Abstract
The International Tamoxifen Pharmacogenomics Consortium was established to address the controversy regarding cytochrome P450 2D6 (CYP2D6) status and clinical outcomes in tamoxifen therapy. We performed a meta-analysis on data from 4,973 tamoxifen-treated patients (12 globally distributed sites). Using strict eligibility requirements (postmenopausal women with estrogen receptor–positive breast cancer, receiving 20 mg/day tamoxifen for 5 years, criterion 1); CYP2D6 poor metabolizer status was associated with poorer invasive disease–free survival (IDFS: hazard ratio = 1.25; 95% confidence interval = 1.06, 1.47; P = 0.009). However, CYP2D6 status was not statistically significant when tamoxifen duration, menopausal status, and annual follow-up were not specified (criterion 2, n = 2,443; P = 0.25) or when no exclusions were applied (criterion 3, n = 4,935; P = 0.38). Although CYP2D6 is a strong predictor of IDFS using strict inclusion criteria, because the results are not robust to inclusion criteria (these were not defined a priori), prospective studies are necessary to fully establish the value of CYP2D6 genotyping in tamoxifen therapy.
Tamoxifen, the pioneering antiestrogenic medicine targeted to the tumor estrogen receptor (ER), is used successfully for long-term adjuvant therapy in breast cancer.1,2 Extensive analyses of clinical trials demonstrate a major increase in patient survivorship in ER-positive patients. In this age of personalized medicine, any opportunity to improve response rates with tamoxifen should be rigorously investigated. Tamoxifen is considered a prodrug, given that hepatic cytochrome P450 2D6 (CYP2D6) metabolizes tamoxifen to metabolites (4-hydroxy tamoxifen and 4-hydroxy-N-desmethyl tamoxifen (endoxifen)) that exhibit significantly greater potency in terms of ER-binding affinity3 and suppression of estradiol-stimulated cell proliferation.4 CYP2D6-mediated metabolism is the rate-limiting enzymatic step for the formation of endoxifen, the most abundant active metabolite.
There has been great inconsistency among studies that have reported the association of known genetic and drug factors influencing CYP2D6 enzyme activity with tamoxifen efficacy. Therefore, the International Tamoxifen Pharmacogenomics Consortium (ITPC) was conceived, and researchers were invited to submit their data—both published and unpublished data sets regarding CYP2D6 genetic variants and clinical outcomes in women treated with tamoxifen in the adjuvant breast cancer setting—to allow a meta-analysis of the potential associations between CYP2D6 and clinical outcomes.
Results
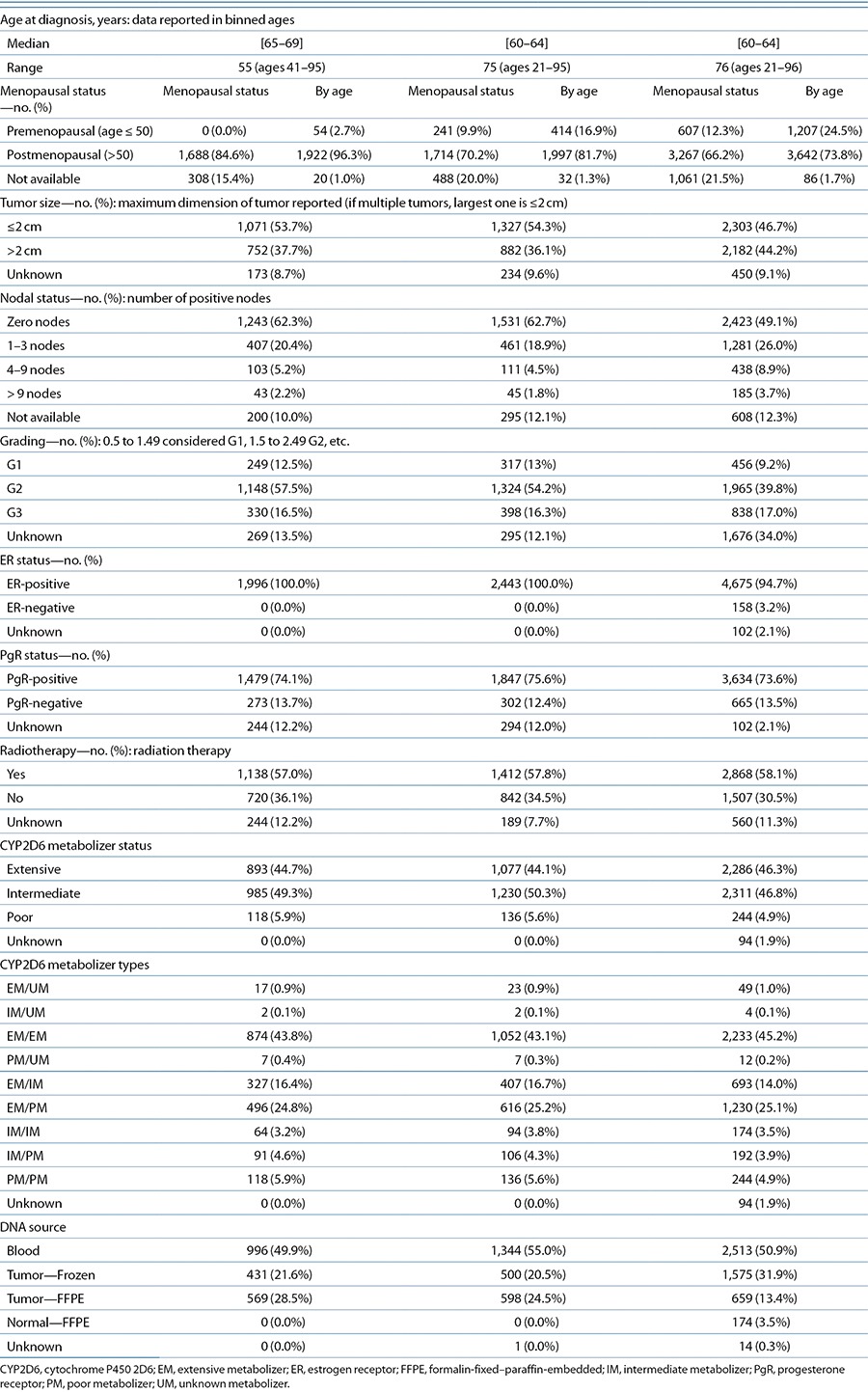
The ITPC comprises 12 research projects from nine countries and three continents that contributed clinical and genetic data for a total of 4,973 breast cancer patients treated with tamoxifen. In Table 1, we show the sample size by site and criteria. Further details for each site are shown in S3c and S5 online. We reported preliminary analyses of these collected cohorts before complete curation by pooling the data from each site.5 For our meta-analyses, three detailed criteria, which ranged from the most restrictive (criterion 1) to the most inclusive (criterion 3), were defined before final curation (see S4 online). In brief, criterion 1, derived from the NCCTG 89-30-52 clinical trial, consisted of postmenopausal women with surgically resected nonmetastatic invasive ER-positive breast cancers who received adjuvant tamoxifen monotherapy at a dose of 20 mg/day for an intended duration of 5 years, and were followed at least annually for recurrence. In addition, analysis of at least CYP2D6*4 was required (detailed in S4a online). Criterion 2 included criterion 1 but allowed both pre- and postmenopausal patients who had received any duration of tamoxifen; moreover, annual follow-up was not required. Criterion 3 included all samples not excluded by any exclusion test for missing data or data inconsistencies (least restrictive). Patient characteristics according to each criterion are provided in Table 2.
Table 1. Sample size by site and criteria.

Table 2. Baseline patient and tumor characteristics.
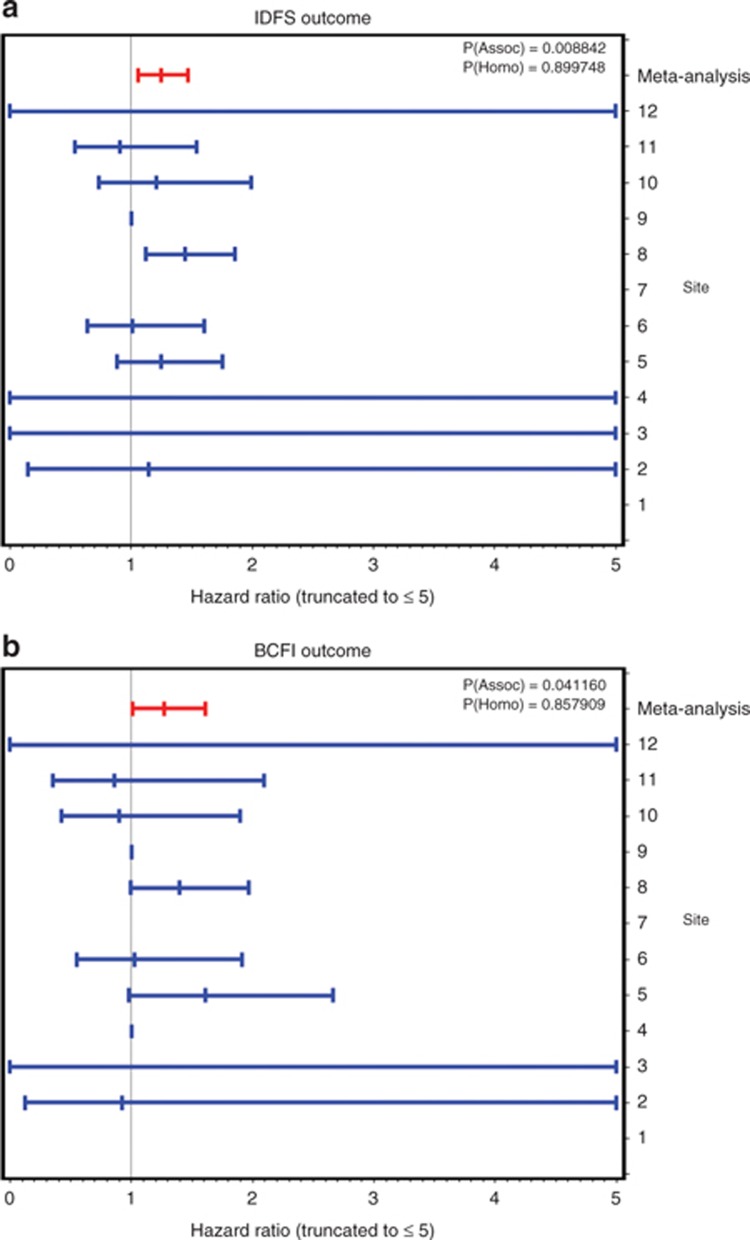
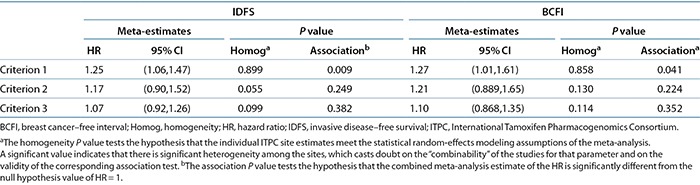
The meta-analysis results combining the hazard ratio (HR) estimates (and the corresponding standard errors (SEs)) from each site are shown for all three criteria groups and both clinical outcomes in Table 3. For each of the six clinical outcome/criteria groups, we give the combined meta-analysis estimate across all 12 sites, its SE, and the results of two statistical tests: a test of the significance that the meta-HR differs from 1 and a test of “homogeneity of the estimates” across sites (a significant value for the latter test indicates that there is more variability than the derSimonian and Laird random-effects model can reasonably accommodate, suggesting that the meta-estimate and its associated P value are suspect). As can be seen for invasive disease–free survival (IDFS), the meta-analyses for criteria 2 and 3 are nearly significantly heterogeneous, whereas there was no indication of heterogeneity for criterion 1 (P = 0.899). For patients meeting criterion 1, the meta-HR for IDFS was 1.25 (95% confidence interval = 1.06, 1.47), and for breast cancer–free interval, it was 1.27 (95% confidence interval = 1.01, 1.61). These are both statistically significant, at P = 0.009 and P = 0.04, respectively. However, for the criterion 2 (P = 0.25) and criterion 3 (P = 0.38) subsets, the CYP2D6 HR was not significant for either outcome.
Table 3. Meta-analyses of CYP2D6 HRs on clinical outcome in inclusion/exclusion criteria subsets.
In Figure 1, we show the individual HRs for each site for subjects meeting criterion 1, assuming an additive genetic model for CYP2D6 (coded 0 = extensive metabolizer (EM), 1 = intermediate metabolizer (IM), and 2 = poor metabolizer (PM)) as estimated from a Cox proportional-hazards model using additional risk covariates to predict clinical outcome. Corresponding figures for criteria 2 and 3 are provided in S6 online. (Note that the list of covariates used in the Cox models included age at primary diagnosis, menopause status at diagnosis, metastatic disease at primary diagnosis, maximum tumor dimension, number of positive nodes, grade, smoking status, ER and progesterone receptor status, intended tamoxifen dose and duration, systemic therapy before surgery, chemotherapy, radiation treatment, adjuvant aromatase inhibitor therapy, and additional hormone therapy. The specific set of covariates used for each site was chosen from this list so as to retain at least 70% of the patients from that site; hence, the exact set of covariates used differs in each site's Cox model. Moreover, several of these covariates were used as inclusion/exclusion items in the basic definitions of the three basic criteria subset groups and thus became irrelevant for those analyses.)
Figure 1.
Individual site estimates of hazard ratios of CYP2D6 genotype on clinical outcome, along with the meta-analyses for the criterion 1 subset. (a) Invasive disease–free survival (IDFS) outcome. (b) Breast cancer–free interval (BCFI) outcome.
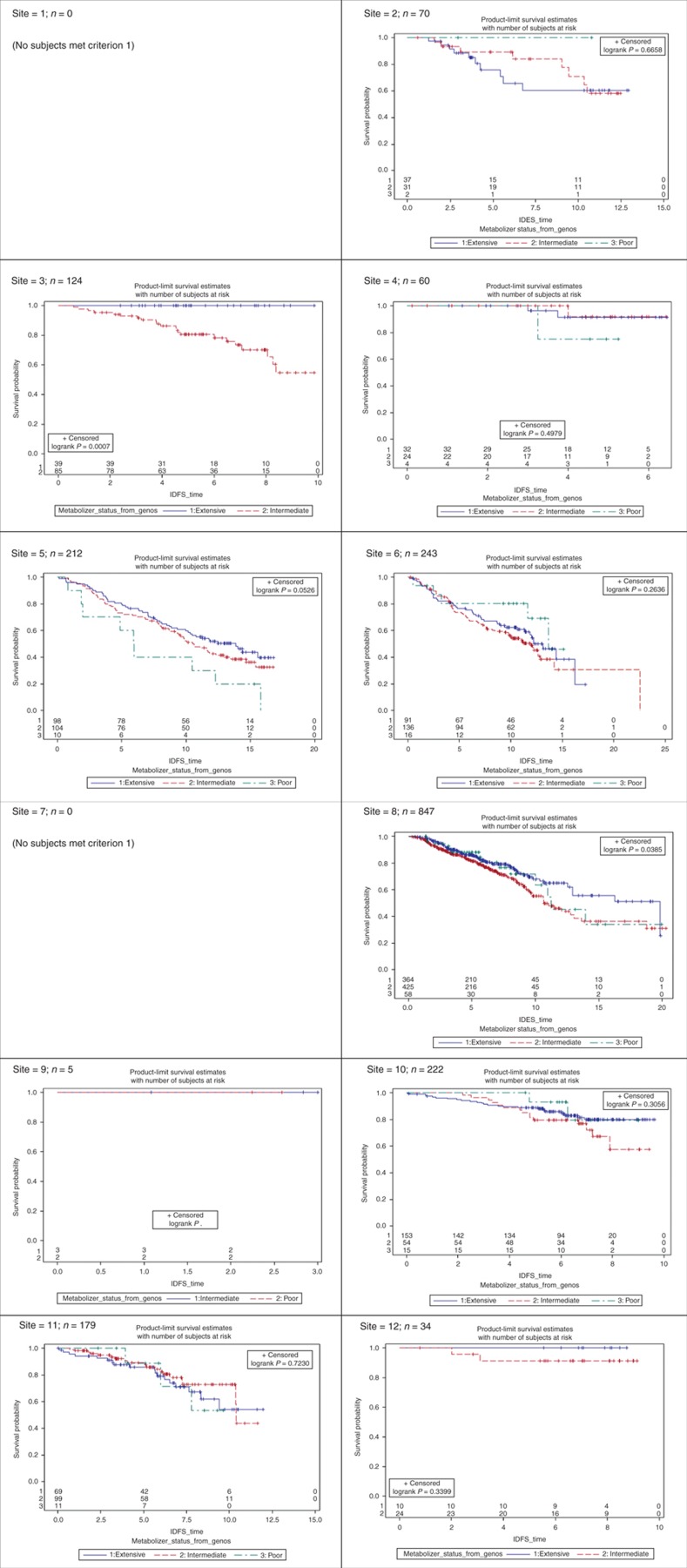
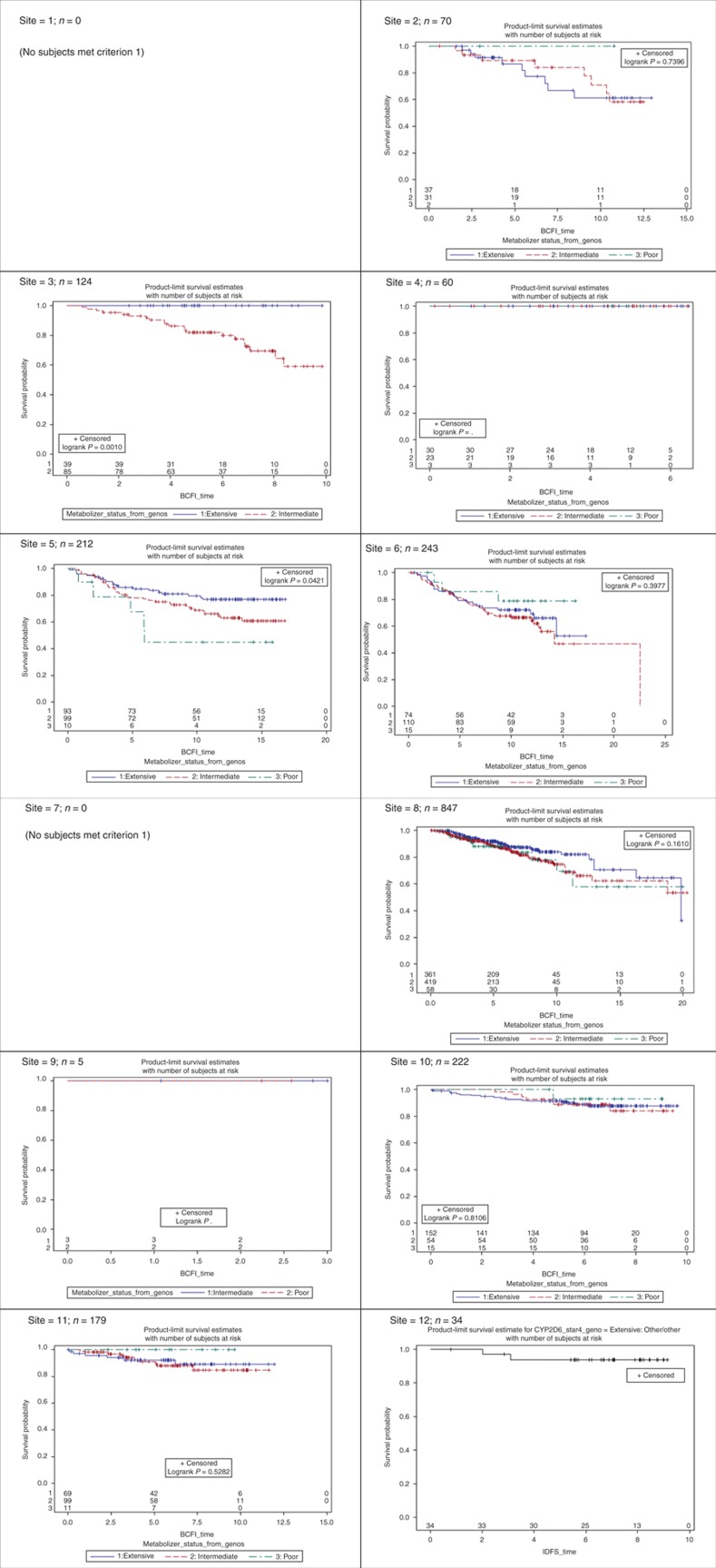
Site-specific product-limit estimates of the three CYP2D6 metabolizer status genotype groups (EM, IM, and PM) are shown in Figures 2 and 3 for criterion 1 patients. Sites 1 and 7 had no subjects who met inclusion/exclusion for criterion 1. The corresponding figures for patients meeting criteria 2 and 3 are shown in S6 online. As seen in Figure 2, for IDFS sites, 3, 5, and 8 show a strong significant effect in the direction expected by the known pharmacokinetic effects of CYP2D6 on endoxifen exposure, namely, a poorer clinical response for the IM and/or PM genotype groups. Other sites show a trend in the expected direction between the IM and EM groups, but the much smaller PM group is often inconsistent with the expectation, and the separation in the three survival curves is not strong enough to reach statistical significance (e.g., sites 6 and 12). For some sites, there is no hint of any significant difference (e.g., sites 2, 4, 10, and 11), and for one of these, site 2, the direction of effect is exactly opposite than expected. There is a danger in overinterpreting such “trends” (either in favor or against expectation) when there is no statistically significant difference, because some level of site-to-site variation is to be expected. The key question is not whether such variation exists but whether it centers over the null hypothesis or over the alternative; this is the question that the meta-analysis is designed to answer. However, these simple product-limit survival curves show great study-to-study heterogeneity, which complicates both the analyses and the interpretation. We have similar heterogeneous results for the breast cancer–free interval outcome, shown in Figure 3. The corresponding figures in S6 online show a similar pattern for the subsets of patients meeting criteria 2 and 3, although the heterogeneity seems to be even more pronounced as the exclusion criteria are loosened. This is not a surprising result, considering that the criteria themselves impose a certain level of homogeneity.
Figure 2.
Site-specific effects of CYP2D6 metabolizer status on clinical outcomes for subjects meeting inclusion criterion 1 (outcome = invasive disease–free survival (IDFS)).
Figure 3.
Site-specific effects of CYP2D6 metabolizer status on clinical outcomes for subjects meeting inclusion criterion 1 (outcome = breast cancer–free interval (BCFI)).
Discussion
Prospective pharmacology studies consistently demonstrate that CYP2D6 genetic variants are associated with variable plasma concentrations of endoxifen.4,6 Endoxifen exposure is related to duration of tamoxifen use and dose, wherein an increase in the tamoxifen dose (from 20 to 40 mg daily) significantly increases endoxifen exposure in patients with reduced or null CYP2D6 metabolism but not in CYP2D6 EMs.7 However, coadministration of CYP2D6-inhibiting drugs4 reduces CYP2D6 enzyme activity, and nonadherence to tamoxifen is more commonly observed in patients with normal or increased CYP2D6 metabolism.8
Despite the consistent pharmacogenetic effects of CYP2D6 on endoxifen exposure, there is considerable controversy regarding the validity of CYP2D6 as a predictor of tamoxifen outcome.9,10 Although recent secondary analyses from the Arimidex, Tamoxifen, Alone or in Combination (ATAC) trial and the Breast International Group (BIG) 1-98 study11,12 did not demonstrate an association between CYP2D6 and tamoxifen outcome, these studies provoked criticism due to concerns regarding genotyping error and the analysis of small subsets of the main trials.13–16
By contrast, a secondary analysis from another large prospective adjuvant tamoxifen trial, the Austrian Breast and Colorectal Cancer Study Group 8 (ABCSG 8), demonstrated that for women treated with 5 years of adjuvant tamoxifen at a dose of 20 mg/day, CYP2D6 PMs had a statistically significant higher odds of recurrence or death as compared with CYP2D6 EMs, and CYP2D6 PMs/IMs and PMs/EMs tended to exhibit a higher odds of recurrence as compared with patients without the PM alleles. However, this effect was not observed for patients who had switched to anastrozole, a drug not metabolized by CYP2D6. These data suggest that the effects of CYP2D6 genotype may be masked if patients receive a shorter duration of tamoxifen or other active drugs besides tamoxifen, which alter the hazard for recurrence.17
We approached the tamoxifen controversy by performing a global meta-analysis of available clinical and CYP2D6 genetic data of tamoxifen-treated breast cancer patients. All groups from across the world with both published and unpublished CYP2D6 data were invited to participate. We initially presented a pooled analysis of these data,5 in which we found no association between CYP2D6 and IDFS. Following this presentation, we developed a new analysis plan (not defined before the initial negative presentation), which included the following: (i) articulation of three criteria to analyze the data according to the quality of the genetic and clinical data, (ii) additional curation to obtain missing clinical and genetic data, and (iii) a new statistical analysis plan, which applied a random-effects meta-analysis strategy instead of a pooled analysis strategy. Notably, Criterion 1 is most stringent, requiring strict control for as many pharmacologic factors as possible known to affect endoxifen exposure, which include use of tamoxifen monotherapy, genotyping of multiple CYP2D6 alleles for accurate CYP2D6 phenotype assignment, use of one tamoxifen dose (20 mg), and intended duration of tamoxifen use for 5 years. In addition, eligibility for this cohort was restricted to women with invasive ER-positive status, postmenopausal breast cancer, and the requirement for annual follow-up, parameters required in any prospective clinical trial and that were requirements of criterion 1 (patients who were knowingly not followed were excluded from criterion 1), but not from criteria 2 and 3. These factors may have contributed to the substantial increase in heterogeneity comparing criterion 1 with criteria 2 and 3. However, it should be noted that these criteria impose a certain bias because the majority of negative studies submitted to the ITPC were observed in criteria 2 and 3.
In general, a substantial number of subjects comprising criterion 3 had misclassification of the predicted drug metabolism phenotype due to the lack of a comprehensive coverage of loss-of-function alleles.18,19 More than 20 loss-of-function alleles out of 100 known CYP2D6 genetic variants contributed to a frequency of ~8% of PMs in a population of European descent. Limiting the analysis to the most common such allele, CYP2D6*4, as was frequently done in the older published literature, will result in misclassification of 35% of PMs, thereby falsely assigning the undetected PMs to the EM or IM groups. Notably, 871/1,996 patients comprising criterion 1 had optimal CYP2D6 phenotype assessment obtained by AmpliChip genotyping, and this may have contributed to the robustness of criterion 1 results, which demonstrated an association between CYP2D6 and tamoxifen treatment outcome (breast cancer–free interval: HR = 1.27, 95% confidence interval = 1.01–1.61).
The ITPC intended to perform a global study including several thousand patient samples; however, the majority of the subjects were not comprehensively genotyped because DNA was not of sufficient quality. We performed a subgroup analysis using patient samples for which full coverage of alleles by the AmpliChip genotyping platform was available using criterion 1 (871/1,635 AmpliChip-genotyped subjects met criterion 1). When confined to the Amplichip subjects, the estimates of the pharmacodynamic HRs for CYP2D6 were similar to what they were for the entire set of subjects meeting criterion 1.
A major source of potential genotyping errors may be related to DNA source. CYP2D6 is one of the most difficult genes to genotype because of the numerous polymorphisms and adjacent pseudogenes. Some platforms cannot detect the presence of the *5 deletion, particularly in DNA derived from formalin-fixed–paraffin-embedded (FFPE) tissue. However, several sites used multiple platforms to validate their genotyping data, reducing potential genotyping errors across the entire data set. Importantly, CYP2D6 genotypes obtained from blood-derived DNA reflect the patients' germ-line genotypes, known to influence endoxifen plasma concentrations. By contrast, CYP2D6 genotypes from tumor-derived DNA may be subject to error due to somatic mutation by loss of heterozygosity, known to affect the CYP2D6 locus at 22q13 in up to 30% of breast tumors.20–22 Thus, when CYP2D6 genotype is derived from tumor samples, an excess number of homozygotes may result as a consequence of loss of heterozygosity. This form of genotyping error is revealed by Hardy–Weinberg Equilibrium (HWE) testing, as was observed in the Breast International Group 1-98 study, in which strong departures from HWE (to a magnitude of 10–92) were observed, leading to a call for retraction of this article.3,12,16
For criterion 1, 49.9% of our patient DNA samples originated from blood, 21.6% from fresh-frozen tissues, and 28.5% from FFPE tissues. For criterion 2, 55.0% samples originated from blood, 20.5% were fresh-frozen tissues, and 24.5% from FFPE tissues. For criterion 3, 50.9% of DNA samples originated from blood, 31.9% from fresh-frozen tumor, 13.4% from FFPE tumor tissues, and 3.5% from FFPE normal tissue. Although we cannot exclude the presence of somatic events leading to misclassification of CYP2D6 genotype, as evident from HWE deviation identified in data from some sites, comprehensive testing for HWE did not reveal significant violations across most sites. Moreover, the extent of deviation from HWE in the *4 allele was not associated with sites that evinced less clinical benefit from tamoxifen in patients who were assessed to be PMs in terms of their CYP2D6 status. This suggests that genotyping errors are unlikely to be a major issue in our analyses.
Our findings are subject to the shortcomings commonly encountered when performing retrospective “biomarker” studies. In our study, most sites were unable to collect or control for the factors known to alter endoxifen exposure, including dose and duration of tamoxifen administration and patients' adherence to the regimen. Although tamoxifen adherence is increasingly recognized as a critical factor for drug efficacy,23 most studies evaluating tamoxifen biomarkers have not controlled for adherence. Other confounders include limited CYP2D6 allele coverage and lack of information regarding the coadministration of CYP2D6 inhibitors, leading to potential misclassification of the CYP2D6 drug metabolism phenotype. Therefore, our meta-analysis results depend heavily on which subgroup of patients we include. If we accept that utmost precautions must be applied to avoid the distortion of results from influences derived from the aforementioned shortcomings, it follows that merely increasing the numbers of subjects without controlling the quality of input data, as done in our preliminary overview analysis,5 may result in heterogeneity that masks the effect of a pharmacokinetic biomarker such as CYP2D6. From this, we conclude that until results from prospective adjuvant studies are available, women who meet criterion 1 as established in this and other independent cohorts (ABCSG 8) should be counseled regarding the potential impact of CYP2D6 on the effectiveness of adjuvant tamoxifen, and potent CYP2D6 inhibitors should be avoided in these patients. Prospective adjuvant studies are needed to determine whether genotype-guided selection of hormonal therapy will improve the outcomes of women with early-stage ER-positive breast cancer, and results from ongoing prospective studies in the metastatic setting are eagerly awaited. A similarly motivated study on warfarin is currently being conducted in the Clarification of Optimal Anticoagulation through Genetics trial.24
By strict clinical and genotype criteria, reduced CYP2D6 metabolism is associated with a higher risk of recurrence (as measured by IDFS) in tamoxifen-treated women. However, the heterogeneity observed across sites contributing data to the ITPC points to the likely influence of critical confounding factors unlikely to be controllable in global retrospective studies. This study demonstrates the complexity of performing a retrospective biomarker study that focuses on the genetic factors that affect exposure to an active metabolite, endoxifen, for a drug, tamoxifen, administered for 5 years. Our observation that <50% of the patients in this study met the basic eligibility criteria—in terms of similar disease, treatment, and control for critical pharmacological factors such as dose and duration of tamoxifen—provides insight into possible reasons for the discrepancies in the literature on CYP2D6 and tamoxifen. Although CYP2D6 is a predictor of IDFS in a subset of patients treated with tamoxifen, the lack of an effect in the entire heterogeneous study population suggests that prospective studies are necessary to finally establish whether genotype-guided selection of hormonal therapy improves clinical outcomes of women with ER-positive breast cancer.
Methods
Data collection and study cohorts. The ITPC invited any research group from across the world that had published or unpublished CYP2D6 data to participate in this meta-analysis. The ITPC comprises 12 research projects for a total of 4,973 breast cancer patients treated with tamoxifen. This retrospective study does not include a control group not treated with tamoxifen. These data were curated at the PharmGKB (Pharmacogenomics Knowledge Base, http://www.pharmgkb.org). Consent for participation in the ITPC and DNA collection, CYP2D6 genetic testing, and submission of data was obtained under local ethical review board permissions.
We collected information on clinical factors previously shown to be associated with breast cancer therapy and prognosis that were available from the information received from the sites. These data included demographic characteristics, cancer history, cancer recurrence, use of other therapies, use of concomitant medications known to affect CYP2D6 phenotype, ER status, and classic prognostic factors such as tumor size and number of affected lymph nodes. Information was also collected regarding the presence of CYP2D6 genetic variants (*2, *3, *4, *5, *6, *10, *17, and *41, categorized by their DNA sources), for which coverage of these alleles varied by site. For 1,635 subjects, CYP2D6 variants assessable from blood DNA using the AmpliChip CYP450 test (Roche) were collected. A complete list of the information collected is detailed in S1–S3 online, including the project-specific CYP2D6 genotype assays used and the DNA source. Independent confirmation of CYP2D6 genotypes was not performed owing to lack of access to subjects' samples. The clinical outcome variable was either breast cancer–free interval or IDFS, as previously defined.25 The complete data set of genotypes and clinical variables is available at http://www.pharmgkb.org.
Statistical analysis. Because the ITPC was not a prospectively defined multicenter study with a common protocol, there is potential for considerable study-to-study heterogeneity. Therefore, we did not analyze the combined data as a single series even though we had access to individual-level data from all studies. Rather, we applied a random-effects meta-analysis strategy. This provided estimates of the effect of CYP2D6 in each study's data separately, allowing us to examine the consistency of the results across sites. The meta-analysis is a two-stage procedure. In the first stage, we fit proportional-hazards models to the data from each of the ITPC sites separately, predicting clinical outcome after surgery from CYP2D6 genotype and other relevant covariates. These analyses produced a set of 12 parameter estimates of the HRs of CYP2D6 genotypes on outcome, along with their corresponding SEs (one for each site). In the second stage, we used a random-effects meta-analysis procedure26 to test for study heterogeneity (i.e., whether the 12 studies met the assumptions of the meta-analysis sufficiently so as to be combinable using that method). When the heterogeneity was not significant, we combined the log-HRs into a single, meta-analysis estimate of the effect of CYP2D6 on tamoxifen-treated recurrence and/or survival outcomes. The DerSimonian and Laird method also provides a penalty in its test of overall association for moderate levels of study-to-study heterogeneity (i.e., for heterogeneity that is not so severe as to be statistically significant). This method is therefore conservative in its conclusions when heterogeneity is a potential issue.
Study Highlights

Acknowledgments
The authors acknowledge useful conversations with Donald A. Berry (The University of Texas MD Anderson Cancer Center). The complete data set of genotypes and clinical variables, analysis codes, and full analyses is available to registered PharmGKB users at http://www.pharmgkb.org. We are grateful to all breast cancer patients for their participation. We thank the physicians and other hospital staff, scientists, research assistants, and study staff who contributed to the patient recruitment, data collection, and sample preparation.
This work was supported by grants from the National Institutes of Health (National Institute of General Medical Sciences, the National Cancer Institute, and the National Institute of Child Health and Human Development R24 GM61374, CA-25224, CA-37404, CA-15083, CA-35113, CA-35269, CA-63849, CA35103, CA-35195, CA-35272, CA-35101, CA-37417, CA-35415, CA-52352, CA-35448, CA-60274, 1R01CA133049-01, U19 GM61388, CA-116201, CA-58207, CA-51008, T32 GM007546, and CA051008; Breast Cancer Research (Scotland); Tayside Tissue Bank; California Breast Cancer Research Program grant 14OB-0166; Cancer Research UK; Deutsches Krebsforschungszentrum, Heidelberg, Germany; Robert Bosch Foundation, Stuttgart, Germany; 7FP EU Marie Curie Initial Training Network “FightingDrugFailure” (GA 238132); GrantStichting Emmanuel van der Schueren (scientific partner of the Vlaamse Liga tegen Kanker); and the National Project for Personalized Genomic Medicine, Ministry for Health & Welfare, Republic of Korea (A111218-PG02). W.S. is supported by grant Deutsche Forschungsgemeinschaft SCHR 1323/2-1, Germany.
The institutions in the International Tamoxifen Pharmacogenomics Consortium are listed below.:
Writing committee:
Michael A. Province, Matthew P. Goetz, David A. Flockhart, Hiltrud Brauch, and Teri E. Klein
Data contributors:
Deutsches Krebsforschungszentrum, Heidelberg, Germany: Ute Hamann, Julia Boländer, and Hans-Ulrich Ulmer, Frauenklinik Städtisches Klinikum Karlsruhe, Germany
Dr. Margarete Fischer-Bosch Institute of Clinical Pharmacology Stuttgart and University of Tuebingen, Tuebingen, Germany: Hiltrud Brauch, Werner Schroth, Matthias Schwab, Stefan Winter, Michel Eichelbaum, Peter Fritz, and Wolfgang Simon
Division of Research, Federal Institute for Drugs and Medical Devices, and University of Bonn Medical Faculty, Bonn, Germany: Julia C. Stingl
Indiana University and University of Michigan, USA: David A. Flockhart and Anne T. Nguyen
Inje University College of Medicine and Department of Clinical Pharmacology, Busan Paik Hospital, Busan, Korea: Jae-Gook Shin and Ji-Yeob Choi
Mayo Clinic in Rochester, Minnesota, USA: Matthew P. Goetz, James N. Ingle, Vera J. Suman, and Richard M. Weinshilboum
Ninewells Hospital and Medical School, Dundee, UK: Colin A. Purdie and Lee B. Jordan
Örebro University, Sweden: Pia Wegman
RIKEN Center for Genomic Medicine and University of Tokyo, Japan: Hitoshi Zembutsu, Taisei Mushiroda, and Kazuma Kiyotani
Roswell Park Cancer Institute, USA: Christine B. Ambrosone
University Breast Center Franconia, University Hospital Erlangen, Erlangen, Germany: Peter A Fasching, Reiner Strick, and Matthias W Beckmann
University Hospitals Leuven, Belgium: Patrick Neven and Anne-Sophie Dieudonné
University Institute of Medical and Chemical Laboratory Diagnostics, Paracelsus Private Medical University, Salzburg, Austria: Elisabeth Haschke-Becher
University of California, San Francisco, San Francisco, USA: Alan H.B. Wu, Wendy Lorizio, and Elad Ziv
University of Dundee, UK: Philip Quinlan, Lee B. Jordan, and Colin A. Purdie
Department of Gynecology and Obstetrics, University of Mainz, Mainz, Germany: Marcus Schmidt and Heinz Koelbl
University of Manchester, Manchester, UK: William G. Newman, Roberta Ferraldeschi, Anthony Howell, and Ayse Latif
Vesalius Research Center, Catholic University of Leuven, Leuven, Belgium: Diether Lambrechts
Yonsei University Health System, Korea: Byeong-Woo Park
Statistical Analysis:
Mayo Clinic in Rochester, MN, USA: Vera J. Suman
Stanford University, Stanford, CA, USA: Teri E. Klein and Ryan Whaley
Washington University in St. Louis, MO, USA: Michael A. Province
Data Curation:
Stanford University, Stanford, CA, USA: Teri E. Klein, Ryan Whaley, Joan M. Hebert, and Li Gong
Site Principal Investigators:
Russ B. Altman and Teri E. Klein (Stanford University), Christine A. Ambrosone (Roswell Park Cancer Institute), Hiltrud Brauch (IKP Stuttgart), Jae-Gook Shin (Inje University), Matthew P. Goetz (Mayo Clinic), William G. Newman (University of Manchester), Patrick Neven (Belgium), Alastair M. Thompson (University of Dundee), Sten Wingren (Örebro University), Hitoshi Zembutsu (University of Tokyo), and Elad Ziv (University of California, San Francisco)
Steering Committee:
Russ B. Altman, Hiltrud Brauch, Michel Eichelbaum, David A. Flockhart, Matthew P. Goetz, James N. Ingle, V. Craig Jordan, Teri E. Klein, Kent Osborne, Yusuke Nakamura, and Richard M. Weinshilboum
Participating sites:
The Christie NHS Foundation Trust, Manchester Academic Health Science Centre, University of Manchester, Manchester, UK
Department of Cancer Prevention and Control, Roswell Park Cancer Institute, Buffalo, NY, USA
Department of Clinical Medicine, Örebro University, Örebro, Sweden
Department of Genetics, Stanford University, Stanford, CA, USA
Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Erlangen, Germany
Department of Gynecology and Obstetrics, University Hospitals Leuven, Leuven, Belgium
Department of Oncology, Catholic University Leuven, Leuven, Belgium
Department of Medicine/GIM and Clinical Pharmacology and Experimental Therapeutics, and Helen Diller Family Comprehensive Cancer Center, University of California, San Francisco, San Francisco, CA, USA
Department of Molecular Pharmacology & Experimental Therapeutics, Mayo Clinic, Rochester, MN, USA
Department of Oncology, Georgetown University, Washington, DC, USA
Department of Oncology, Mayo Clinic, Rochester, MN, USA
Department of Oncology and Pharmacology, Mayo Clinic, Rochester, MN, USA
Department of Pharmacology; Mayo Clinic, Rochester, MN, USA
Department of Pharmacology and Pharmacogenomics Research Center, Inje University College of Medicine and Department of Clinical Pharmacology, Inje University, Busan Paik Hospital, Busan, Korea
Department of Surgery, Yonsei University Health System, Seoul, Korea
Deutsches Krebsforschungszentrum, Heidelberg, Germany
Division of Biomedical Statistics and Informatics, Mayo Clinic, Rochester, MN, USA
Division of Clinical Pharmacology, School of Medicine, Indiana University, IN, USA
Dr. Margarete Fischer-Bosch Institute of Clinical Pharmacology Stuttgart, and University of Tuebingen, Germany
Dundee Cancer Centre, Dundee, UK
Genetic Medicine, Manchester Academic Health Science Centre, University of Manchester, Manchester, UK
Laboratory for Pharmacogenetics, RIKEN Center for Genomic Medicine, Yokohama, Japan
Laboratory of Molecular Medicine, Human Genome Center, Institute of Medical Science, The University of Tokyo, Tokyo, Japan
Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum, Heidelberg, Germany
Ninewells Hospital and Medical School, Dundee, UK
A.M.T. and W.G.N. report Roche funding for genotyping H.B. and M. Schwab report that they have initiated scientific collaborations in 2009 with Roche Molecular Diagnostics and Siemens Healthcare Diagnostics Products, respectively P.A.F. and M.W.B. report Novartis research funding. M.-T.M.L. is a paid consultant of YongLin Health Foundation. R.B.A. is a founder, equity holder, and consultant for Personalis. The other authors declared no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/cpt
Supplementary Material
References
- Jordan V.C. Tamoxifen: a most unlikely pioneering medicine. Nat. Rev. Drug Discov. 2003;2:205–213. doi: 10.1038/nrd1031. [DOI] [PubMed] [Google Scholar]
- Davies C., et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–784. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.D., et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res. Treat. 2004;85:151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- Stearns V., et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J. Natl. Cancer Inst. 2003;95:1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- Goetz M.P., Berry D.A., Klein T.E. 31136 Findings from the International Tamoxifen Pharmacogenomics Consortium. Proceedings of the San Antonio Breast Cancer Symposium, San Antonio, Texas. 9–13 December. 2009. Abstract 33
- Mürdter T.E., et al. German Tamoxifen and AI Clinicians Group Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasma. Clin. Pharmacol. Ther. 2011;89:708–717. doi: 10.1038/clpt.2011.27. [DOI] [PubMed] [Google Scholar]
- Irvin W.J., Jr, et al. Genotype-guided tamoxifen dosing increases active metabolite exposure in women with reduced CYP2D6 metabolism: a multicenter study. J. Clin. Oncol. 2011;29:3232–3239. doi: 10.1200/JCO.2010.31.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae J.M., et al. COBRA investigators Cytochrome P450 2D6 activity predicts discontinuation of tamoxifen therapy in breast cancer patients. Pharmacogenomics J. 2009;9:258–264. doi: 10.1038/tpj.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroth W., et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302:1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M.P., et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J. Clin. Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- Rae J.M., et al. ATAC trialists CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J. Natl. Cancer Inst. 2012;104:452–460. doi: 10.1093/jnci/djs126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan M.M., et al. Breast International Group (BIG) 1-98 Collaborative Group CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the breast international group 1-98 trial. J. Natl. Cancer Inst. 2012;104:441–451. doi: 10.1093/jnci/djs125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauch H., et al. Tamoxifen use in postmenopausal breast cancer: CYP2D6 matters. J. Clin. Oncol. 2013;31:176–180. doi: 10.1200/JCO.2012.44.6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Ratain M.J., Cox N.J., McLeod H.L., Kroetz D.L., Flockhart D.A. Re: CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 trial. J. Natl. Cancer Inst. 2012;104:1264; author reply 1266–1268. doi: 10.1093/jnci/djs304. [DOI] [PubMed] [Google Scholar]
- Stanton V., Jr Re: CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 trial. J. Natl. Cancer Inst. 2012;104:1265–6; author reply 1266–8. doi: 10.1093/jnci/djs305. [DOI] [PubMed] [Google Scholar]
- Goldberg P. Experts claim errors in breast cancer study, demand retraction of practice-changing paper. The Cancer Letter. 2012;38:1–11. [Google Scholar]
- Goetz M.P., et al. CYP2D6 metabolism and patient outcome in the Austrian Breast and Colorectal Cancer Study Group trial (ABCSG) 8. Clin. Cancer Res. 2013;19:500–507. doi: 10.1158/1078-0432.CCR-12-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroth W., et al. CYP2D6 polymorphisms as predictors of outcome in breast cancer patients treated with tamoxifen: expanded polymorphism coverage improves risk stratification. Clin. Cancer Res. 2010;16:4468–4477. doi: 10.1158/1078-0432.CCR-10-0478. [DOI] [PubMed] [Google Scholar]
- Zanger U.M., Turpeinen M., Klein K., Schwab M. Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal. Bioanal. Chem. 2008;392:1093–1108. doi: 10.1007/s00216-008-2291-6. [DOI] [PubMed] [Google Scholar]
- Castells A., Gusella J.F., Ramesh V., Rustgi A.K. A region of deletion on chromosome 22q13 is common to human breast and colorectal cancers. Cancer Res. 2000;60:2836–2839. [PubMed] [Google Scholar]
- Hirano A., et al. Allelic losses of loci at 3p25.1, 8p22, 13q12, 17p13.3, and 22q13 correlate with postoperative recurrence in breast cancer. Clin. Cancer Res. 2001;7:876–882. [PubMed] [Google Scholar]
- Loo L.W., et al. Differential patterns of allelic loss in estrogen receptor-positive infiltrating lobular and ductal breast cancer. Genes. Chromosomes Cancer. 2008;47:1049–1066. doi: 10.1002/gcc.20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.M., et al. Comprehensive CYP2D6 genotype and adherence affect outcome in breast cancer patients treated with tamoxifen monotherapy. Breast Cancer Res. Treat. 2011;125:279–287. doi: 10.1007/s10549-010-1139-x. [DOI] [PubMed] [Google Scholar]
- French B., et al. COAG (Clarification of Optimal Anticoagulation through Genetics) Investigators Statistical design of personalized medicine interventions: the Clarification of Optimal Anticoagulation through Genetics (COAG) trial. Trials. 2010;11:108. doi: 10.1186/1745-6215-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudis C.A., et al. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J. Clin. Oncol. 2007;25:2127–2132. doi: 10.1200/JCO.2006.10.3523. [DOI] [PubMed] [Google Scholar]
- DerSimonian R., Laird N. Meta-analysis in clinical trials. Control. Clin. Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.