Abstract
Pegylated interferon-α (PEG-IFN-α or PEG-IFN 2a and 2b)– and ribavirin (RBV)-based regimens are the mainstay for treatment of hepatitis C virus (HCV) genotype 1. IFNL3 (IL28B) genotype is the strongest baseline predictor of response to PEG-IFN-α and RBV therapy in previously untreated patients and can be used by patients and clinicians as part of the shared decision-making process for initiating treatment for HCV infection. We provide information regarding the clinical use of PEG-IFN-α- and RBV-containing regimens based on IFNL3 genotype.
The purpose of this guideline is to provide information regarding the clinical use of IFNL3 (IL28B) genotyping to guide the use of pegylated interferon-α (PEG-IFN-α or PEG-IFN 2a and 2b) and ribavirin (RBV) combination therapy, including treatment with direct-acting antivirals approved for hepatitis C virus (HCV) genotype 1 infection. Demographic and other clinical variables, such as adherence to psychological or pharmacological therapy, concomitant use of other drugs that may influence efficacy of antiviral treatment, or patient-specific disease characteristics, are not the focus of this guideline. The Clinical Pharmacogenetics Implementation Consortium develops peer-reviewed guidelines that are published and updated regularly at http://www.pharmgkb.org on the basis of emerging evidence.
Focused Literature Review
A systematic literature review focused on IFNL3 (IL28B) genotype and PEG-IFN-α use was conducted (see Supplementary Material online). This guideline was developed on the basis of interpretation of the literature by the authors and experts in the field.
Drugs: PEG-IFN-α
Background
Infection with HCV affects >150 million people worldwide and is one of the leading causes of cirrhosis and hepatocellular carcinoma (hepatitis C fact sheet, Geneva, Switzerland: World Health Organization; accessed 30 September 2012 at http://www.who.int/mediacentre/factsheets/fs164/en/index.html). Before 2011, treatment for chronic HCV infection consisted of combination PEG-IFN-α and RBV therapy for 24 weeks for HCV genotypes 2 and 3 and for 48 weeks for other HCV genotypes.1 In 2011, two first-generation HCV protease inhibitors, boceprevir and telaprevir, were approved to treat HCV genotype 1 infection in many countries, including the United States and the countries of the European Union. These direct-acting antiviral agents are indicated in combination with PEG-IFN-α and RBV therapy for patients with HCV genotype 1 infection, and this regimen is preferred over PEG-IFN-α and RBV alone in countries where direct-acting antivirals have been approved.
The primary goal of treatment is eradication of HCV as measured by sustained virologic response (SVR, defined by undetectable serum viral RNA 12–24 weeks after the end of treatment), which equates with cure of the infection and leads to lower morbidity and mortality.2 However, combination therapy for HCV treatment has a number of limitations. The treatment is expensive, is associated with many side effects, and lasts 24–48 weeks. SVR rates are low for patients with HCV genotypes 1 and 4 treated with PEG-IFN-α and RBV combination.3 The addition of the first-generation protease inhibitors to PEG-IFN-α and RBV has led to SVR rates of 69–75% for patients with HCV genotype 1 infection naive to HCV treatment, but SVR rates are lower for patients who failed previous treatment with PEG-IFN-α and RBV. The SVR rate varies drastically among different races and ethnicities, with patients of African ancestry having much lower response rates as compared with Caucasians and Asians, supporting genetic predisposition to response.4,5 Both host and viral factors have been previously reported to be associated with SVR. Predictors of SVR include HCV genotypes 2 and 3, lower baseline serum HCV RNA level, younger age, female sex, lower hepatic fibrosis stage, lack of insulin resistance, and lower body mass index.3
Gene: IFNL3
Background
IFNL3, also known as IL28B, encodes interferon-λ 3 (IFN-λ 3), a member of the type 3 IFN-λ family with antiviral, antiproliferative, and immune-modulatory activities.6 IFN-λs can be induced by viruses and inhibit HCV replication in vitro.6 Similar to type 1 interferons (IFN-αs), IFN-λs signal through the common janus kinase–signal transducer and activator of transcription and mitogen-activated protein kinase pathways and induce IFN-stimulated gene expression.6 However, IFN-λs engage a unique heterodimeric receptor complex, which is composed of the IL-10R2 (IL10RB gene) and IFN-λR1 (IL28RA gene) receptor chains,6 and are highly expressed only in hepatocytes and epithelial cells, distinct from the ubiquitously expressed IFN-α receptor.
IFNL3 variation is the strongest established pretreatment predictor of treatment response for previously untreated (naive) patients with HCV genotype 1 (see Supplementary Table S1 online for additional genes associated with HCV treatment response). Four genome-wide association studies have independently associated IFNL3 genetic variation with treatment-induced clearance of HCV following PEG-IFN-α and RBV therapy,7,8,9,10 which was later validated in candidate gene studies.11,12,13 These variants are located on chromosome 19, near the IFNL3 gene. The two most commonly tested single -nucleotide polymorphisms are rs12979860 and rs8099917, which are in close proximity and in strong linkage disequilibrium. Favorable response IFNL3 genotypes (CC for rs12979860 and TT for rs8099917) are associated with an approximate two-fold increase in SVR for HCV genotype 1 patients. However, rs12979860 has been found to be less reliable for predicting response to PEG-IFN-α and RBV therapy in Japanese patients as compared with rs8099917.14 The rs12979860 allele frequency varies among different ethnic groups (Supplementary Tables S2 and S3 online), and largely explains the differences in treatment response rates among East Asians, Caucasians, African Americans, and Hispanics with chronic HCV infection.10,15 The rs12979860 favorable C allele is most frequently present in East Asians (allele frequency nearly 0.9), followed by Caucasians and Hispanics, and is the least common among individuals of African origin (allele frequencies of 0.63, 0.55, and 0.39, respectively).10,16,17
The mechanisms by which IFNL3 variations affect antiviral response are still poorly understood. Many of the IFNL3 variants associated with PEG-IFN-α and RBV treatment outcomes are not within the coding region but only in close proximity to the IFNL3 gene. The effects of this variation on IFNL3 gene expression or intrahepatic IFN-stimulated gene expression remain controversial,8,10,18,19,20 and the impact on protein stability or receptor-binding potency of these variants has not been reported. Recently, multiple studies demonstrated that the favorable rs12979860 genotype may affect treatment outcomes via improved innate immune response to IFN therapy and more rapid viral kinetics independent of IFN-stimulated gene expression.21,22,23,24 This variant is also associated with better early viral kinetics during IFN-free treatment of patients with chronic hepatitis C.25
Genetic test interpretation
Laboratory results for IFNL3 genotype are typically reported as reference single-nucleotide polymorphism identification number (rs) followed by the specific genotype (i.e., rs12979860 CC, CT, or TT) with accompanying interpretation (i.e., favorable genotype vs. unfavorable genotype). The assignment of the likely IFNL3 phenotype, based on diplotypes, is summarized in Table 1.
Table 1. Assignment of probable IFNL3 phenotypes based on genotypes.
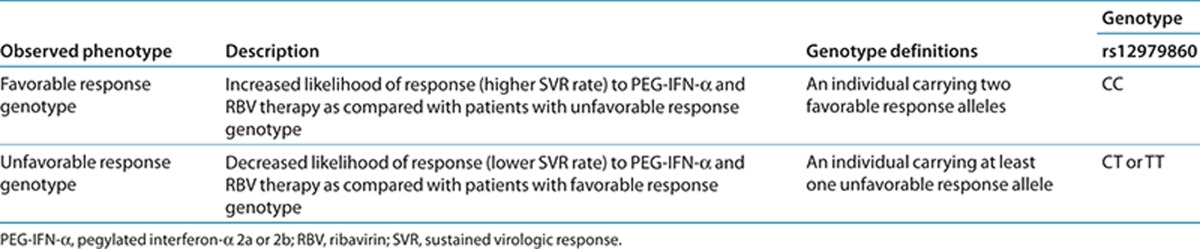
Available genetic test options
See the Supplementary Material online and http://www.PharmGKB.org for more information on commercially available clinical testing options.
Linking genetic variability to variability in drug-related phenotypes
There is substantial evidence linking IFNL3 genotype to phenotypic variability (see Supplementary Table S4 online). Among patients treated for chronic HCV infection, IFNL3 genotype is associated with early viral kinetics and improved SVR. As early as week 4 of therapy, patients with the favorable IFNL3 genotype are more likely to have undetectable HCV RNA.
Application of a grading system to the evidence linking genotypic to phenotypic variability indicates a high quality of evidence in the majority of cases (Supplementary Table S4 online). This body of evidence provides the basis for the therapy recommendations in Table 2.
Table 2. Recommendations for use of PEG-IFN-α–containing regimens based on IFNL3 genotype .
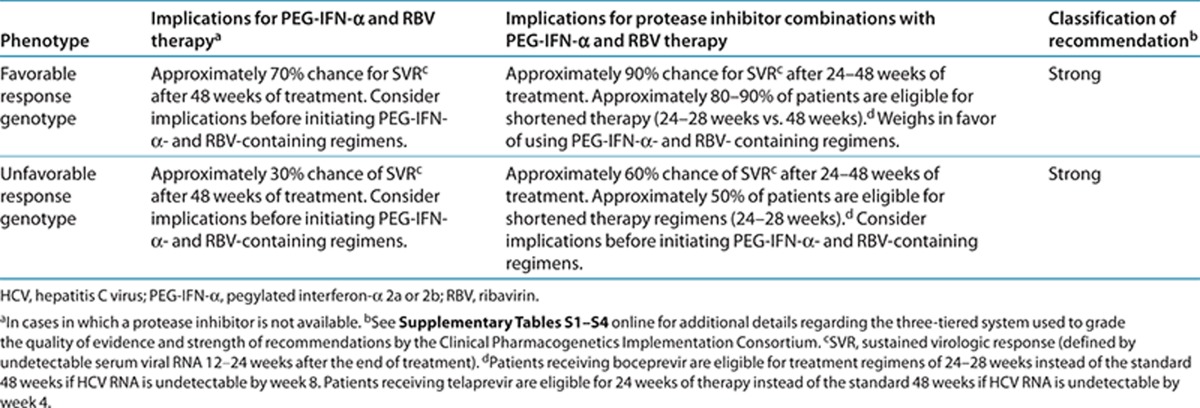
Therapeutic recommendations
Table 2 summarizes the therapeutic recommendations for PEG-IFN-α and RBV therapy based on IFNL3 genotype. Treatment of HCV genotype 1 infection varies throughout the world currently because some regions have access to the new direct-acting antivirals in combination with PEG-IFN-α and RBV, whereas other regions have access only to PEG-IFN-α and RBV. The role of IFNL3 genotyping depends on treatment selection. IFNL3 genotype is only one factor that can influence response rates to PEG-IFN-α and RBV therapy in HCV genotype 1 infection and should be interpreted in the context of other clinical and genetic factors.
PEG-IFN-α and RBV. For patients treated with PEG-IFN-α and RBV alone, IFNL3 genotype is the strongest pretreatment predictor of HCV treatment response. In the intention-to-treat analysis of the original discovery cohort with rs12979860, Caucasian patients with CC genotype were more likely than those with CT or TT genotype to have undetectable serum viral RNA by week 4 (28 vs. 5 and 5%, respectively; P < 0.0001) and to achieve SVR (69 vs. 33 and 27%, respectively; P < 0.0001).17 Similar patterns were observed in Hispanic and African-American patients in this cohort. HCV treatment is associated with significant side effects, and the likelihood of response treatment influences shared decision making between clinicians and patients about initiating treatment.
Protease inhibitor combination regimens—treatment naive. For treatment-naive patients with genotype 1 infection who are treated with protease inhibitor combinations, all IFNL3 genotypes have improved response rates as compared with patients treated with PEG-IFN-α and RBV only. However, patients with the favorable IFNL3 genotype still have higher response rates with the protease inhibitor combination in treatment-naive patients, and these response rates may guide patients and clinicians in their treatment decisions. In the boceprevir phase III treatment naive study of combination with PEG-IFN-α and RBV, SVR rates for rs12979860 CC patients receiving boceprevir ranged from 80 to 82% as compared with 65–71% for CT patients and 59–65% for TT patients.26 Moreover, multivariate regression analysis revealed that rs12979860 CC genotype was a predictor of SVR as compared with CT (odds ratio = 2.6, 95% confidence interval = 1.3–5.1) and TT genotypes (odds ratio = 2.1, 95% confidence interval = 1.2–3.7).
Role of the lead-in. Although IFNL3 genotype is the strongest pretreatment predictor of response to IFN-α-based therapy, the use of the early on-treatment antiviral response has also been extensively evaluated.27 The IFNL3 genotype is a marker for IFN responsiveness, and patients with the favorable IFNL3 genotype are more likely to have significant reductions in HCV RNA during the first 4 weeks of therapy. The boceprevir combination regimen starts with 4 weeks of PEG-IFN-α and RBV, and boceprevir is added in the fifth week. In the analysis of the boceprevir phase III studies, SVR models that considered only baseline characteristics found that the IFNL3 genotype was a predictor of SVR.26,27 When the lead-in response was added to these models, the IFNL3 genotype was no longer a predictor. It has been proposed that these early kinetics minimize the value of the IFNL3 genotype, but the lead-in response is known only for patients who have initiated therapy. For the patient who is considering whether or not to undergo HCV therapy with the boceprevir regimen, IFNL3 genotype remains the most helpful predictor of likelihood of response.27
Duration of therapy. Duration of treatment is another important factor for clinicians and patients to consider before initiating PEG-IFN-α and RBV therapy because patients with favorable IFNL3 genotypes are more likely to respond to shorter treatment courses. Patients receiving boceprevir are eligible for 24- to 28-week regimens instead of the standard 48-week regimen if HCV RNA is undetectable by week 8.27 In the boceprevir phase III clinical trial for treatment-naive patients, rs12979860 CC patients were more likely to have undetectable HCV RNA at week 8 (89%) than CT (53%) or TT (42%) patients.26 SVR rates ranged from 81 to 100% for all patients in whom HCV RNA was undetectable by week 8, regardless of IFNL3 genotype. With telaprevir therapy, patients with undetectable HCV RNA by week 4 are eligible for a treatment regimen of only 24 weeks.27 Given the side-effect burden of PEG-IFN-α and RBV, the possibility of shorter treatment course may influence treatment choice for some patients.
Past treatment. In general, patients who have failed previous IFN-α-based therapies are enriched for the unfavorable IFNL3 genotype; therefore, IFNL3 genotype is less likely to influence clinical decisions. Analysis of phase III boceprevir trial results for patients who were treatment experienced found that IFNL3 genotype did not predict SVR.26 In patients with unclear records of their previous treatment or with questions about the quality of care received in a previous course of therapy, the IFNL3 genotype can be considered a marker of IFN responsiveness that contributes to HCV treatment response.
IFNL3 single-nucleotide polymorphism concordance. Given that both rs12979860 and rs8099917 tests are available, clinicians may receive both pieces of data. The boceprevir phase III program conducted an analysis and found that combining rs12979860 and rs8099917 test results did not improve the strength of the association between the IFNL3 genotype and SVR as compared with the results using rs12979860 genotype alone.26 This analysis also found instances of discordance between rs12979860 and rs8099917. Most rs12979860 CC patients had the favorable TT pattern at the rs8099917 locus. However, of the 426 patients with the favorable TT genotype at the rs8099917 locus, only 208 (48.8%) also had the favorable CC genotype at the rs12979860 locus. This analysis of the boceprevir program reported that both rs12979860 and rs8099917 predict SVR, but rs12979860 is more reliable in this group of patients.
Other considerations
The original discovery of IFNL3 genotype came from the analysis of treatment-naive patients with HCV genotype 1 treated with PEG-IFN-α and RBV, and subsequent studies have evaluated IFNL3 genotype in other HCV patient groups.
Acute infection. With initial exposure to HCV, some patients are able to spontaneously clear the infection. Moreover, multiple well characterized cohort studies demonstrated that patients with the favorable IFNL3 CC genotype are more likely to spontaneously clear the acute infection.7,28,29,30,31 In the original analysis, the effect of rs12979860 variation was evaluated in six well-characterized cohorts of patients with acute HCV infection.28 Patients with the favorable IFNL3 CC genotype were more likely to clear infection spontaneously than patients with the CT/TT genotypes (53 vs. 28%), and results were similar in patients of European and African ancestry. Thus, IFNL3 genotype testing may be informative for the treatment considerations for the acutely infected patient.
HCV genotypes 2 and 3. Outcomes for the treatment of chronic HCV genotypes 2 and 3 have consistently been better than those for HCV genotypes 1 and 4 with IFN-α-based regimens. SVR rates are higher for HCV genotypes 2 and 3 and can be achieved with a shorter therapy duration of 24 weeks.3,32 Studies of patients with HCV genotypes 2 and 3 have not found that the IFNL3 genotype is a predictor of response. The report by Mangia et al. did find that among patients in whom HCV RNA was not undetectable by week 4, SVR was achieved more often in rs12979860 CC patients as compared with CT and TT patients (87 vs. 67 and 29%, respectively; P = 0.0002).33
HIV/HCV coinfection. For patients with HIV/HCV coinfection, IFNL3 genotype has been associated with spontaneous clearance and improved response to treatment with IFN-α-based regimens.7,33 Rallon et al. evaluated a cohort of 164 treated HIV/HCV–coinfected patients, and 74% of patients with rs12979860 CC genotype achieved SVR as compared with 38% of patients with CT or TT genotypes (P ≤ 0.0001).
Incidental findings
The IFNL3 rs12979860 polymorphism has also been linked to HCV-induced hepatocellular carcinoma and graft fibrosis,34 allergic disease in children,35 liver fibrosis,36 viral cirrhosis due to HCV,37 and greater likelihood of HCV persistence, particularly in HCV genotypes 1 and 4.38 The favorable rs12979860 CC genotype is associated with lower frequency of hepatic steatosis in patients with chronic HCV.39 Carriers of the T allele of this variant have also been found to have increased susceptibility to chronic hepatitis B virus (HBV) infection and hepatocellular carcinoma as compared with noncarriers.40
Potential Benefits and Risks for the Patient
Patients considering HCV therapy are confronted with medications with significant side effects and varying response rates. As a result, patients and clinicians must weigh the risks and benefits of treatment. Previous models of baseline characteristics had limited ability to accurately predict outcomes, and the best models incorporated early viral kinetics that required the patient to initiate treatment. At present, IFNL3 genotype is the strongest baseline predictor of treatment response in patients receiving HCV therapy with PEG-IFN-α and RBV. With protease inhibitor regimens, IFNL3 genotype predicts response and also predicts eligibility for the shorter durations of therapy. No clear risks have been determined with IFNL3 genotype testing. Although not studied formally, knowledge of a reduced likelihood of response may result in fewer patients receiving HCV therapy that might have been effective.
Caveats: Appropriate Use AND/OR Potential Misuse of Genetic Tests
IFNL3 genotype is a strong predictor of treatment response for patients receiving treatment for chronic HCV infection. However, genotyping alone does not provide the basis for the decision to treat or not to treat HCV infection. Patients with all IFNL3 genotypes can respond to HCV therapy, and the differences in outcome are reduced with the addition of protease inhibitors. IFNL3 genotype is one of several factors to be considered when estimating the likelihood of treatment response. In addition, the side-effect profile of HCV regimens should be considered independently from the likelihood of response according to IFNL3 genotype.
Acknowledgments
We acknowledge the critical input of members of the Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network, funded by the National Institutes of Health (NIH), particularly Mary V Relling (St Jude Children's Research Hospital). This work was funded by NIH grants GM61374 and U01 GM092666.
The Clinical Pharmacogenetics Implementation Consortium guidelines reflect expert consensus based on clinical evidence and peer-reviewed literature available at the time they are written and are intended only to assist clinicians in decision making and to identify questions for further research. New evidence may have emerged since the time a guideline was submitted for publication. Guidelines are limited in scope and are not applicable to interventions or diseases not specifically identified. Guidelines do not account for all individual variations among patients and cannot be considered inclusive of all proper methods of care or exclusive of other treatments. It remains the responsibility of the health-care provider to determine the best course of treatment for a patient. Adherence to any guideline is voluntary, with the ultimate determination regarding its application to be made solely by the clinician and the patient. The Clinical Pharmacogenetics Implementation Consortium assumes no responsibility for any injury to persons or damage to persons or property arising out of or related to any use of the guidelines of the Clinical Pharmacogenetics Implementation Consortium or for any errors or omissions.
A.J.M. reports research grants from Abbott, Achillion, BMS, GSK, Merck, Roche, and Vertex, in addition to consulting fees from Achillion, BMS, GSK, Merck, and Vertex. D.R.N. reports research grants from Abbott, BI, BMS, Genentech, Gilead, Merck, and Vertex. The other authors declared no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/cpt
Supplementary Material
References
- Kronenberger B., Zeuzem S. Current and future treatment options for HCV. Ann. Hepatol. 2009;8:103–112. [PubMed] [Google Scholar]
- van der Meer A.J., et al. Association between sustained virological response and all-cause mortality among patients with chronic hepatitis C and advanced hepatic fibrosis. JAMA. 2012;308:2584–2593. doi: 10.1001/jama.2012.144878. [DOI] [PubMed] [Google Scholar]
- Ghany M.G., Strader D.B., Thomas D.L., Seeff L.B. American Association for the Study of Liver Diseases Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335–1374. doi: 10.1002/hep.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conjeevaram H.S., et al. Virahep-C Study Group Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology. 2006;131:470–477. doi: 10.1053/j.gastro.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Muir A.J., Bornstein J.D., Killenberg P.G. Atlantic Coast Hepatitis Treatment Group Peginterferon alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N. Engl. J. Med. 2004;350:2265–2271. doi: 10.1056/NEJMoa032502. [DOI] [PubMed] [Google Scholar]
- Kotenko S.V. IFN-λs. Curr. Opin. Immunol. 2011;23:583–590. doi: 10.1016/j.coi.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A., et al. Swiss Hepatitis C Cohort Study; Swiss HIV Cohort Study Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–45, 1345.e1. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- Suppiah V., et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- Ge D., et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- Montes-Cano M.A., et al. Interleukin-28B genetic variants and hepatitis virus infection by different viral genotypes. Hepatology. 2010;52:33–37. doi: 10.1002/hep.23624. [DOI] [PubMed] [Google Scholar]
- Abe H., et al. Common variation of IL28 affects gamma-GTP levels and inflammation of the liver in chronically infected hepatitis C virus patients. J. Hepatol. 2010;53:439–443. doi: 10.1016/j.jhep.2010.03.022. [DOI] [PubMed] [Google Scholar]
- McCarthy J.J., et al. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 2010;138:2307–2314. doi: 10.1053/j.gastro.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., et al. The rs8099917 polymorphism, when determined by a suitable genotyping method, is a better predictor for response to pegylated alpha interferon/ribavirin therapy in Japanese patients than other single nucleotide polymorphisms associated with interleukin-28B. J. Clin. Microbiol. 2011;49:1853–1860. doi: 10.1128/JCM.02139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satapathy S.K., Lingisetty C.S., Proper S., Chaudhari S., Williams S. Equally poor outcomes to pegylated interferon-based therapy in African Americans and Hispanics with chronic hepatitis C infection. J. Clin. Gastroenterol. 2010;44:140–145. doi: 10.1097/MCG.0b013e3181ba9992. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., et al. Association of two polymorphisms of the IL28B gene with viral factors and treatment response in 1,518 patients infected with hepatitis C virus. J. Gastroenterol. 2012;47:596–605. doi: 10.1007/s00535-012-0531-1. [DOI] [PubMed] [Google Scholar]
- Thompson A.J., et al. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120–9.e18. doi: 10.1053/j.gastro.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Dill M.T., et al. Interferon-induced gene expression is a stronger predictor of treatment response than IL28B genotype in patients with hepatitis C. Gastroenterology. 2011;140:1021–1031. doi: 10.1053/j.gastro.2010.11.039. [DOI] [PubMed] [Google Scholar]
- Urban T.J., et al. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888–1896. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M., et al. Hokuriku Liver Study Group Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 2010;139:499–509. doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- Naggie S., et al. Dysregulation of innate immunity in hepatitis C virus genotype 1 IL28B-unfavorable genotype patients: impaired viral kinetics and therapeutic response. Hepatology. 2012;56:444–454. doi: 10.1002/hep.25647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell C.D., et al. Single nucleotide polymorphism upstream of interleukin 28B associated with phase 1 and phase 2 of early viral kinetics in patients infected with HCV genotype 1. J. Hepatol. 2012;56:557–563. doi: 10.1016/j.jhep.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindh M., et al. Interleukin 28B gene variation at rs12979860 determines early viral kinetics during treatment in patients carrying genotypes 2 or 3 of hepatitis C virus. J. Infect. Dis. 2011;203:1748–1752. doi: 10.1093/infdis/jir193. [DOI] [PubMed] [Google Scholar]
- Bochud P.Y., et al. IL28B polymorphisms predict reduction of HCV RNA from the first day of therapy in chronic hepatitis C. J. Hepatol. 2011;55:980–988. doi: 10.1016/j.jhep.2011.01.050. [DOI] [PubMed] [Google Scholar]
- Chu T.W., et al. Effect of IL28B genotype on early viral kinetics during interferon-free treatment of patients with chronic hepatitis C. Gastroenterology. 2012;142:790–795. doi: 10.1053/j.gastro.2011.12.057. [DOI] [PubMed] [Google Scholar]
- Poordad F., et al. SPRINT-2 and RESPOND-2 Investigators Factors that predict response of patients with hepatitis C virus infection to boceprevir. Gastroenterology. 2012;143:608–18.e1. doi: 10.1053/j.gastro.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Muir A.J. IL28B in the era of direct-acting antivirals for hepatitis C. J. Clin. Gastroenterol. 2013;47:222–227. doi: 10.1097/MCG.0b013e3182680221. [DOI] [PubMed] [Google Scholar]
- Thomas D.L., et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S., et al. A polymorphism in IL28B distinguishes exposed, uninfected individuals from spontaneous resolvers of HCV infection. Gastroenterology. 2011;141:320–5, 325.e1. doi: 10.1053/j.gastro.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Extremera A., et al. Genetic variation in interleukin 28B with respect to vertical transmission of hepatitis C virus and spontaneous clearance in HCV-infected children. Hepatology. 2011;53:1830–1838. doi: 10.1002/hep.24298. [DOI] [PubMed] [Google Scholar]
- Tillmann H.L., et al. German Anti-D Study Group A polymorphism near IL28B is associated with spontaneous clearance of acute hepatitis C virus and jaundice. Gastroenterology. 2010;139:1586–92, 1592.e1. doi: 10.1053/j.gastro.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Hadziyannis S.J., et al. PEGASYS International Study Group Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 2004;140:346–355. doi: 10.7326/0003-4819-140-5-200403020-00010. [DOI] [PubMed] [Google Scholar]
- Mangia A., et al. An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology. 2010;139:821–7, 827.e1. doi: 10.1053/j.gastro.2010.05.079. [DOI] [PubMed] [Google Scholar]
- Eurich D., et al. Role of IL28B polymorphism in the development of hepatitis C virus-induced hepatocellular carcinoma, graft fibrosis, and posttransplant antiviral therapy. Transplantation. 2012;93:644–649. doi: 10.1097/TP.0b013e318244f774. [DOI] [PubMed] [Google Scholar]
- Gaudieri S., et al. Genetic variations in IL28B and allergic disease in children. PLoS ONE. 2012;7:e30607. doi: 10.1371/journal.pone.0030607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz P., et al. Progression of liver fibrosis in HIV/HCV genotype 1 co-infected patients is related to the T allele of the rs12979860 polymorphism of the IL28B gene. Eur. J. Med. Res. 2011;16:335–341. doi: 10.1186/2047-783X-16-8-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabris C., et al. IL-28B rs12979860 C/T allele distribution in patients with liver cirrhosis: role in the course of chronic viral hepatitis and the development of HCC. J. Hepatol. 2011;54:716–722. doi: 10.1016/j.jhep.2010.07.019. [DOI] [PubMed] [Google Scholar]
- Falleti E., et al. Role of interleukin 28B rs12979860 C/T polymorphism on the histological outcome of chronic hepatitis C: relationship with gender and viral genotype. J. Clin. Immunol. 2011;31:891–899. doi: 10.1007/s10875-011-9547-1. [DOI] [PubMed] [Google Scholar]
- Tillmann H.L., et al. Beneficial IL28B genotype associated with lower frequency of hepatic steatosis in patients with chronic hepatitis C. J. Hepatol. 2011;55:1195–1200. doi: 10.1016/j.jhep.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S., et al. Genetic variation in IL28B is associated with the development of hepatitis B-related hepatocellular carcinoma. Cancer Immunol. Immunother. 2012;61:1433–1439. doi: 10.1007/s00262-012-1203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


