in one or a few cell divisions (22–25), which could greatly accelerate progression to cancer. Such rapid somatic genomic evolution, which has been called “punctuated evolution”, could have profound implications for early detection of EA and other cancers by narrowing the time window of opportunity for screening and surveillance. EAs have high levels of somatic chromosome copy number alterations compared to other cancers such as gastric and colorectal (26, 27), and there is evidence that chromosome abnormalities develop in BE before EA (28–30). This high level of genome derangement is believed to be due to exposure of BE to gastroduodenal reflux of bile acids, nitrosamines, reactive oxygen species, and inflammatory responses to the injury that are genotoxic, as reviewed in Reid et al (17). Yet, most BE do not progress to EA (12, 17).
Much of the focus of cancer genomic research has understandably been on genes and gene pathways in advanced cancers to improve patient treatment strategies (31, 32). However, relatively little investigation has been devoted to understanding the dynamics of somatic genomic evolution that leads to cancer and underlies clinical observations that some BE rapidly progress to EA while the great majority do not progress over a lifetime. Because endoscopic surveillance for early detection has been a standard of care for BE for more than two decades, it provides an opportunity to investigate somatic genomic evolution over space and time in patients who do and do not progress to EA. Here we present a study of dynamic somatic evolution in BE using a case-cohort design, which unlike case-control or nested case-control studies preserves the characteristics of the entire cohort, allowing evaluation of the temporal occurance of somatic chromosomal alterations (SCA) and EA outcomes while permitting a cost-effective approach for genomic investigations (33).
Materials and Methods
Human Subjects
The Seattle Barrett’s Esophagus Study has been approved by the University of Washington Human Subjects Review Committee since 1983 with reciprocity from the Fred Hutchinson Cancer Research Center (FHCRC) Institutional Review Board since 1994. FHCRC has an approved Federal Wide Assurance (#FWA000019200) with the Department of Health and Human Services (DHHS). The IRB Registration number is IRB00005619. Five hundred and sixteen patients with BE without EA at the baseline endoscopy met the inclusion criteria for the study and were placed into periodic endoscopic biopsy surveillance for early detection of EA. Endoscopic biopsies were obtained using a jumbo forceps and a four-quadrant biopsy protocol for histology and research purposes every one to two centimeter intervals along the entire length of the BE (34). Histologic evaluation for diagnosis of cancer was performed by an expert GI pathologist (RDO). Blood samples collected at endoscopy were processed for use as constitutive genome controls.
Study design
The Seattle Barrett’s Esophagus Study is a dynamic cohort study (35). For this study, eligible participants (n=516) included all cohort members who were enrolled between 8/3/88 and 3/25/09 with at least two endoscopic visits and sufficient tissue for analyses at each time point. No eligible participants were excluded. We then performed a case-cohort study (33) of 248 patients, including all 79 who progressed to EA (progressors) and 169 who did not progress during surveillance follow-up (nonprogressors). A random sample of 197 patients from the eligible cohort (Table S1) was initially selected regardless of follow-up time. All remaining cases (progressors) who were not initially selected were then added to the study group giving a total of 248 patients who were followed for 20,425 person-months. Thus, the case-cohort study included all research participants who developed cancer by the end of follow-up (3/25/09) according to the case-cohort design. No patients were lost due to testing failures. This study design is cost effective as it preserves the characteristics of the cohort while assaying samples with relatively expensive SNP arrays from only a subset of individuals. Samples from two endoscopies were evaluated per individual. For progressors these were the biopsies from the baseline endoscopy and the penultimate endoscopy just prior to the diagnosis of EA (n=63) or at the time of diagnosis of EA (n=16). For nonprogressors, the baseline and the endoscopy prior to the final endoscopy were used.
Sample processing
Previous studies have shown that EAs arise in fields of genomic abnormalities that are larger than the cancers themselves and can be detected using one biopsy every two cm in the BE segment (36–39). One research biopsy obtained at each two centimeter interval from flat mucosa without visible evidence of EA in the BE segment was evaluated by SNP arrays to assure uniform, unbiased sampling of Barrett’s epithelium (Figure S1A). The mean number of biopsies per endoscopy evaluated by 1M SNP arrays was 2.4 (range 1–9) in nonprogressors and 3.1 (range 1–9) in progressors. The number of biopsies reflects a 2cm sampling protocol based on varying segment length. Mean segment length was 5.2 cm (range 1–20 cm) in nonprogressors and 7.0 cm (range 0–19 cm) in progressors, consistent with previous results indicating a trend for increased progression with increased segment length (40). Biopsies were collected in MEM with 10% DMSO, 5% fetal calf serum, 5mM Hepes and frozen at −70°C. Biopsies were incubated 45 minutes in 30mM EDTA in HBSS at room temperature and the epithelium was isolated by peeling the epithelium away from the underlying stroma (Figure S1B) (41). DNA was extracted using Puregene DNA Isolation Kit (Gentra Systems, Inc.) and quantitated with Picogreen (Quant-iT dsDNA Assay, Invitrogen). DNA from 177 blood samples and 71 gastric biopsies were plated as constitutive controls. Eight samples failed resulting in 1,272 BE biopsies with a genotype call rate 99.4%. Genotyping was performed at the FHCRC Genomics Laboratory using 200ng on the Illumina HumanOmni1-Quad beadchip utilizing Infinium HD Super Assay methods, scanned using an Illumina iScan and intensity data extracted with software (GenomeStudio v2010.2 with Genotyping Module v1.7) with a GenCall cutoff of 0.15. Input reports were generated using the Partek Report Plug-in v2.13.1.
HumanOmni1-Quad v1.0 DNA Analysis BeadChip SNP array data processing and SCA calling
GenomeStudio was used to calculate chromosome copy number (logR ratio) of each BE sample paired with its constitutive control and Partek (v6.5) software was used for allele specific copy number estimation. Allele-specific copy number, b-allele frequency and flow cytometric ploidy data (available from previously utilized biopsies in 239 individuals from a previous study (37)) were used to inform manual copy number baseline adjustment in 38/1,272 samples (Figure S2). All samples then underwent segmentation analysis using Partek (v6.5). Additional methods details are provided in Supplementary Materials.
Genetic divergence to assess spatial dynamics in time windows
To determine if heterogeneity of SCA between samples increases with physical distance in the esophagus in different time windows we measured genetic divergence (42) to evaluate differences in SCA between all biopsies within an endoscopy for each individual. Genetic divergence is defined as the proportion of 1 Mb genomic segments across the genome with different SCA calls between any two comparison biopsies. The higher the value the more divergent are the SCA patterns between the two comparision samples. Kruskal-Wallis test was used to compare divergence in each spatial category (2cm, 4cm, 6cm, and 8+cm apart) between nonprogressors and progressors. Mann-Kendall trend test was used to test for a trend of divergence to increase with physical distance within each time window.
Temporal dynamics of SCA
Using SCA data from progressor and nonprogressors biopsies, we assessed odds ratios using genome-wide SCA during evolution to EA in each of the three time windows (0–24, 24–48 and >48 months). Specifically, in each time window, the odds ratio (OR) of SCA in each 1 Mb genomic segment along the whole genome for each type of SCA (yes/no binary variable) was calculated. The p-value for significance of odds ratio was adjusted for multiple comparisons using a false discovery threshold such that the number of falsely discovered 1 Mb genomic segments is 1 or less. The case-cohort study design allowed further quantification of the risk of evolution to EA for various types of SCA across the genome with consideration of individual patient follow-up time. Specifically, the hazard ratio of SCA in each 1 Mb genomic segment along the whole genome for each type of SCA (yes/no binary variable) was estimated based on the case-cohort design (43). P-values were adjusted for multiple comparisons at q= 0.01 level. The hazard ratio quantified the cancer risk for a group that had a specific SCA versus another group that did not have the same SCA. Thus, a value equal to 1 indicated a particular region of SCA had no altered risk of EA; a value larger than 1 indicated the SCA had an increased risk for progression to EA. Therefore, the hazard ratio estimated for a specific location of SCA is the risk for cancer development estimated for those who carry the SCA relative to those who do not carry the SCA in the same location.
Results
Global assessment of dynamic evolution of SCA in nonprogressor and progressor populations
Using a unique longitudinal case-cohort study of individuals with BE, we measured levels and timing of SCA in nonprogressor and progressor populations as they approach the final endoscopy in the study or the diagnosis of a surveillance detected cancer, respectively. The case-cohort study design, which is described in detail in methods, considers the temporal relationship of SCA and EA outcomes and preserves the characteristics of the entire cohort (Table S1) (33, 43). To ascertain an unbiased temporal and spatial sampling from progressors and nonprogressors, we analyzed one biopsy from endoscopically unremarkable BE mucosa without visible evidence of cancer obtained from every two centimeter interval of the BE segment from two endoscopies in each individual, a baseline endoscopic biopsy procedure and the procedure prior to their final endoscopy (169 nonprogressors) or their last endoscopy prior to or at the time of cancer diagnosis (79 progressors). Epithelium isolated from 1,272 mapped esophageal biopsies from the Barrett’s segment were evaluated using Illumina 1M SNP arrays and compared to normal constitutive controls.
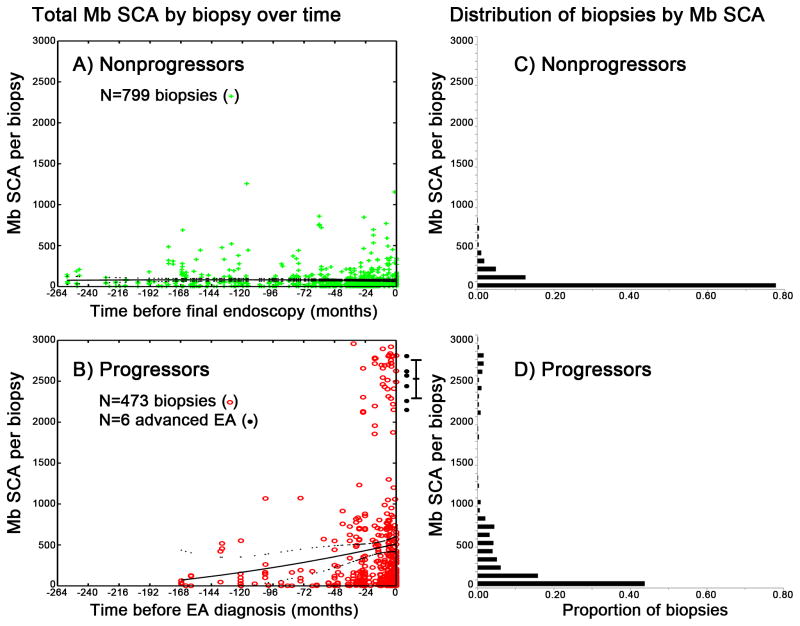
We first assesed total megabases (Mb) of SCA per genome as a metric of whole genome integrity in each biopsy including homozygous deletions, copy loss, copy neutral loss of heterozygosity (cnLOH), balanced copy gain of both alleles and allele-specific copy gain, as a global measure of SCA over time in the population of progressors and nonprogressors as they approach the diagnosis of EA or the last endoscopy in the study (Figure 1A–D). Almost all nonprogressor biopsies have SCA by 1M SNP arrays, but biopsies from nonprogressors had low levels of total SCA that remained low (95% <283 Mb) over prolonged follow-up (range = 0.14–21.4 years, mean = 8.6 years) (Figure 1A, 1C).
Figure 1. Total SCA dynamics in nonprogressor and progressor populations over time.
Panels A, B) Total SCA in megabases (Mb) (y-axis) per biopsies obtained at times (x-axis) before the last endoscopy in nonprogressors (Panel A) or before diagnosis of EA in progressors (Panel B. Solid black lines = trend in mean SCA, dotted black lines = 95% CI of the means fitted by quadratic polynomial regression. Six black points right of Panel B are SCA levels from advanced EA surgical resection specimens, with 95% CI of the mean. Panel C) In nonprogressors, many biopsies 105/799 (13%) have <1 Mb SCA per biopsy, and 759/799 (95%) of all nonprogressors biopsies have SCA <283 Mb, indicating that somatic genomic alterations are present in most Barrett’s biopsies but the level of SCA is generally low in nonprogressors. All 799 nonprogressors biopsies are below 1300 Mb SCA. Panel D). In contrast, only 28/473 (6%) progressor biopsies have <1 Mb SCA, and 168/473 (36%) of biopsies sampled before EA in progressors have SCA above 283 Mb (Panel D). There is a significant gap between 1300–1850 Mb, then in progressors we see 43/473 (9%) biopsies above 1850 Mb SCA, all with significant amounts of copy gain, balanced gain, and cnLOH indicative of genome doubling.
Beyond 48 months, the population of progressors had slightly increased SCA compared to nonprogressors (mean SCA per biopsy = 299.7 Mb and 178.6 Mb, respectively, p=0.007). In the 24–48 month time window, the population of progressors had markedly increased SCA compared to nonprogressors (474.9 Mb vs. 136.8 Mb, p=7.9×10−8), and this difference became even more pronounced within 0–24 months (602.5 Mb vs. 179.8 Mb, p=5.4×10−7). A similar result was obtained when using percentage of abnormal markers among all the covered markers on the SNP array (Table S2) and when time was divided into 12-month windows. In the 0–24 month time window there is a highly significant gap in SCA with no intermediates (p=0.0049, Monte Carlo simulation test). These populations include biopsies with up to ~1300 Mb SCA and biopsies with very high SCA > ~1850 Mb with evidence of genome doubling not found in nonprogressors (Figure 1B, 1D). Because this study was part of a program of early detection, surveillance detected cancers arising in the progressors were too small to be sampled for array analysis (7). Therefore, we examined SCA levels in six surgical resection specimens from patients with advanced symptomatic EA who were not part of our program of early detection, all of which showed these strikingly high levels of SCA (right side of Figure 1B).
As a population, nonprogressors have abnormal genomes, but the overall spectrum of SCA remains qualitatively similar across all time windows (Figure 2A, Figure S3A–C). Progressors develop in the genetic background that characterizes nonprogressors, and therefore frequently create mosaic BE segments in which some areas of the esophagus have low SCA similar to nonprogressors and others have large scale genomic alterations (Figure 2B, Figure S3D–F).
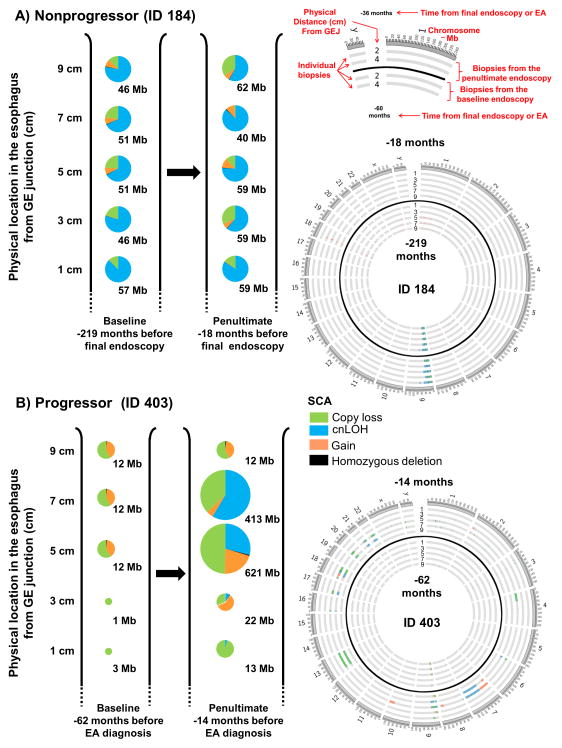
Figure 2. Proportion of different types of SCA in spatially separated biopsies over time.
SCA amount (Mb) and SCA type measured in spatially separated biopsies in two time points in a nonprogressor (Panel A) and a progressor (Panel B). At each timepoint, one biopsy is depicted as a single pie chart at each two-centimeter (cm) interval from the gastroesophageal junction (GEJ) with each pie chart showing the distribution of SCA types in each biopsy. The size of each pie chart is representational of different amounts of SCA measured in a single biopsy. A schematic is provided for interpretation of the Circos plots (66) with SCA in each biopsy at both timepoints. ID 184 shows the persistence of relatively genomically stable cell populations over a period of almost 16 years in a nonprogressing individual (Panel A). ID 403 shows the development of a cell population with significantly increased SCA during the approximately four years between the two endoscopies. Note the persistence of a cell population at the top of the figure having 12 Mb of SCA (Panel B). GEJ– gastroesophageal junction.
Evolution of genetic diversity in nonprogressors and progressors over space and time
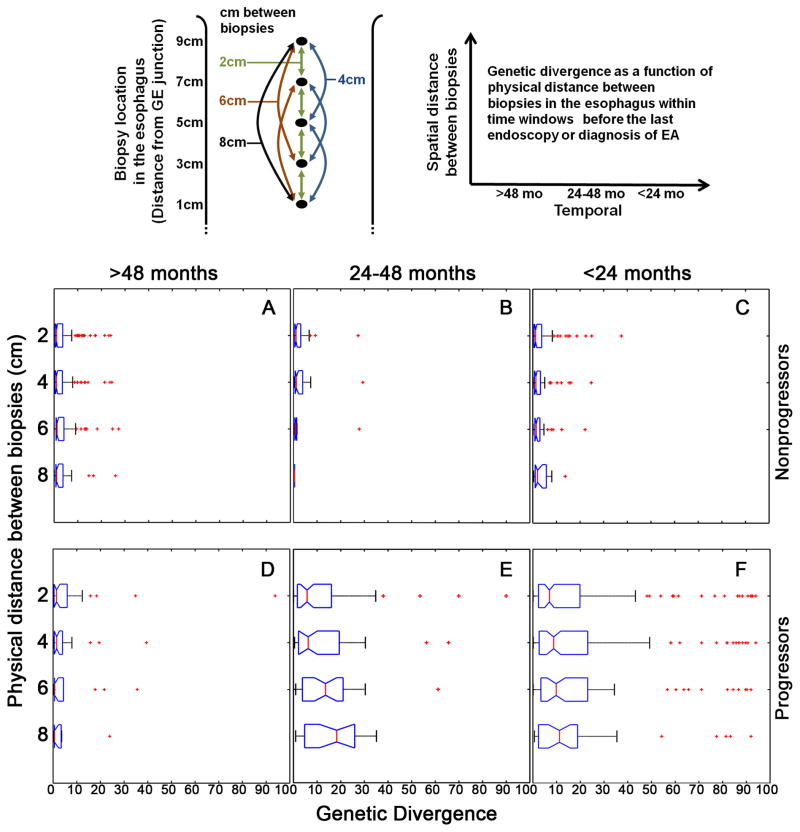
Genomic instability generates diverse cellular populations on which selection acts to promote cancer (16). Our previous investigations have been limited to measuring genetic diversity at a single point in time using low density microsatellite repeats on two chromosomes and DNA content flow cytometry (44, 45), but how diversity evolves over space and time in patients with BE who do and do not progress to EA is unknown. Here, we report genetic divergence over space in the esophagus at different time windows in nonprogressors and progressors (Figure 3A–F) and in individuals over time (Figure 4). In the population of nonprogressors, patterns of SCA remain similar between biopsies regardless of their spatial relationship in the esophagus and do not significantly change in any of the time windows (Figure 3A–C). Prior to 48 months, there was no significant difference in diversity between progressors and nonprogressors (Figure 3A and 3D). In the 24–48 month time window before cancer, progressors became more divergent over space in the BE segment (Figure 3E) whereas within 24 months of the diagnosis of EA divergence is high regardless of physical distance within the BE segment (Figure 3F). To test if there was an effect of BE segment length on genetic diversity, we evaluated diversity between adjacent biopsies in short (<3cm), medium (3–6cm) and long (>6cm) BE segments. We found no significant difference in genetic diversity in biopsies separated by 2cm between these groups across all time windows in either nonprogressors or progressors (data not shown).
Figure 3. Evolution of genetic diversity over space and time in progressors and nonprogressors.
Research biopsies used for this study were obtained according to a systematic predetermined protocol with one biopsy every two centimeters (cm) in the Barrett’s segment without any visible evidence of cancer. The top left example shows all pairwise divergence comparisons between biopsies. Comparisons include four 2cm (green), three 4cm (blue), two 6cm (brown) and one 8cm (black) in the example shown. (Panels A–F) Population level genetic divergence of spatially separated biopsies in each endoscopy within time windows in nonprogressors before the final endoscopy (Panels A–C) and in progressors before or at diagnosis of EA (Panels D–F). For each individual, the proportion of 1 Mb genomic segments with a different SCA call (present or absent) was calculated for all possible paired biopsy comparisons at each endoscopy. The X-axis is the percent of 1 Mb genomic segments with different SCA calls between any two biopsies, for all pairwise comparisons within an endoscopy for each individual and reflects the distribution of the level of heterogeneity among all pairs of biopsies within an individual. The Y-axis is the physical spatial distance between biopsies. For each time window, divergence between biopsies in each spatial category (2cm, 4cm, 6cm, 8+cm apart) was compared. In the >48 month time window there was no significant difference in divergence between nonprogressors and progressors in any spatial category (p>0.313 among all four comparisons, Kruskal-Wallis test, Figure 3A vs. 3D). In contrast, for all spatial categories in both the 24–48 month and <24 month time windows, progressor biopsies had significantly higher divergence than in nonprogressors (p<0.0014 among all eight comparisons, Kruskal-Wallis test, Figure 3B vs. 3E and Figure 3C vs. 3F). In nonprogressors, there was no trend for increasing SCA divergence between biopsies regardless of physical distance apart in the esophagus in >48, 24–48, and <24 month time windows (p=0.149, p=0.692, and p=0.222, respectively, Mann-Kendall trend test, Figure 3A, 3B, 3C). Similarly, in progressors, divergence remains low in the >48 month time window regardless of physical distance (p=0.28, Mann-Kendall trend test, Figure 3D). In contrast, in progressors the 24–48 month time window had a significant trend of increasing divergence with increasing physical distance (p<0.0051, Mann-Kendall trend test, Figure 3E), with divergence remaining equally high in all physical distance categories in the <24 month time window (p=0.279, Mann-Kendall trend test, Figure 3F).
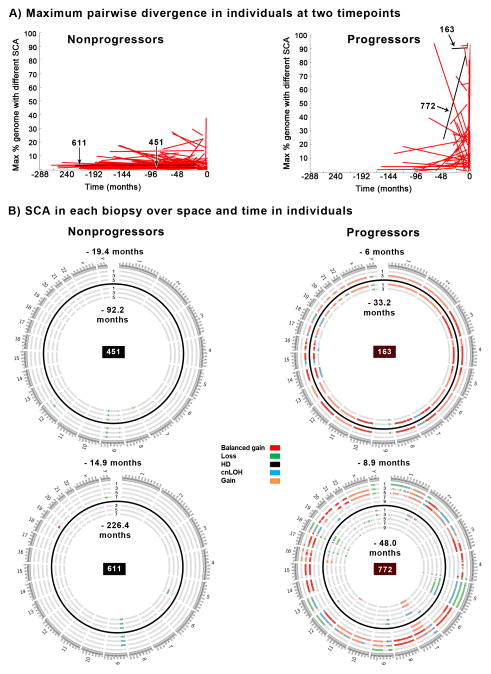
Figure 4. Individual level diversity over time.
Maximum genetic divergence of spatially separated biopsies in both endoscopies from the 104 nonprogressors and 57 progressors in whom SCA had been measured in at least two biopsies separated by two centimeters at both timepoints (Panel A). The maximum pairwise divergence between any two biopsies within the same endoscopy is plotted for the baseline and last endoscopy. Typically nonprogressors show selection of a small number of SCA events that have expanded throughout the BE segment and remain stable over long periods of time. Four patient examples are highlighted as black lines in Panel A, and and the SCA for each biopsy for these examples is shown in Panel B. Each concentric ring of the Circos plot shows the SCA for a single biopsy, with the baseline timepoint biopsies shown in the inner rings and the last endoscopy shown in the outer rings, with the two timepoints separated by a black ring. Location of each biopsy every 2 cm in the esophagus is labeled as cm from the GE junction. ID 451 and 611 are typical nonprogressors with expansion of 9p loss or cnLOH and typically 2–5 other small, sub-chromosomal or highly localized regions of SCA. In addition, the typical nonprogressor has small, random SCA events that are detected in one or a few samples but are not selected in the initial expansion and do not persist over time (e.g. ID 451, −92.2 mo, 5 cm, ch 8, and −19.4 mo, 3 cm, ch 12; ID 611 −226 mo, 5 cm, ch 5, and −14.9 mo, 7 cm, ch 19 (Panel B). Progressors with high diversity typically had samples with co-selected events that represent the punctuated stage of neoplastic evolution, and spatially localized genome doubling events such as in IDs 163 and 772 (Panel B). HD=homozygous deletion.
We then evaluated maximum pairwise divergence between all pairs of spatially separated biopsies within the same endoscopy in all individuals who have at least two biopsies in both a baseline and last endoscopy (104 nonprogressors and 57 progressors). Nonprogressors have relatively homogenous genomes that remain comparatively stable over long periods of time with only a small minority having any two biopsies at a single timepoint with different SCA calls in more than 15% of 1 Mb genomic segments throughout the genome (Figure 4A, nonprogressors). In contrast, 32/57(56%) of progressors have biopsies within the same timepoint with different SCA in >15% of the genome, and 14/57 (25%) have different SCA patterns in more than 40% of the genome (Figure 4A, progressors). Typical nonprogressors genomes have a small number of SCA events that expand to most of the BE segment prior to the baseline endoscopy with only small, random events arising in one or a few biopsies over time (Figure 4B Circos IDs 451 and 611). Many progressors show high levels of diversity over space throughout the esophagus that does not significantly change over time (Figure 4B Circos ID 163) or increases over time (Figure 4B Circos ID 772). In a subset of cases, there is a reduction in diversity over time as large genomic changes are selected and expand throughout the esophagus.
Temporal dynamics of SCA
At times >48 months before the final endoscopy or EA, both nonprogressors and progressors have similar frequency of SCA involving ~12% of the genome (Figure 5A odds ratio (OR) >48mo, Table S3). This BE SCA landscape includes various regions of small homo- and hemizygous deletions often at known fragile sites and 9p arm loss or cnLOH (Table S3), and a low frequency of larger gains on chromosomes 8 and 18. Many of the SCA shared between nonprogressors and progressors are also evident at time windows closer to EA (Figure 5B, 5C, Table S3). No regions of SCA were detected in any time window that had a significantly higher frequency of SCA in nonprogressors. These results identify regions of SCA that are common across time in both nonprogressors and progressors.
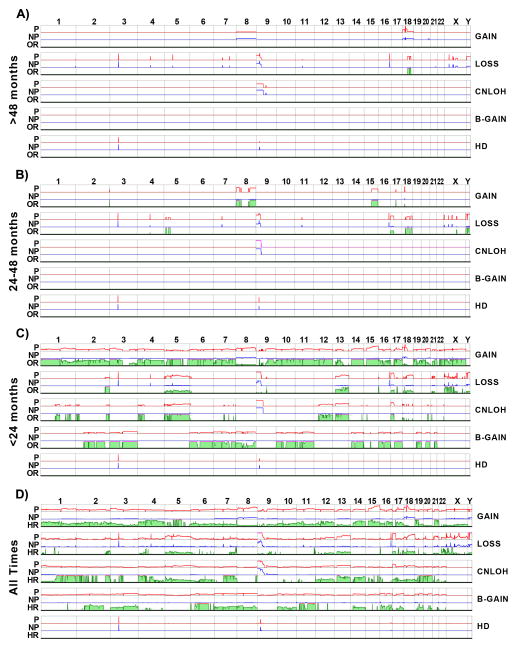
Figure 5. SCA frequency, odds ratios in time windows, and hazard ratio considering all times.
Panels A–C) Frequency of SCA in progressors (P, red line) and nonprogressors (NP, blue line) and odds ratio (OR, green) of SCA for EA risk based on data from A) >48 months, B) 24–48 months and C) <24 months before final endoscopy in nonprogressors or detection of EA in progressors. Panel D) Frequency of SCA and hazard ratio (HR, green) for SCA events across the genome based on case-cohort design. Frequency of each SCA event was determined in progressors (P, red line) and nonprogressors (NP, blue line). The HR was calculated for each 1 Mb window across the genome, considering follow-up time and censoring status of each individual. Chromosome number (1 through Y) is indicated along top and SCA type on right. SCA frequencies are shown from 0 and truncated at 0.5; OR and HR range from 0 to 20, with a pink line at the top indicating a ratio greater than 20. HD=homozygous deletion.
In contrast, progressors evolved additional SCAs that were not shared with nonprogressors over time. Two small losses on chromosome 18q were the only regions with statistically significantly higher frequency of SCA in progressors at >48 months before EA (Figure 5A, green, Table S4). Within 24–48 months of EA, ~8% of the genome had developed larger regions of copy loss and gain at increased frequency in progressors, including gains on chromosome 8 and 15q and losses on 5q, 17p, and 18 (Figure 5B OR 24–48mo, Table S4). Within 24 months of EA diagnosis, we observed genome doublings with extensive balanced gains, cnLOH and gains involving more than the 90% of the genome showing statistically higher frequencies of SCA in progressors (Figure 5C OR <24 mo, Tables S4). However, the majority of these genome-wide events were detected at a relatively low frequency of 10–20% in the population of progressors. We also observed some events that appeared to be co-selected within individual biopsies beginning in the 24–48 month time window and continuing in the <1500 Mb SCA population in the 0–24 month time window that were not dominated by the genome doublings (Supplementary Figure S4). These results provide a timescale on which to anchor multi-stage neoplastic evolution that distinguishes those in the population with BE who progress to EA from those who do not.
The case-cohort study design takes into account the temporal relationship of SCA and EA outcomes within individuals in the cohort (33, 43) and allowed quantitative mapping of EA risk of different SCA types by estimating hazard ratio genome-wide (Figure 5D). Considering follow-up times for each individual, progressors had varying low frequencies of SCA with high hazard ratios across large segments of the genome. Much of this SCA occurred significantly more often or nearly exclusively in progressors in biopsies with genome doublings that resulted in large regions of the genome being potent discriminators of EA risk. Thus, when genomic instability and diversity were high, we detected a large number of infrequent alterations that are found almost entirely in progressors and thus represent low-frequency, high-risk alterations.
Discussion
Endoscopic screening for, and surveillance of, BE is recommended by professional guidelines (46). Yet, multiple studies from academic and community practices have consistently reported near-total failure of screening and surveillance of BE to reduce the mortality of EA (14, 17, 47). Here, we address a fundamental property of neoplastic evolution that is rarely assessed: the window of opportunity for detection of genomic abnormalities that will evolve to EA.
BE and EA arise in a highly genotoxic reflux environment, and both have been reported to have SCA detectable by SNP arrays (26, 28–30, 48). Yet most BE do not progress to EA (12, 14, 15). Here, we present a longitudinal case-cohort study of BE that permits an innovative investigation of somatic chromosomal evolution over space and time in a cohort of patients who do and do not progress to EA. As the population of individuals with BE approach the diagnosis of cancer or the final endoscopy in this study, somatic genomes of nonprogressors remain relatively stable throughout the Barrett’s segment during long-term follow-up, whereas progressors developed chromosome instability, genomic diversity, selection and co-selection of SCA and genome doublings that result in rapid progression to EA, primarily within a four year time interval before cancer detection. Because our case-cohort study design preserves the characteristics of the entire cohort, we were able to investigate SCA from patients who have different follow-up times based on their rates of neoplastic evolution.
Our spatial and temporal data support a model of nonlinear dynamic chromosomal evolution in BE (Figure 6) (49). Nonprogressor somatic genomes were characterized by low levels of SCA, including deletions at fragile sites most commonly involving FHIT and WWOX, localized CDKN2A hemi- and homozygous deletions, and 9p arm loss and cnLOH, events also frequently detected in BE and many cancers including EA (27, 29, 50)]. These changes appear to have originated in an initial BE expansion that occurred before the baseline endoscopy in which Barrett’s epithelium has a selective advantage over squamous epithelium in the harsh, mutagenic environment of acid and bile reflux (17, 51, 52). Importantly, the somatic genomes of nonprogressors remain remarkably stable across space and time with relative homogeneity of SCA at the level of resolution of 1M SNP arrays.
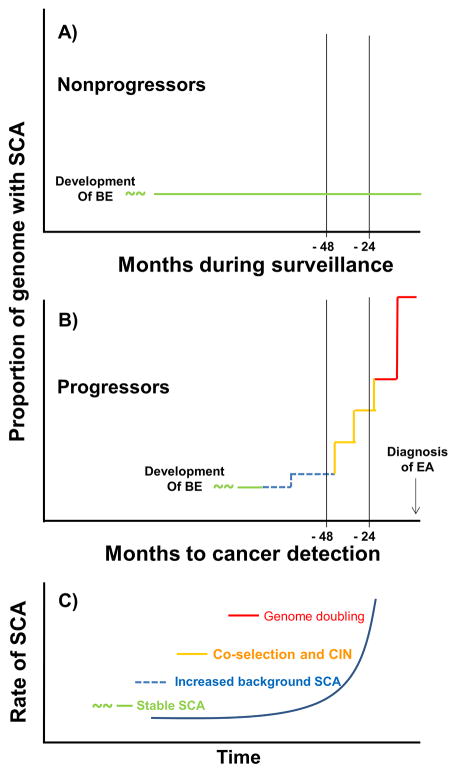
Figure 6. Model of somatic evolutionary dynamics of SCA in nonprogressors and progressors.
Somatic chromosomal evolution in nonprogressors and progressors begins at an unknown time prior to detection of BE in the clinic. By the time of diagnosis of BE, nonprogressors are characterized by an initial expansion of SCA primarily at fragile sites, including FHIT, CDKN2A and WWOX, and 9p loss or cnLOH that developed prior to clinical detection, which typically remains relatively stable without generating progressive chromosome instability, diversity, or further selection of SCA over prolonged follow-up (green line) (Panel A). Progressor genomes evolve in this background of low SCA (green line) with an increase in SCA compared to nonprogressors that becomes detectable 48 months prior to the diagnosis of EA, with only 18q loss detected at a significantly higher frequency than in nonprogressors before this time (blue dashed line). In the 24–48 months prior to EA diagnosis, total SCA increases markedly (yellow line) with selected larger gains on 8 and 15q and losses on 5p, 17p, 18 and Y conferring increased risk of progression as well as a background of co-selection of large regions of gains and/or losses on different chromosomes that are detected in individual biopsies. This background of co-selection continues to be detected in individual biopsies with SCA <1500 Mb in the 0–24 month window. This is followed by catastrophic genome doublings that result in leaps in SCA to >1850 Mb within 24 months of EA diagnosis (brown line) (Panel B). This model, based upon longitudinal data in humans in vivo, proposes that the rate of SCA accumulation increases in the population of progressors through selection and co-selection of large regions of SCA and catastrophic genome doublings as they approach the diagnosis of EA (Panel C).
Progressors have evidence of accelerating genomic alterations that occur on the background of the BE SCA landscape beginning with an initial phase more than 48 months before EA diagnosis characterized by an increased background of gains or losses of whole chromosomes or chromosome arms, although only 18q loss is consistently detected at a higher frequency in progressors. This region includes over 136 genes, but only SMAD4 was found to be significantly mutated in a recent large whole exome sequencing study of 149 esophageal adenocarcinomas (32). This increase in CIN is followed by increasing genetic diversity over space in the BE segment and selection of whole chromosome and chromosomal arm gains and losses that are associated with increased risk of progression in the 24–48 month time window before diagnosis of EA. This was followed by sudden catastrophic genome doublings and high genomic diversity that were detected within 24 months of the diagnosis of surveillance detected EA. Some progressors had low SCA at the baseline endoscopy but either developed high SCA after baseline or had a small number of somatic alterations that carry high hazard ratios for progression to cancer. Although CIN and genome doublings have been well documented in the majority of EAs, some EAs may develop by other mechanisms; for example microsatellite instability is believed to underlie the development of a small subset of EAs (32). A subset of low SCA progressors did not undergo the initial expansion of Barrett’s epithelium and presented as short segment BE. These may represent a subclass of cancers similar to recently reported copy-number devoid breast cancers (53). Alternatively, it is possible that in short segment BE with little opportunity for clonal expansion, genomic abnormalities could not be detected because they were removed in biopsies taken for clinical management, which are taken before research biopsies for ethical considerations.
Our results are supported by a recent cross-sectional study of advanced cancers by Carter et al, who reported that approximately 70% of advanced EAs had evidence of one or more genome doublings and that these doublings appeared to be preceded by specific arm-level copy number alterations (54). Their computational simulations showed that the genome doublings could not readily be explained as multiple sequential events on different chromosomes but were likely single doubling events; however, their study design could not determine whether these doublings arose before or after the onset of cancer. Further evidence for this concept is provided by earlier DNA content flow cytometry studies in BE reporting that increased 4N fractions are an unstable intermediate that develop in cells with 17p LOH and are followed rapidly by progression to aneuploidy detected on average 17 months later (55). Thus, the evidence in several studies support the concept that genome doublings arise by a single or very few episodes of doubling that occur suddenly rather than gradually over time and have the potential to rapidly evolve to cancer. Somatic evolutionary dynamics of progression, including genomic instability, generation of diversity, selection, co-selection and genome catastrophes, may drive the process of neoplastic progression and be generalizable across other tumor types. In this regard, it is notable that similar sequences of CIN followed by genome doublings have also been inferred based on data from a single point in space and time in advanced cancers of the breast, ovary, lung and colon (54). A recent study of ovarian cancer reported a 5-year window of opportunity for early detection, which is similar to our results showing increased SCA within four years of the diagnosis of early EA (56).
A number of previous studies have examined somatic chromosome copy number in BE and/or EA samples using SNP arrays (26, 28–30, 48). The majority of these studies examined alterations in EA samples taken at a single point in space and time. Gu et al contributed substantially to our understanding that biopsies from BE in patients who developed EA had increased chromosomal aberrations (29). Our study builds on this knowledge, but it is unique from previous studies in several aspects: i) a case-cohort study design including progressors and nonprogressors with patient samples collected longitudinally according to a systematic biopsy protocol (7) and a strong EA endpoint rather than surrogate markers such as dysplasia; ii) highly enriched epithelium separated from the underlying stroma, eliminating confounding signals from normal cells; and iii) high density (1 million) SNP arrays providing dense coverage across the genome, allowing smaller lesions to be determined more accurately (57). These unique aspects allow comparison of how indolent BE and BE that progresses to EA evolve over time and space in humans in vivo.
We have used EA as the outcome rather than dysplasia for a number of reasons. Formal statistical criteria for evaluating surrogate biomarkers were developed two decades ago (17). Valid surrogate markers need to accurately represent the true end point, in this case EA, and need to be easily and objectively measured. Neither high-grade dysplasia nor any other grade of dysplasia in BE has been demonstrated to be a valid surrogate for EA. In fact, diagnoses of dysplasia, including high-grade dysplasia are not reproducible as reviewed in Odze, 2011 (58) and they have high misclassification rates resulting in false positive rates for progression to EA ranging from 42% to 84% (13, 59). Unfortunately, use of irreproducible measures as outcomes has impaired the ability to compare results from different centers; the multiple limitations of using dysplasia as a surrogate marker for EA risk have been thoroughly reviewed recently (17).
We have included all EA detected by surveillance in this study (patients who did not have EA detected at the baseline endoscopy) in contrast to many studies that have excluded a number of EAs from analysis based on an arbitrary designation as “prevalent” if they were detected during variable follow-up ranging from 6–12 months (12, 13, 47). The inclusion of all surveillance detected EAs is reasonable because the cancers were endoscopically invisible, detected only by biopsy, and arose in a setting with genome rearrangements that occur suddenly, potentially within a single cell division (54).
The goal of this study was to elucidate somatic chromosomal evolutionary dynamics during progression to EA compared to nonprogressors, which are the majority of patients seen clinically. Our results revealed previously unsuspected rapid evolutionary dynamics in BE prior to the development of EA in contrast to prolonged relative genomic stability in nonprogressors at the level of 1M SNP arrays. We also found some aspects of rapid somatic evolution, including genome doublings, which are shared across a number of other malignancies. A recent whole exome sequencing study of 149 EAs reported that EA has one of the highest mutation frequencies among cancers, exceeded only by lung and melanoma (32). Yet selected mutations were found in only 26 genes out of more than 8,000 that were mutated and only four genes other than TP53 (72%) had mutation frequencies greater than 10% suggesting that it is unlikely that point mutations in specific genes will provide robust discrimination as predictors of progression to EA. However, the high mutation frequencies they observed raise the possibility that an increased mutation rate develops early in progression to EA as a consequence of the genotoxic environment (32). Perhaps mutation rate, an evolutionary measure, may be able to differentiate progressors and nonprogressors before the development of CIN, thereby increasing the window of opportunity for early detection of patients with BE at high risk of progression to EA.
Stringent controls on proliferation in multicellular organisms constrain individual cells to a fitness valley on an adaptive landscape from the perspective of their individual proliferative potential (60). Chromosome instability, selection and co-selection of large chromosome regions, and genome doubling provide redundant copies of genes that may facilitate further evolution of gene copy number and can be viewed as large-effect mutations in which phenotypic leaps can be achieved in a few mutational steps allowing cells to gain higher fitness and a selective proliferative advantage (54, 60). We observed co-selection of regions from different chromosomes in individual biopsies beginning in the 24–48 month time window (Supplemental Figure S4). This co-selection of genomic regions from different chromosomes may result from structural rearrangements that have been reported to occur more frequently in EA than colorectal cancer (32), but whole genome sequencing will be required to determine the extent to which such rearrangements contribute to rapid progression. Alternatively, co-selection of unlinked SCAs may occur under strong selection in the toxic gastroduodenoesophageal reflux environment of BE by affecting phenotypic variation that negatively or positively regulates proliferation (60, 61).
The cancer paradigm has been built around the concept that neoplastic evolution to cancer is a gradual, deterministic process (62). However, evolution of SCA is a dynamic, stochastic process. The concepts of sudden, punctuated evolutionary events that lead to rapid progression to cancer in cross-sectional studies are relatively new (24, 25, 54). Our study is unique in that we used SCA to explore rate of progression to EA compared to nonprogressing controls. The temporal dynamics of somatic genomic evolution and the windows of opportunity for early detection are poorly defined in BE/EA. Until these windows are better understood, simply identifying additional biomarkers will not decrease EA mortality given current screening and surveillance approaches that fail to detect rapid evolution to a lethal EA and selectively detect indolent BE.
Overdiagnosis is defined as diagnosis of “disease” that will never cause symptoms or death during a patient’s lifetime (63, 64). Our findings may explain clinical cohort observations that the great majority of patients with BE (90–95%) do not progress to EA, but die of other causes and that those who do progress to EA while in surveillance can develop rapidly progressive disease without evidence of EA at the index endoscopy. Our results may also inform population observations that 95% of EAs are detected at advanced stages when patients become symptomatic without a prior diagnosis of BE even though the evidence indicates that they arose in BE (65). There is abundant evidence of overdiagnosis of BE, but there is no evidence for overdiagnosis of indolent EA in the population because there has been little change in the proportion of patients found with in early stage EA while EA mortality has continued to increase rapidly despite current screening strategies (1). However, screening may occasionally detect BE that rapidly progresses to EA that was not diagnosed at the index endoscopy. In a retrospective Danish database study of 11,028 patients with BE, 66% of new cases of EAs were diagnosed within one year of the index endoscopy (12). Similarly, in a large cohort study of 1,099 veterans with BE, 56% of EAs were diagnosed within 25 months of the index endoscopy (13). Our study provides a somatic genomic evolutionary explanation for these clinical observations that the majority of EAs are detected within a short time after the diagnosis of BE. Our findings of significantly increased SCA and somatic genomic diversity within four years of a cancer diagnosis suggests that rapid somatic genomic evolution may underlie underdiagnosis of rapidly progressing life-threatening disease that presents as an advanced malignancy, whereas the majority of BE maintains relative somatic chromosomal stability at the level of 1M SNP arrays over prolonged periods underlying overdiagnosis of nonprogressing BE.
Supplementary Material
Acknowledgments
Financial support: X. Li, P. Galipeau, T. Paulson, C. Sanchez, J. Arnaudo, K. Liu, R. Odze, P. Blount, C. Maley, T. Vaughan and B. Reid. were supported by National Cancer Institute (NCI) P01CA091955. X Li and B. Reid were also supported by NCI RC1 CA 146973. T. Vaughan was also supported by NCI K05CA124911. C. Maley and R. Kostadinov were supported by NCI R01 CA140657 and Research Scholar #117209-RSG-09-163-01-CNE from the American Cancer Society. S. Self and C. Sather were supported by NCI P30 CA015704. C. Sather and B. Reid were also supported by Fred Hutchinson Cancer Research Center Institutional Funds. M. Kuhner was supported by the University of Washington, Department of Genome Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank all the research participants who have made this study possible, Christine Karlsen, Valerie Cerera, Heather Kissel and Patricia Christopherson for patient care and research biospecimen coordination, and David Cowan and Terri Watson for database support.
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Author contributions:
Conception and design - XL PCG TGP CAS CCM SGS TLV PLB BJR
Development of methodology - XL PCG TGP CAS CCM SGS TLV PLB BJR
Acquisition of data - TGP CAS JA CLS
Analysis and interpretation of data - XL PCG TGP CAS JA KL RLK MKK CCM SGS TLV PLB BJR
Writing, review and/or revision of the manuscript - XL PCG TGP PLB BJR
Administrative, technical, or material support - XL PCG KL RLK RDO MKK CCM PLB TLV BJR
Study supervision – CAS PLB BJR
References
- 1.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142–6. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 2.Dubecz A, Gall I, Solymosi N, Schweigert M, Peters JH, Feith M, et al. Temporal trends in long-term survival and cure rates in esophageal cancer: a SEER database analysis. J Thorac Oncol. 2012;7:443–7. doi: 10.1097/JTO.0b013e3182397751. [DOI] [PubMed] [Google Scholar]
- 3.Li Z, Rice TW, Liu X, Goldblum JR, Williams SJ, Rybicki LA, et al. Intramucosal esophageal adenocarcinoma: primum non nocere. J Thorac Cardiovasc Surg. 2013;145:1519–24. 24 e1–3. doi: 10.1016/j.jtcvs.2012.10.027. [DOI] [PubMed] [Google Scholar]
- 4.Rice TW, Zuccaro G, Jr, Adelstein DJ, Rybicki LA, Blackstone EH, Goldblum JR. Esophageal carcinoma: depth of tumor invasion is predictive of regional lymph node status. Ann Thorac Surg. 1998;65:787–92. doi: 10.1016/s0003-4975(97)01387-8. [DOI] [PubMed] [Google Scholar]
- 5.Prenzel KL, Holscher AH, Drebber U, Agavonova M, Gutschow CA, Bollschweiler E. Prognostic impact of nodal micrometastasis in early esophageal cancer. Eur J Surg Oncol. 2012;38:314–8. doi: 10.1016/j.ejso.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Wang KK, Sampliner RE. Updated guidelines 2008 for the diagnosis, surveillance and therapy of Barrett’s esophagus. Am J Gastroenterol. 2008;103:788–97. doi: 10.1111/j.1572-0241.2008.01835.x. [DOI] [PubMed] [Google Scholar]
- 7.Reid BJ, Blount PL, Feng Z, Levine DS. Optimizing endoscopic biopsy detection of early cancers in Barrett’s high-grade dysplasia. Am J Gastroenterol. 2000;95:3089–96. doi: 10.1111/j.1572-0241.2000.03182.x. [DOI] [PubMed] [Google Scholar]
- 8.van Sandick JW, van Lanschot JJ, Kuiken BW, Tytgat GN, Offerhaus GJ, Obertop H. Impact of endoscopic biopsy surveillance of Barrett’s oesophagus on pathological stage and clinical outcome of Barrett’s carcinoma. Gut. 1998;43:216–22. doi: 10.1136/gut.43.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weston AP, Sharma P, Topalovski M, Richards R, Cherian R, Dixon A. Long-term follow-up of Barrett’s high-grade dysplasia. Am J Gastroenterol. 2000;95:1888–93. doi: 10.1111/j.1572-0241.2000.02234.x. [DOI] [PubMed] [Google Scholar]
- 10.Bytzer P, Christensen PB, Damkier P, Vinding K, Seersholm N. Adenocarcinoma of the esophagus and Barrett’s esophagus: a population- based study. Am J Gastroenterol. 1999;94:86–91. doi: 10.1111/j.1572-0241.1999.00776.x. [DOI] [PubMed] [Google Scholar]
- 11.Corley DA, Levin TR, Habel LA, Weiss NS, Buffler PA. Surveillance and survival in Barrett’s adenocarcinomas: a population-based study. Gastroenterology. 2002;122:633–40. doi: 10.1053/gast.2002.31879. [DOI] [PubMed] [Google Scholar]
- 12.Hvid-Jensen F, Pedersen L, Drewes AM, Sorensen HT, Funch-Jensen P. Incidence of adenocarcinoma among patients with Barrett’s esophagus. N Engl J Med. 2011;365:1375–83. doi: 10.1056/NEJMoa1103042. [DOI] [PubMed] [Google Scholar]
- 13.Schnell TG, Sontag SJ, Chejfec G, Aranha G, Metz A, O’Connell S, et al. Long-term nonsurgical management of Barrett’s esophagus with high-grade dysplasia. Gastroenterology. 2001;120:1607–19. doi: 10.1053/gast.2001.25065. [DOI] [PubMed] [Google Scholar]
- 14.Macdonald CE, Wicks AC, Playford RJ. Ten years’ experience of screening patients with Barrett’s oesophagus in a university teaching hospital. Gut. 1997;41:303–7. doi: 10.1136/gut.41.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sikkema M, de Jonge PJ, Steyerberg EW, Kuipers EJ. Risk of esophageal adenocarcinoma and mortality in patients with Barrett’s esophagus: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2010;8:235–44. doi: 10.1016/j.cgh.2009.10.010. quiz e32. [DOI] [PubMed] [Google Scholar]
- 16.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reid BJ, Li X, Galipeau PC, Vaughan TL. Barrett’s oesophagus and oesophageal adenocarcinoma: time for a new synthesis. Nat Rev Cancer. 2010;10:87–101. doi: 10.1038/nrc2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–8. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 21.Rajagopalan H, Nowak MA, Vogelstein B, Lengauer C. The significance of unstable chromosomes in colorectal cancer. Nature Reviews Cancer. 2003;3:695–701. doi: 10.1038/nrc1165. [DOI] [PubMed] [Google Scholar]
- 22.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–77. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navin N, Krasnitz A, Rodgers L, Cook K, Meth J, Kendall J, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20:68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dulak AM, Schumacher SE, van Lieshout J, Imamura Y, Fox C, Shim B, et al. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012;72:4383–93. doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goh XY, Rees JR, Paterson AL, Chin SF, Marioni JC, Save V, et al. Integrative analysis of array-comparative genomic hybridisation and matched gene expression profiling data reveals novel genes with prognostic significance in oesophageal adenocarcinoma. Gut. 2011;60:1317–26. doi: 10.1136/gut.2010.234179. [DOI] [PubMed] [Google Scholar]
- 29.Gu J, Ajani JA, Hawk ET, Ye Y, Lee JH, Bhutani MS, et al. Genome-wide catalogue of chromosomal aberrations in barrett’s esophagus and esophageal adenocarcinoma: a high-density single nucleotide polymorphism array analysis. Cancer Prev Res (Phila) 2010;3:1176–86. doi: 10.1158/1940-6207.CAPR-09-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Galipeau PC, Sanchez CA, Blount PL, Maley CC, Arnaudo J, et al. Single nucleotide polymorphism-based genome-wide chromosome copy change, loss of heterozygosity, and aneuploidy in Barrett’s esophagus neoplastic progression. Cancer Prev Res (Phila Pa) 2008;1:413–23. doi: 10.1158/1940-6207.CAPR-08-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agrawal N, Jiao Y, Bettegowda C, Hutfless SM, Wang Y, David S, et al. Comparative Genomic Analysis of Esophageal Adenocarcinoma and Squamous Cell Carcinoma. Cancer Discov. 2012;2:899–905. doi: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–86. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prentice RL. A case-cohort design for epidemiologic cohort studies and disease prevention trials. Biometrika. 1986;73:1–11. [Google Scholar]
- 34.Levine DS, Blount PL, Rudolph RE, Reid BJ. Safety of a systematic endoscopic biopsy protocol in patients with Barrett’s esophagus. Am J Gastroenterol. 2000;95:1152–7. doi: 10.1111/j.1572-0241.2000.02002.x. [DOI] [PubMed] [Google Scholar]
- 35.Vaughan TL, Dong LM, Blount PL, Ayub K, Odze RD, Sanchez CA, et al. Non-steroidal anti-inflammatory drugs and risk of neoplastic progression in Barrett’s oesophagus: a prospective study. Lancet Oncol. 2005;6:945–52. doi: 10.1016/S1470-2045(05)70431-9. [DOI] [PubMed] [Google Scholar]
- 36.Barrett MT, Sanchez CA, Prevo LJ, Wong DJ, Galipeau PC, Paulson TG, et al. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Gen. 1999;22:106–9. doi: 10.1038/8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galipeau PC, Li X, Blount PL, Maley CC, Sanchez CA, Odze RD, et al. NSAIDs modulate CDKN2A, TP53, and DNA content risk for future esophageal adenocarcinoma. PLoS Med. 2007;4:e67. doi: 10.1371/journal.pmed.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prevo LJ, Sanchez CA, Galipeau PC, Reid BJ. p53-mutant clones and field effects in Barrett’s esophagus. Cancer Research. 1999;59:4784–7. [PubMed] [Google Scholar]
- 39.Rabinovitch PS, Reid BJ, Haggitt RC, Norwood TH, Rubin CE. Progression to cancer in Barrett’s esophagus is associated with genomic instability. Lab Invest. 1989;60:65–71. [PubMed] [Google Scholar]
- 40.Avidan B, Sonnenberg A, Schnell TG, Chejfec G, Metz A, Sontag SJ. Hiatal hernia size, Barrett’s length, and severity of acid reflux are all risk factors for esophageal adenocarcinoma. Am J Gastroenterol. 2002;97:1930–6. doi: 10.1111/j.1572-0241.2002.05902.x. [DOI] [PubMed] [Google Scholar]
- 41.Cheng H, Bjerknes M, Amar J. Methods for the determination of epithelial cell kinetic parameters of human colonic epithelium isolated from surgical and biopsy specimens. Gastroenterology. 1984;86:78–85. [PubMed] [Google Scholar]
- 42.Mountain JL, Cavalli-Sforza LL. Multilocus genotypes, a tree of individuals, and human evolutionary history. Am J Hum Genet. 1997;61:705–18. doi: 10.1086/515510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Self SG, Prentice RL. Asymptotic distribution theory and efficiency results for case-cohort studies. Annals Statistics. 1988;16:64–81. [Google Scholar]
- 44.Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–73. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 45.Merlo LM, Shah NA, Li X, Blount PL, Vaughan TL, Reid BJ, et al. A Comprehensive Survey of Clonal Diversity Measures in Barrett’s Esophagus as Biomarkers of Progression to Esophageal Adenocarcinoma. Cancer Prev Res (Phila) 2010 doi: 10.1158/1940-6207.CAPR-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spechler SJ, Sharma P, Souza RF, Inadomi JM, Shaheen NJ. American Gastroenterological Association medical position statement on the management of Barrett’s esophagus. Gastroenterology. 2011;140:1084–91. doi: 10.1053/j.gastro.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 47.Corley DA, Mehtani K, Quesenberry C, Zhao W, de Boer J, Weiss NS. Impact of Endoscopic Surveillance on Mortality From Barrett’s Esophagus-Associated Esophageal Adenocarcinomas. Gastroenterology. 2013;145:312–9. e1. doi: 10.1053/j.gastro.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nancarrow DJ, Handoko HY, Smithers BM, Gotley DC, Drew PA, Watson DI, et al. Genome-wide copy number analysis in esophageal adenocarcinoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2008;68:4163–72. doi: 10.1158/0008-5472.CAN-07-6710. [DOI] [PubMed] [Google Scholar]
- 49.Li X, Blount PL, Vaughan TL, Reid BJ. Application of biomarkers in cancer risk management: evaluation from stochastic clonal evolutionary and dynamic system optimization points of view. PLoS Comput Biol. 2011;7:e1001087. doi: 10.1371/journal.pcbi.1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai LA, Kostadinov R, Barrett MT, Peiffer DA, Pokholok D, Odze R, et al. Deletion at fragile sites is a common and early event in Barrett’s esophagus. Mol Cancer Res. 2010;8:1084–94. doi: 10.1158/1541-7786.MCR-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nancarrow DJ, Clouston AD, Smithers BM, Gotley DC, Drew PA, Watson DI, et al. Whole genome expression array profiling highlights differences in mucosal defense genes in Barrett’s esophagus and esophageal adenocarcinoma. PLoS One. 2011;6:e22513. doi: 10.1371/journal.pone.0022513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ostrowski J, Mikula M, Karczmarski J, Rubel T, Wyrwicz LS, Bragoszewski P, et al. Molecular defense mechanisms of Barrett’s metaplasia estimated by an integrative genomics. J Mol Med. 2007;85:733–43. doi: 10.1007/s00109-007-0176-3. [DOI] [PubMed] [Google Scholar]
- 53.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galipeau PC, Cowan DS, Sanchez CA, Barrett MT, Emond MJ, Levine DS, et al. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett’s esophagus. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:7081–4. doi: 10.1073/pnas.93.14.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown PO, Palmer C. The preclinical natural history of serous ovarian cancer: defining the target for early detection. PLoS Med. 2009;6:e1000114. doi: 10.1371/journal.pmed.1000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, Self SG, Galipeau PC, Paulson TG, Reid BJ. Direct inference of SNP heterozygosity rates and resolution of LOH detection. PLoS Comput Biol. 2007;3:e244. doi: 10.1371/journal.pcbi.0030244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Odze RD. What the gastroenterologist needs to know about the histology of Barrett’s esophagus. Curr Opin Gastroenterol. 2011;27:389–96. doi: 10.1097/MOG.0b013e328346f551. [DOI] [PubMed] [Google Scholar]
- 59.Reid BJ, Levine DS, Longton G, Blount PL, Rabinovitch PS. Predictors of progression to cancer in Barrett’s esophagus: baseline histology and flow cytometry identify low- and high-risk patient subsets. American Journal of Gastroenterology. 2000;95:1669–76. doi: 10.1111/j.1572-0241.2000.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pavelka N, Rancati G, Li R. Dr Jekyll and Mr Hyde: role of aneuploidy in cellular adaptation and cancer. Curr Opin Cell Biol. 2010;22:809–15. doi: 10.1016/j.ceb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 62.Croswell JM, Ransohoff DF, Kramer BS. Principles of cancer screening: lessons from history and study design issues. Semin Oncol. 2010;37:202–15. doi: 10.1053/j.seminoncol.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Welch HG, Black WC. Overdiagnosis in cancer. J Natl Cancer Inst. 2010;102:605–13. doi: 10.1093/jnci/djq099. [DOI] [PubMed] [Google Scholar]
- 64.Esserman LJ, Thompson IM, Jr, Reid B. Overdiagnosis and overtreatment in cancer: An opportunity for improvement. JAMA. 2013 doi: 10.1001/jama.2013.108415. [DOI] [PubMed] [Google Scholar]
- 65.Theisen J, Stein HJ, Dittler HJ, Feith M, Moebius C, Kauer WK, et al. Preoperative chemotherapy unmasks underlying Barrett’s mucosa in patients with adenocarcinoma of the distal esophagus. Surg Endosc. 2002;16:671–3. doi: 10.1007/s00464-001-8307-3. [DOI] [PubMed] [Google Scholar]
- 66.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.