Abstract
Thin-basement-membrane nephropathy (TBMN) and Alport syndrome (AS) are progressive collagen IV nephropathies caused by mutations in COL4A3/A4/A5 genes. These nephropathies invariably present with microscopic hematuria and frequently progress to proteinuria and CKD or ESRD during long-term follow-up. Nonetheless, the exact molecular mechanisms by which these mutations exert their deleterious effects on the glomerulus remain elusive. We hypothesized that defective trafficking of the COL4A3 chain causes a strong intracellular effect on the cell responsible for COL4A3 expression, the podocyte. To this end, we overexpressed normal and mutant COL4A3 chains (G1334E mutation) in human undifferentiated podocytes and tested their effects in various intracellular pathways using a microarray approach. COL4A3 overexpression in the podocyte caused chain retention in the endoplasmic reticulum (ER) that was associated with activation of unfolded protein response (UPR)–related markers of ER stress. Notably, the overexpression of normal or mutant COL4A3 chains differentially activated the UPR pathway. Similar results were observed in a novel knockin mouse carrying the Col4a3-G1332E mutation, which produced a phenotype consistent with AS, and in biopsy specimens from patients with TBMN carrying a heterozygous COL4A3-G1334E mutation. These results suggest that ER stress arising from defective localization of collagen IV chains in human podocytes contributes to the pathogenesis of TBMN and AS through activation of the UPR, a finding that may pave the way for novel therapeutic interventions for a variety of collagenopathies.
Alport syndrome (AS) and thin-basement-membrane nephropathy (TBMN) are genetic diseases of the glomerular basement membrane (GBM), an important key component of the glomerular filtration barrier. They are caused by pathogenic changes in the genes encoding the α3/α4/α5 chains of type IV collagen, an abundant constituent of the GBM, and can lead to total or partial loss of its network.1,2 Mutations have been identified in all three genes that encode the collagen IV chains in patients with AS and TBMN, while the number of these mutations exceeds 500 to date (Human Gene Mutation Database). AS is invariably associated with CKD and ESRD usually before 30 years of age, often accompanied by sensorineural deafness and/or ocular abnormalities. TBMN is characterized primarily by microscopic hematuria and was classically thought to be a benign disease with excellent prognosis. Still, our group and others have described particular COL4A3/COL4A4 mutations in patients with the dual diagnosis of TBMN and FSGS who developed CKD/ESRD, thus establishing TBMN as a far-from-benign condition, at least in a subset of patients.3,4 Nonetheless, the exact molecular mechanisms by which these mutations exert their pathogenic effect remain unclear.
Physiologically, the collagen IV α3, α4, and α5 chains assemble through recognition of their carboxy-terminal NC1 domains and form helical heterotrimers that undergo several enzymatic post-translational modifications in the cell’s endoplasmic reticulum (ER) before being secreted into the GBM. Once in the extracellular space, they form a multistructural network that has a dual role in providing strength to the membrane and participating in dynamic biological processes by interacting with many other proteins.5,6 Previous studies showed that numerous COL4A3/A4/A5 mutations in the NC1 domains share destructive effects on heterotrimer formation and/or secretion of the heterotrimer from cells.7–9 Therefore, mutant collagen IV heterotrimers could potentially cause deleterious extracellular effects due to their absence from the GBM, as well as putative intracellular effects due to accumulation of misfolded protein inside the producing cells. Importantly, it has been shown that the adult GBM collagen IV chains are solely produced by the podocyte and not by the endothelial or adjacent cells.10 This fact confers the podocyte a central role as the collagen IV producing and secreting cell. Of note, Heidet et al. demonstrated that in the AS podocyte, the COL4A3 protein, even when absent from the GBM, is detected within the cell, a finding that is distinct among patients with AS.11
Herein, we addressed the putative effect of overexpression of both normal and mutant COL4A3 chains on various intracellular pathways of the human podocyte. To target this, we profiled mRNA on human cultured podocytes that overexpressed the wild-type (WT) or mutant COL4A3-G1334E chain for panoramic assessment of gene expression. One of the pathways that emerged as highly deregulated in our analysis is the unfolded protein response pathway (UPR). Inside the cell, an imbalance between protein load and proper protein folding is called ER stress. As a defense mechanism against this imbalance, the cells have at hand a robust mechanism termed the UPR and activated by intracellular retention of unfolded protein.12 UPR allows cells to recover from stress by temporarily halting protein translation and activating signaling pathways that lead to the production of molecular chaperones involved in protein folding. Once activated, this adaptive pathway may protect the cell from future insults.13 However, in cases of prolonged or deregulated ER stress owing to genetic or other factors, this pathway could become maladaptive and cytotoxic and lead to apoptosis.14
We further examined this pathway by testing whether chaperone binding immunoglobulin protein (BiP), a central marker of UPR activation, is upregulated in renal biopsy specimens from patients with a confirmed COL4A3-G1334E mutation. Finally, we examined UPR activation using a relevant knockin mouse model we recently generated in our laboratory carrying the Col4a3-G1332E mutation (the equivalent of the COL4A3-G1334E mutation in patients with AS and TBMN). To our knowledge, this is the first mouse model carrying a missense glycine COL4A3 mutation, which causes AS and TBMN in humans.4,15,16
Results
Overexpression of COL4A3-WT and COL4A3-G1334E Chains in AB8/13 Podocyte Cells
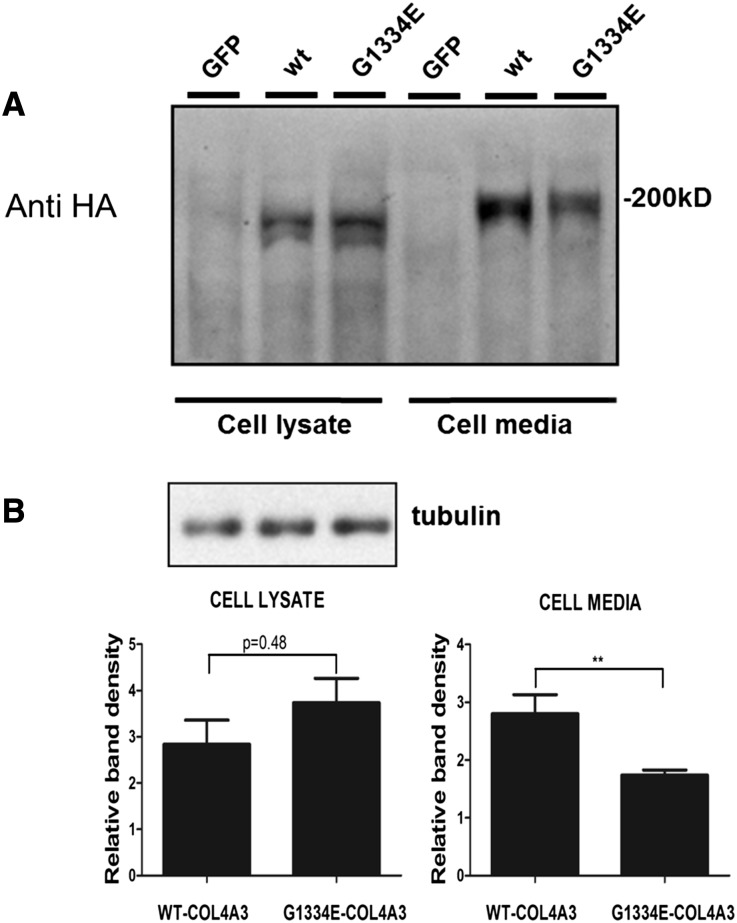
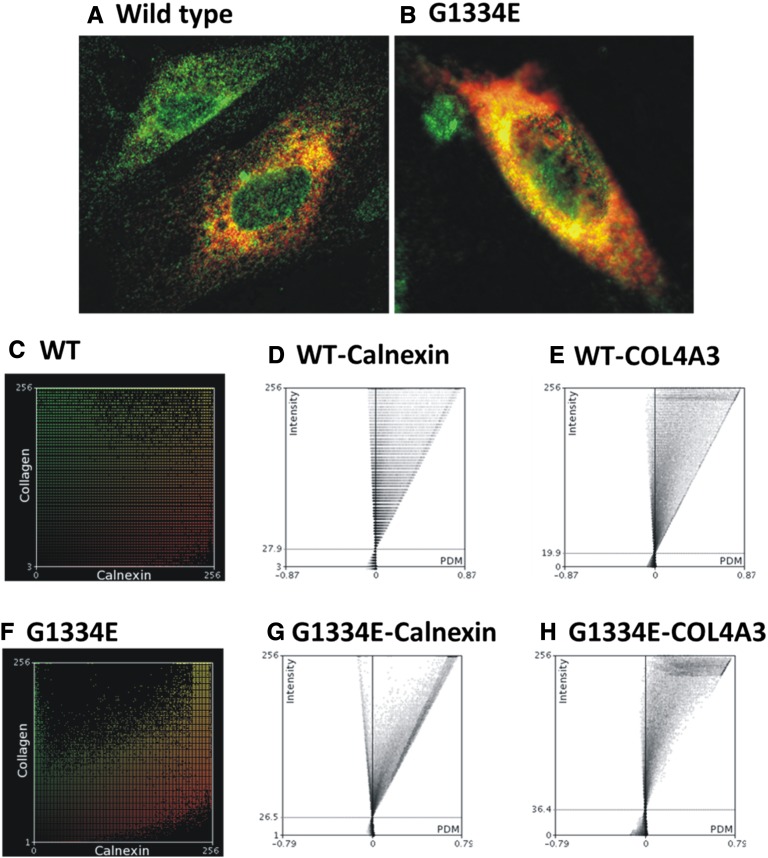
Transient transfection of podocytes with equal amounts of normal COL4A3-WT and mutant COL4A3-G1334E constructs resulted in equal expression of genes as tested in each transfection experiment by quantitative real-time PCR (qPCR) analysis (not shown). Cells expressing the mutant chain showed significantly reduced secretion of the mutant protein, as revealed by Western blotting of the cellular medium 48 hours after transfection (P<0.01) (Figure 1, A and B). The mutant COL4A3-G1334E demonstrated more accumulation into the cellular lysate compared with the WT chain, but this finding did not reach significance at the 48-hour time point (Figure 1B). To determine the cellular location of the expressed chains, we performed immunocytochemistry experiments on the same cells grown on coverslips. Results strongly suggest that overexpressed collagen chains were mainly retained into the cell’s ER and were colocalized with the well-established ER marker, calnexin (Figure 2, A and B). This colocalization was more evident for the mutant COL4A3-G1334E chain. Intensity correlation analysis plots indicated that the colocalization of collagen and calnexin in the WT is more random, as opposed to the improved association seen in the mutant (Figure 2, C–H).
Figure 1.
Mutant COL4A3 chain is secreted less efficiently when expressed in AB8/1 cells. (A) AB8/13 cells were transiently transfected with expression vectors containing WT (COL4A3-WT) collagen chain or mutated COL4A3 (COL4A3-G1334E) cDNAs, which included an HA epitope at C-terminus of the protein. Single chain expression and secretion were measured via Western blot analysis of the cell lysate and the cell medium, respectively, 48 hours after transfection. No HA antigen was detected in AB8/13 cells transfected with a vector expressing the green fluorescent protein (GFP). (B) The extracellular levels of the COL4A3-G1334E chain were significantly lower, reaching 70% of WT COL4A3 levels. A shows a representative Western blot and B shows relative band density that corresponds to HA tag immunoreactivity (i.e., COL4A3) that was normalized to tubulin expression for the cell lysate. Data are represented as means±SEM of n≥3 independent experiments; **P<0.01.
Figure 2.
Mutant collagen IV chain is more prominently localized in the ER when expressed in AB8/13 cells. Equal expression of the WT (A) or the G1334E mutant protein (B) is retained in the cells’ ER, as shown by colocalization (yellow) of the ER marker calnexin (green) with COL4A3 chain (red). Intensity correlation analysis of the immunocytochemistry images C and F show the color scatter plots of the red (collagen) and green (calnexin) channels. (D, E, G, and H) Intensity correlation analysis plots where for each pixel the product of differences from the mean value is plotted with respect to its intensity. As indicated, the co-localization of collagen and calnexin in the WT is more random as opposed to the improved association seen in the mutant. The intensity correlation analysis was performed using the ImageJ software.
Identification of Significantly Deregulated Genes Using Microarrays
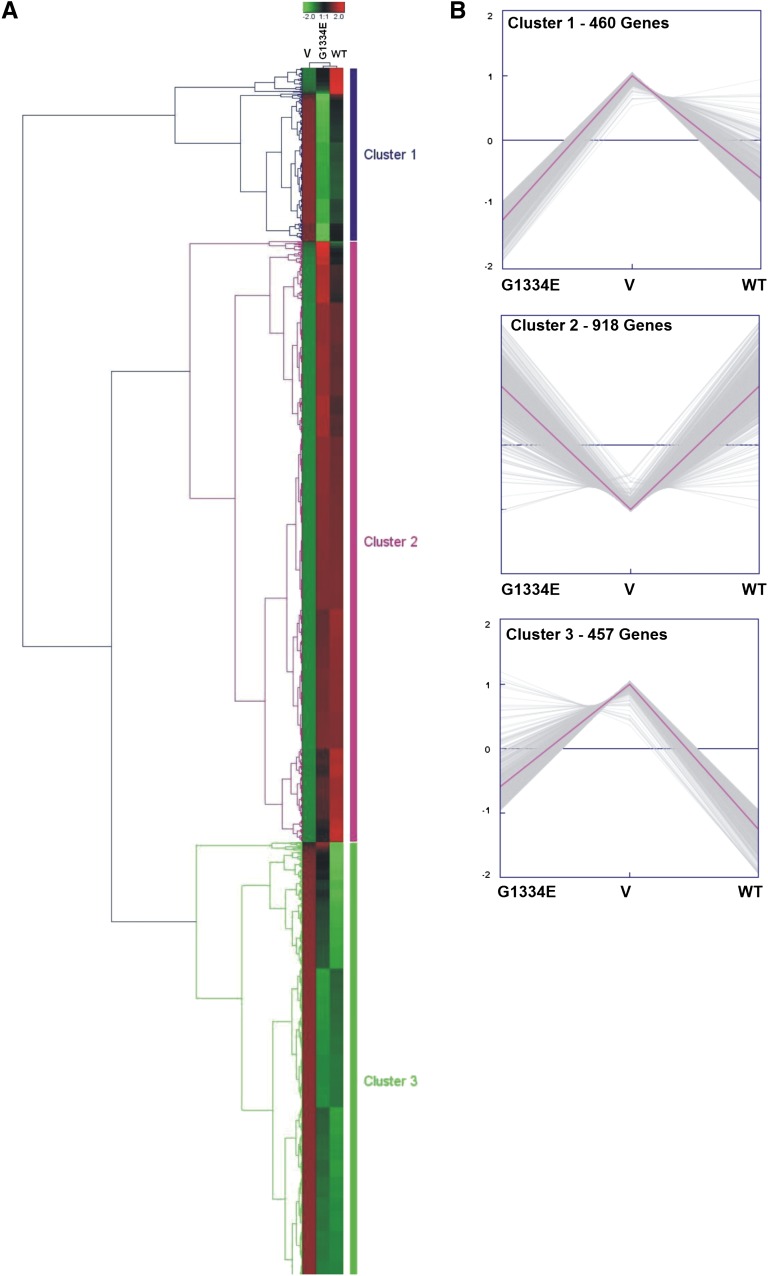
To study in a high-throughput manner which cellular pathways are affected by collagen overexpression in the podocyte, we performed microarray profiling and identified 1835 differentially expressed genes among the empty vector, the COL4A3-WT– and the COL4A3-G1334E–expressing cells. Of these, 1512 genes were significantly deregulated between the WT and vector and 1553 between the G1334E and vector, respectively. The top 40 (20 top upregulated and 20 top downregulated) significantly deregulated genes in the vector versus the COL4A3-WT are depicted in Supplemental Table 1, A and B. A similar pattern was observed for the vector versus the COL4A3-G1334E–expressing cells (Supplemental Table 1, C and D). No genes were significantly deregulated, under the strict criteria we applied, between the cells that overexpressed the WT or the mutant chains. The deregulated genes were further clustered using unsupervised two-way hierarchical clustering with Euclidian distance and were separated into three main groups (Figure 3A). Gene clustering with the K-means algorithm verified the existence of three main gene clusters (Figure 3B).
Figure 3.
Unsupervised hierarchical cluster illustrating differentially expressed genes between vector only (V), WT, and COL4A3-G1334E (G1334E)–expressing cells. (A) Unsupervised hierarchical cluster based on 1835 probe sets with highest variation in three different transfections in podocytes: with empty pcDNA6-HA vector (V) and vectors expressing the WT COL4A3 chain or the mutant COL4A3-G1334E chain (G1334E). Three main gene clusters were noticed. Color saturation is directly proportional to the measured expression ratio magnitude. Euclidian distance was used as the metric. Rows represent individual probe sets. Columns represent the three experimental conditions. Red bars indicate high expression, and green bars indicate low expression. (B) K-means clustering verified the existence of three main gene clusters: cluster 1 (460 genes), cluster 2 (918 genes), and cluster 3 (457 genes).
Enrichment Analysis on Significantly Deregulated Pathways: Deregulation of the UPR Pathway
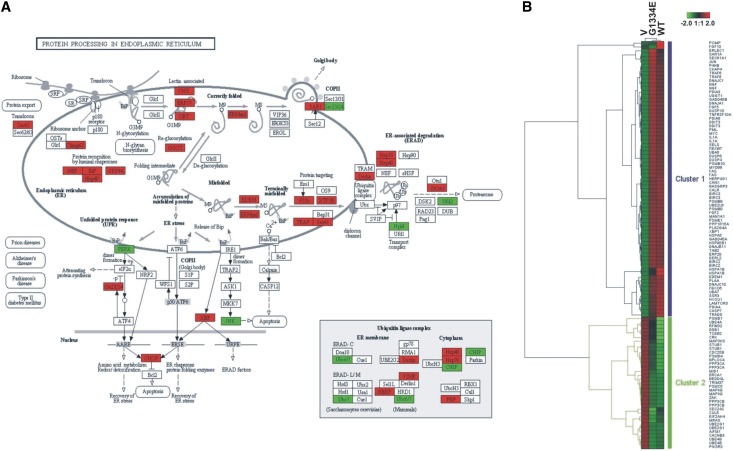
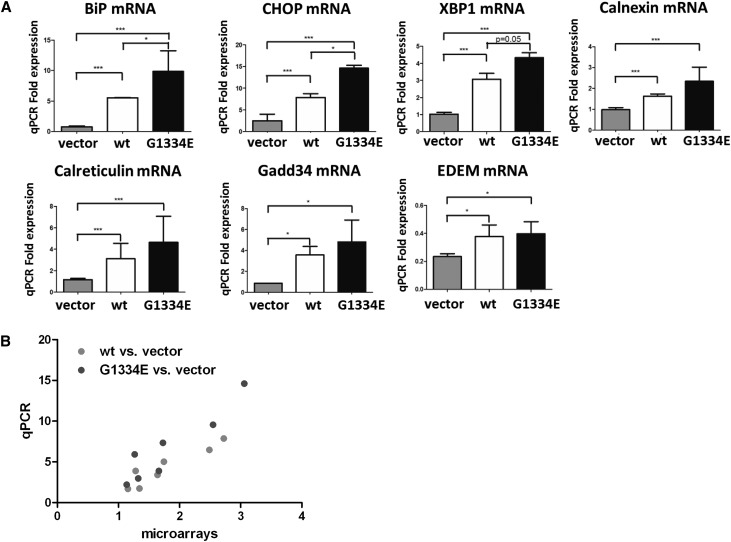
The significantly deregulated genes identified above were separated into two groups: upregulated and downregulated in the vector versus the COL4A3-WT. The pattern was the same for the vector versus COL4A3-G1334E. We found 10 pathways linked with deregulated genes, with P<0.05 after Benjamini correction (Supplemental Table 2). Among them, the top three pathways were the UPR (P<0.001), the p53 (P=0.0023), and the glyoxylate and dicarboxylate metabolism pathway (P=0.0048). Because the UPR pathway was the top linked pathway with the deregulated genes, we used the “protein processing in the endoplasmic reticulum” pathway from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to depict the most deregulated genes that play a significant role in it (P=1.7E-9; Fisher exact test) (Figure 4A). Furthermore, we focused on 118 genes involved directly in the protein processing in the ER and we clustered them, as described above (Figure 4B). Thirty-one and seven genes were upregulated or downregulated, respectively, in the “protein processing in endoplasmic reticulum” pathway (Supplemental Table 3). Among them, the genes selected for further validation were the ones encoding the protein chaperones BiP, calnexin, and calreticulin; the downstream transcription factor X-box binding protein 1 (XBP1); the UPR target genes Gadd34 and EDEM; and the proapoptotic marker C/EBP homologous protein (CHOP).
Figure 4.
(A) Enrichment analysis showing that the protein processing in the endoplasmic reticulum pathway is greatly deregulated in transfected cells. The KEGG pathway depicts the molecules that take part in the protein processing in the ER. The highlighted molecules were significantly deregulated in the cells transfected with the mutant COL4A3-G1334E or the COL4A3-WT chain versus those transfected with an empty vector. Significantly upregulated genes in the pathway are shown in red, whereas significantly downregulated genes are green (see also Supplemental Table 3). (B) Unsupervised hierarchical cluster based on 118 genes involved directly in the protein processing in the endoplasmic reticulum recognizes two main gene clusters. Color saturation is directly proportional to the measured expression ratio magnitude. Euclidian distance was used as the metric. Rows represent individual probe sets. Columns represent the three experimental conditions: empty vector (V), WT, or G1334E. Red bars indicate high expression, and green bars indicate low expression.
Verification of UPR Activation Using qPCR and Western Blotting
Use of qPCR confirmed the results obtained by the microarray experiments showing significantly increased expression of all UPR markers identified by the arrays in cells expressing WT or the COL4A3-G1334E mutant versus the empty vector (Figure 5A, gray versus white and gray versus black bars). Remarkably, the qPCR method revealed statistically significant differences in comparing the variable expression of the BiP, CHOP, and XBP1 genes between the WT versus vector on one hand and between the G1334E versus vector-expressing cells on the other (Figure 5A, white versus black bars). This trend was also evident in the microarray data but did not reach significance. Therefore, it was interesting to observe that expression of the G1334E mutation into human podocytes results in significantly increased expression of the mRNA levels of particular UPR markers compared with equal expression of the WT chain.
Figure 5.
The deregulation of selected genes based on microarray data is verified by qPCR. (A) Graph plots of qPCR data in showing fold increase in mRNAs of various UPR genes in cells expressing vector only (gray bars), WT COL4A3 (white bars), or COL4A3-G1334E mutant chain (black bars) compared with cells expressing vector only. All mRNAs of UPR genes were significantly upregulated in cells expressing WT or mutant COL4A3 compared with cells expressing vector only, thus verifying the microarray data. BiP, CHOP, and XBP1 mRNAs are significantly increased in mutant expressing cells compared with controls (Data are means±SEM, n≥3 independent experiments; *P<0.05; **P<0.01. (B) Positive correlation between microarrays and qPCR experimentation. Good agreement is identified between the microarrays and the qPCR results both for the WT versus vector (R2=0.87; 95% confidence interval [95% CI], 0.62 to 0.99; P=0.0020) and for the G1334E versus vector (R2=0.88; 95% CI, 0.62 to 0.99; P=0.0019) experiments. Results are presented as scatterplot. EDEM, ER degradation enhancer, mannosidase α-like.
The ratios representing gene expression changes are shown as fold-change and are correlated with the microarray data in Figure 5B. The validation experiments showed that the expression patterns of the genes tested were similar to the microarray data. There was very good agreement between the microarrays and the qPCR results both for the WT versus vector (R2=0.87; 95% confidence interval, 0.62 to 0.99; P=0.0020) and for the G1334E versus vector experiments (R2=0.88; 95% confidence interval, 0.62 to 0.99; P=0.0019) (Figure 5B). This supported the reliability of the data obtained from global microarray analysis.
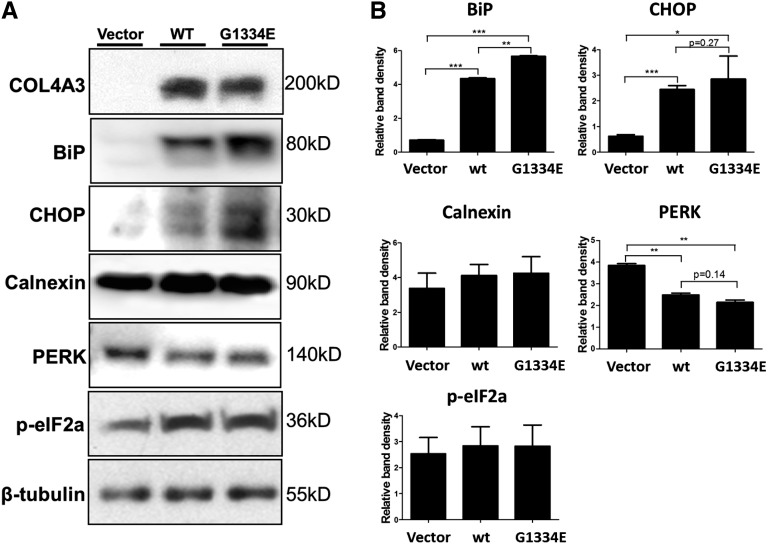
Additionally, we tested protein expression of the UPR markers BiP, CHOP, calnexin, and PERK, as well as phosphorylation of the eIF2a protein, the latter being an important player in protein synthesis shutdown in the UPR cascade.17 Results show a significant increase in BiP protein levels between the vector and the COL4A3-WT as well as between the vector and the G1334E-COL4A3–expressing cells. Interestingly, results show a significant increase in BiP protein levels between the WT- and the mutant-expressing cells as well (Figure 6). Furthermore, a significant increase is evident in CHOP protein levels between the vector and the collagen (WT or mutant)-expressing cells. A significant decrease is observed in the total protein levels of the PERK protein in cells expressing the WT or the mutant COL4A3 chain compared with vector-only–expressing cells, in agreement with the microarray analysis. No significant change was observed in the calnexin and phosphorylated p-eIF2a levels (Figure 6).
Figure 6.
UPR proteins are deregulated in AB8/13 cells transfected with wild type or mutant COL4A3. (A) Undifferentiated cells are transfected with equal amounts of COL4A3-WT (WT), COL4A3-G1334E (G1334E), or the empty pcDNA6/HA vector (Vector; used as a negative control). Protein expression of the UPR markers BiP, CHOP, calnexin, PERK, and p-eIF2a was measured 48 hours after transfection via Western blotting. β-Tubulin expression in the same samples was used as an equal loading control. Shown is a representative blot, with differential levels of the various proteins. (B) Western blotting is quantified via densitometric analysis. Data are means±SEM of three independent experiments. While calnexin and p-eIF2a remain unaltered, BiP and CHOP are upregulated in cells overexpressing the WT or mutant collagen IV chain, and PERK is downregulated. *P<0.05; **P<0.01; ***P<0.001.
Overexpression of WT and Mutant COL4A3 Chains Induce ER Stress, as Shown by XBP1 Splicing
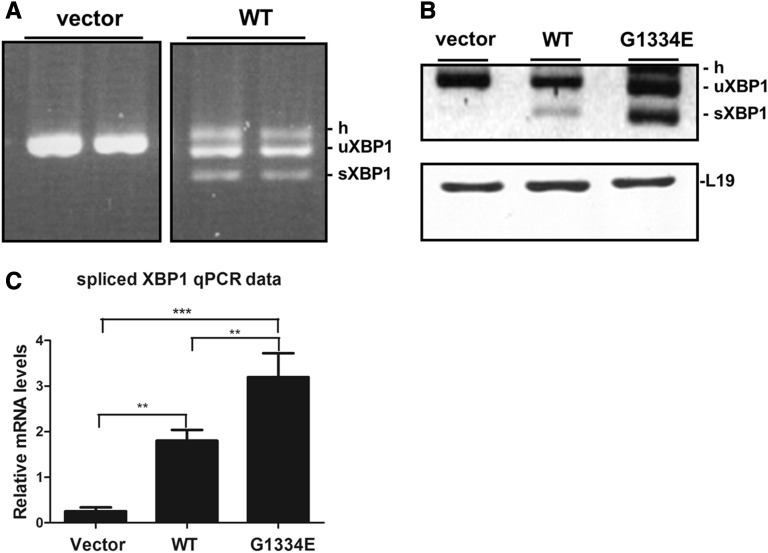
An established assay to examine UPR activation is the widely used XBP1 splicing assay.18 Accumulation of misfolded protein within the cell’s ER causes the dissociation of BiP from IRE1, resulting in its dimerization and subsequent activation. Full-length XBP1 requires the endoribonuclease domain of active IRE1 for processing into active sXBP1; thus, the splicing of XBP1 is a key marker for UPR activation. When podocytes expressed the COL4A3-WT chain, there was obvious splicing of the XBP1 mRNA compared with cells expressing vector-only cDNA (Figure 7A). Additionally, cells overexpressing the GFP protein or the slit diaphragm protein filtrin (NEPH3), cloned in the same vector, were examined in parallel experiments to assess whether the splicing effect is specific for the collagen protein or a general effect. No splicing was observed in GFP or the filtrin-expressing cells (results not shown). Surprisingly, expression of the COL4A3-G1334E mutant into podocytes resulted in significantly more XBP1 splicing compared with cells expressing the WT and under identical experimental conditions (Figure 7B). Densitometric analysis of the spliced versus the unspliced product in gels like the one in Figure 7B revealed statistical significance in the results obtained from WT versus the mutant cells (not shown). The latter was more directly confirmed by qPCR of the spliced XBP1 isoform using primers in the spliced-unspliced interphase of XBP1’s mRNA, as described by Cawley et al.19 Spliced XBP1 was significantly increased in COL4A3-G1334E– versus COL4A3-WT–expressing cells (Figure 7C).
Figure 7.
Single-chain expression of WT or COL4A3-G1334E induces XBP1 splicing in AB8/13 cells. (A) Representative experiment of RT-PCR of the XBP1 mRNA in AB8/13 cells transiently expressing COL4A3-WT collagen chain or the empty pcDNA6/HA vector (vector). PCR products were run on 3% agarose gel. It is apparent that overexpression of WT chain induces XBP1 splicing, as evidenced by the appearance of the smaller spliced form (sXBP1) and the heteroduplex species formed between unspliced and spliced chains (h). The spliced molecules are smaller by 26 bp. (B) Representative experiment of RT-PCR of the XBP1 mRNA in AB8/13 cells transiently expressing COL4A3-WT, mutant COL4A3-G1334E chain, or the empty pcDNA6/HA vector. PCR products were run on 3% agarose gel. It is evident that overexpression of both WT and mutant chains results in XBP1 splicing. L19 was used as an internal PCR control. (C) Real-time PCR for direct quantitation of the spliced XBP1 isoform using primers in the spliced-unspliced interface. Notice the highly statistical difference in the spliced form between vector-only–, WT-, and mutant-expressing cells. Especially important is the differential induction of XBP1 splicing comparing WT with mutant cells. Data are means±SEM of three independent experiments; **P<0.01; ***P<0.001. uXBP1, unspliced XBP1.
Knockdown of Endogenous COL4A3 Chains Using Small Interfering RNA Induces ER Stress in AB8/13 Cells
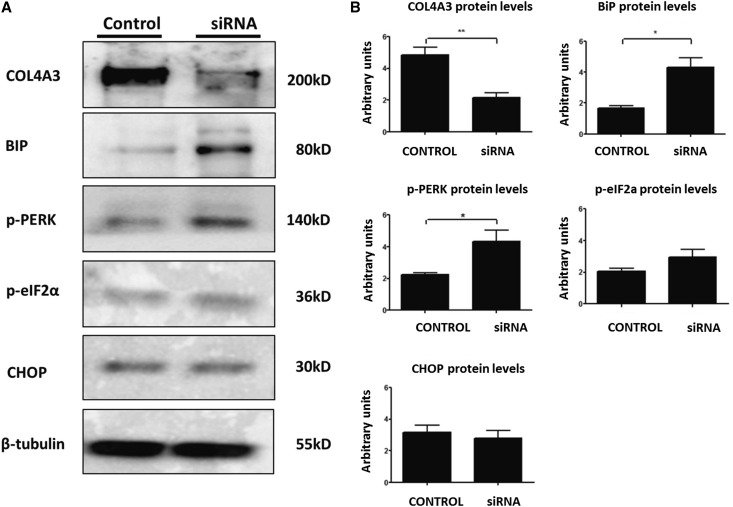
Both undifferentiated and differentiated AB8/13 cells express the COL4A3, COL4A4, and COL4A5 chains, with expression increasing upon differentiation (Supplemental Figure 1). In differentiated podocytes, when the COL4A3 chain is knocked down using small interfering RNA (siRNA), there is activation of the UPR pathway, as shown by upregulation of BiP and increased phosphorylation of protein kinase–like endoplasmic reticulum kinase (PERK), as well as a trend for an increase in p-eIF2a (Figure 8). Interestingly, the proapoptotic marker CHOP remains unaltered.
Figure 8.
Knockdown of endogenous COL4A3 in differentiated podocytes activates the UPR pathway. Differentiated cells are treated with COL4A3 siRNA to knock down endogenous COL4A3 expression. (A) Protein expression of the UPR markers BiP, CHOP, p-PERK, and p-eIF2a was measured 72 hours after transfection via Western blotting. β-Tubulin expression in the same samples was used as equal loading control. Scrambled siRNA was used as a negative control in all experiments. Shown is a representative blot, with differential levels of the various proteins. Note the effective downregulation of the target gene and the upregulation of UPR markers. (B) Western blotting in A was quantified via densitometric analysis. Data are means±SEM of three independent experiments. While CHOP remained unaltered, BiP and p-PERK were upregulated in cells being treated with COL4A3 siRNA. *P<0.05; **P<0.01.
BiP Expression Is Increased in Kidney Biopsy Specimens from COL4A3-G1334E Mutation Carriers
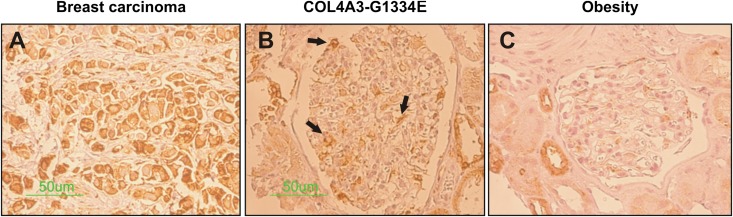
To examine whether the UPR pathway is involved in the pathogenic process among patients with TBMN, we examined BiP protein staining in biopsy samples from two patients who were confirmed heterozygous carriers of the COL4A3-G1334E mutation. BiP expression was increased in the glomeruli of these patients (Figure 9B) compared with expression in biopsy specimens from obese individuals who were used as controls (Figure 9C). BiP expression in tubular epithelial cells was similar in both the COL4A3-G1334E patients and obese individuals. A biopsy sample from a patient with breast carcinoma was stained as a positive control (Figure 9A).
Figure 9.
BiP protein expression is increased in renal biopsies of COL4A3-G1334E heterozygous carriers. Detection of BiP expression in the glomerulus from renal biopsy specimens of two patients diagnosed with TBMN and confirmed heterozygous carriers of the COL4A3-G1334E mutation, using immunohistochemical staining (B). Kidney biopsy specimens from a patient with breast carcinoma (A) and six obese persons (C) were used as positive and negative controls, respectively. In the patients with confirmed COL4A3-G1334E mutation, serial sections show strong perinuclear BiP immunoreactivity in the glomerulus compared with the obese controls. Tubular cells of both the patient with COL4A3-G1334E and the obese persons are stained positive for BiP. All samples are sex and age matched.
Activation of the UPR Pathway in a Mouse Carrying the Col4a3-G1332E Missense Mutation
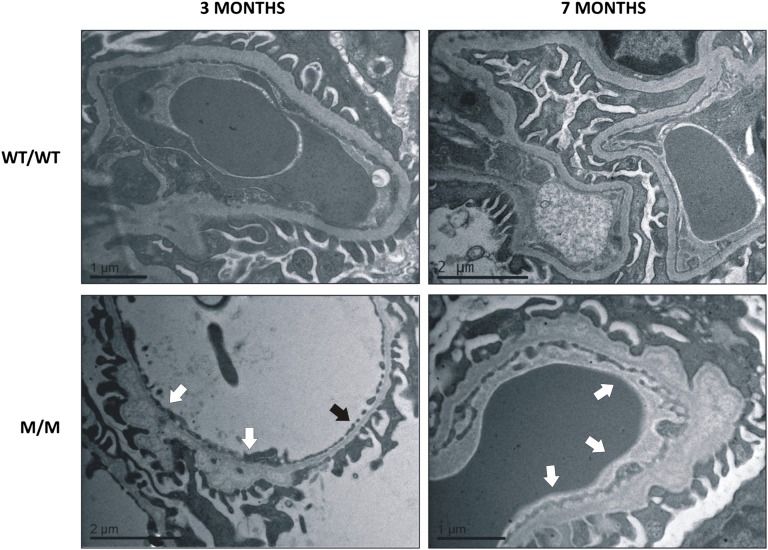
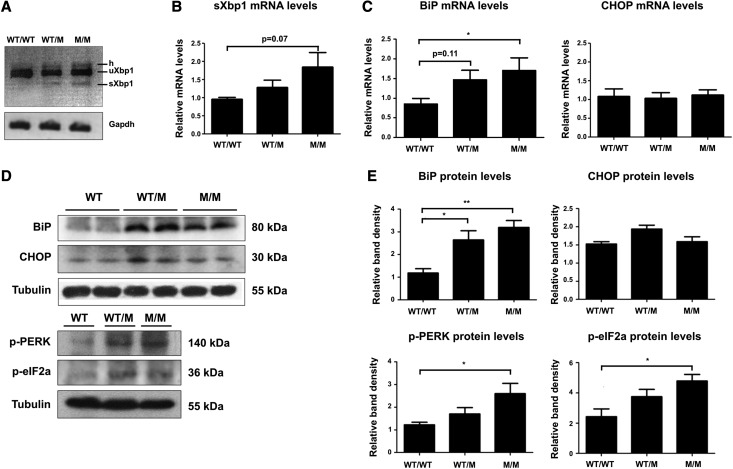
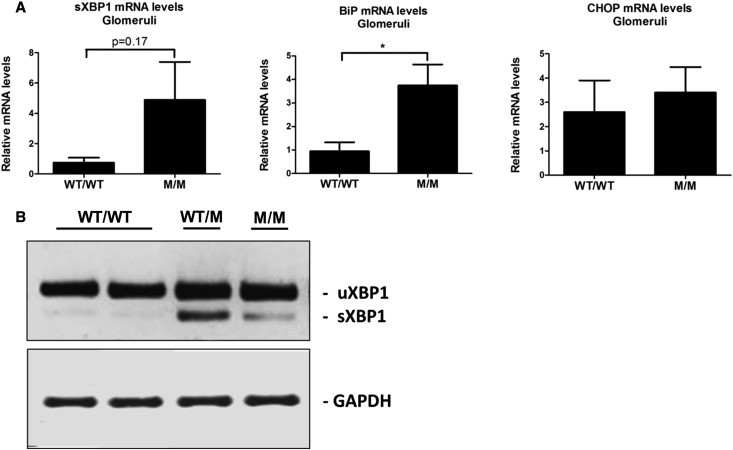
To investigate the mechanisms whereby the COL4A3-G1334E mutation causes pathology in vivo, we generated a relevant mouse model. To achieve this, we used gene-targeting technology to successfully generate a knockin mouse for the Col4a3-G1332E mutation, the equivalent of the COL4A3-G1334E mutation in patients with AS and TBMN (Supplemental Figure 2). These mice express the COL4A3 chain normally and in equal amounts, as evidenced by both qPCR (results not shown) and Western blot experiments (Supplemental Figure 3). Additionally, the ultrastructural analysis of homozygous Col4a3-G1332E mice demonstrated thin GBM with areas of mild (3-month-old mice) or severe (7-month-old mice) thickening, consistent with AS nephritis (Figure 10). In these mice we assessed for evidence of UPR activation in the kidney of mutant heterozygous (WT/M) and homozygous (M/M) 3-month-old animals versus their WT littermates (WT/WT). In contrast to the WT, reverse transcription PCR revealed XBP1 splicing in both heterozygous and homozygous mice (Figure 11A). qPCR analysis using primers on the spliced-unspliced interphase suggested increased XBP1 splicing in homozygous mice versus control (Figure 11B). Also, BiP mRNA was significantly upregulated in the homozygous mouse versus the WT littermates, whereas CHOP mRNA levels remained unaltered (Figure 11C). Additionally, BiP, p-PERK, and p-eIF2α protein levels were significantly upregulated in homozygous mice versus their control littermates (Figure 11, D and E). Finally, we isolated glomeruli from mutant homozygous mice and their WT littermates and examined UPR activation specifically in glomerular cells. The UPR marker BiP was significantly upregulated in mRNA isolated from mutant mice glomeruli (M/M) versus controls (WT/WT) (Figure 12A). We also observed an increased trend in spliced XBP1 mRNA as measured by qPCR, while the XBP1 splicing assay showed the presence of a spliced band in mutant mice (Figure 12B).
Figure 10.
Ultrastractural pathology of the mutant knockin mice is consistent with Alport Syndrome nephritis. WT mice (WT/WT) display normal GBM thickness, 280–300 nm range, while G1332E/G1332E (M/M) homozygous mice demonstrate thin GBMs, 140–160 nm range (black arrow), with areas of mild (3-month-old mice) or severe (7-month-old mice) irregular thickening (white arrows), consistent with AS nephritis.
Figure 11.
The UPR pathway is activated in whole kidney lysates of Col4a3-G1332E mice. (A) XBP1 splicing assay using RNA from whole kidney lysate from WT, Col4a3-G1332E heterozygous (WT/M), and Col4a3-G1332E homozygous (M/M) mice. Splicing is evident by the presence of the lower-molecular-weight band on 2% agarose gel. Mouse glyceraldehyde 3-phosphate dehydrogenase is used as control. Notice the presence of the spliced band in the heterozygous (WT/M) and homozygous (M/M) mice. (B) Real-time PCR for direct quantitation of the spliced XBP1 isoform using primers in the spliced–unspliced interface. Significance is not reached, but there is an obvious trend toward spliced XBP1 increase in homozygous (M/M) mice. Data are means±SEM of three independent experiments. (C) Examination of BiP and CHOP mRNA levels by qPCR. RNAs were extracted from whole kidney tissue of WT, Col4a3-G1332E heterozygous (WT/M), and Col4a3-G1332E homozygous (M/M) mice. Shown is significant upregulation of the BiP but not CHOP mRNA levels. (D) Representative Western blot to demonstrate protein expression level change in the homogenates of whole kidney lysate from 3-month-old WT or Col4a3-G1332E heterozygous (WT/M) or homozygous (M/M) knock-in mice. Equal amounts of protein homogenates were resolved by SDS-PAGE followed by Western blot. The blots in each panel came from the same gel. Notice differential expression of proteins between WT and mutant mice. (E) Quantification of representative blots as in D is shown in graphic form. The expression levels of BiP, CHOP, p-PERK, and p-eIF2α were normalized to β-tubulin levels (*P<0.05; **P<0.01, n≥3); M/M, homozygous for the Col4a3-G1332E mutation; WT/M, heterozygous for the Col4a3-G1332E mutation. Three mice were used for each condition. All UPR genes except CHOP were significantly overexpressed. h, hybrid; sXBP1, spliced XBP1; uXBP1, unspliced XBP1.
Figure 12.
Upregulation of UPR marker mRNA in glomeruli isolated from Col4a3-G1332E knock-in mice. (A) Examination of BiP and CHOP mRNA levels by qPCR. RNAs were extracted from isolated mouse glomeruli of WT and Col4a3-G1332E homozygous mutant (M/M) mice. Shown is significant upregulation of the BiP but not CHOP mRNA levels. Spliced XBP1 mRNA levels show an obvious increase trend. (B) Representative experiment of reverse transcription PCR of the XBP1 mRNA in RNA isolated from WT or mutant mice glomeruli. PCR products were run on 3% agarose gel. It is apparent that there is induction of XBP1 splicing in mutant mice, as evidenced by the appearance of the smaller spliced form (sXBP1). The spliced molecules are smaller by 26 bp. Mouse glyceraldehyde 3-phosphate dehydrogenase is used as control. M/M, homozygous mouse; WT/M, heterozygous mouse; WT/WT, WT mice. *P<0.05; n=3.
Discussion
Mutations in the COL4A3/A4/A5 genes result in heritable kidney diseases, such as AS and TBMN, collectively referred to as collagen IV nephropathies.20 The cellular pathomechanisms of these diseases are complex and clearly include extracellular effects due to reduced or no secretion of collagen IV heterotrimers in the GBM. Additionally, one cannot rule out effects caused by collagen mutations on intracellular pathways, something that to our knowledge has not been previously tested in human podocytes. Because collagen IV α3/α4/α5 chains originate solely from podocytes,10 one can anticipate that the glomerular phenotypes of AS/TBMN are caused by defects that concern specifically or mainly the podocyte. Interestingly, it has been shown via biopsy immunofluorescence that in the patients with X-linked AS, the COL4A3 chain is still expressed and appears to accumulate within the podocyte.11
Collagen IV Secretion and UPR Phenotype in a Podocyte Cellular Model
We first focused on the putative intracellular effects of WT or mutant (G1334E) COL4A3 chain overexpression on podocyte function. In experiments using transiently transfected undifferentiated podocytes, we showed reduced secretion of the single G1334E mutant chain in the cellular medium compared with the WT chain (Figure 1). Immunocytochemistry experiments revealed that the COL4A3 single chains are retained in the cell’s ER, both in the WT- and mutant-overexpressing cells, but this fact is more evident in the latter (Figure 2). This differential behavior of the cells could explain the reduced secretion of the mutant in the cellular medium. Collagen IV is known to be secreted as a trimer;21–23 therefore, we should point out that it is questionable whether the single-chain secretion actually takes place in vivo or is a phenomenon observed only in an artificial in vitro system after overexpression of the single chains. Here, secretion of the COL4A3 chain could be due to association of the overexpressed chain with the endogenous COL4A4 and COL4A5 chains that have been shown to be expressed, even at lower levels, in the undifferentiated podocytes (Supplemental Figure 1). The association of the transfected COL4A3 mutant chains with normal endogenous chains could also account for and explain the formation of misfolded trimers that triggered the UPR activation upon quality control in the ER. Nonetheless, secretion of single collagen chains was previously reported in both human embryonic kidney 293 and Chinese hamster ovary cells overexpressing single collagen IV chains.8,24,25
Furthermore, global mRNA profiling identified and verified the UPR pathway, among others, as a downstream prime aftermath of overexpression of WT or mutant COL4A3 chains in human undifferentiated podocytes. The UPR pathway arose as the most highly deregulated pathway associated with COL4A3-WT or COL4A3-G1334E overexpression (Supplemental Table 2). Importantly, we validated the microarray data for several UPR genes using qPCR (Figure 5). During the qPCR verification we demonstrated significant upregulation of the genes BiP and CHOP between cells expressing the WT or the COL4A3-G1334E single chains in equal amounts (Figure 5A). A similar differential increased expression was evident in the activated form of XBP1, as tested by the splicing assay in cells expressing the WT or mutant collagen chain (Figure 7). These results were also supported by over-representation of the respective proteins, although not always reaching statistical significance (Figure 6). It is not clear from these experiments why the PERK gene appears downregulated in the cells and upregulated in the mice (see below).
We recognize that in our experimental set-up we used singly transfected cells, which expressed either a WT or a mutant collagen IV chain. This resulted in intracellular overexpression (nearly 30-fold compared with the endogenous at the mRNA level and 10-fold at the protein level), which was adequate on its own to activate the UPR signaling cascade. An explanation as to why overexpression of the mutant chain results in higher UPR upregulation compared with the WT is not obvious at first sight. There could be two complementary explanations: One is that this differential UPR upregulation occurs because the cell can apply quality control on the nascent single collagen chain level. Another is that the mutant COL4A3 chain associates poorly with the endogenous COL4A4 and COL4A5 chains, resulting in more misfolded protein being retained in the ER and subsequent higher activation of particular UPR genes due to a dose effect.
Notably, previous studies showed that intracellular accumulation of mutant COL4A1 and COL4A2 chains, causative for hemorrhagic stroke in humans, are retained in the cell’s ER and activate the UPR pathway.26
Importantly, the UPR pathway is triggered both when podocytes overexpress a collagen chain and when the endogenous COL4A3 chain is knocked down by siRNA. Activation is evident in both cases by the increase of the key UPR marker BiP, demonstrating that the cell senses this protein chain imbalance and exerts its quality control in the ER. Of note, the pathway is differentially activated in these two contrasting conditions that result in similar outcomes. COL4A3 overexpression results in activation of the proapoptotic branch of the UPR with upregulation of the proapoptotic marker CHOP. However, in COL4A3 downregulation, CHOP remains unaltered and the PERK UPR branch is activated with increase of eIF2a phosphorylation. These results could reflect the variable activation of the pathway shifting toward the cytoprotective or proapoptotic direction, depending on the nature and/or severity of the stress the cell is facing.
UPR Phenotype on Human Renal Biopsy Specimens
We next tested the activation of the UPR pathway in human biopsy material because we are mindful that extrapolation of the cell culture data obtained herein should be done with caution. This is because in vitro experiments are based on overexpression or downregulation and a relative brief experimental time frame, whereas AS and TBMN are disease processes that take many years to become established. We demonstrated UPR activation, as shown by increased expression of BiP, a sensitive indicator of UPR activation27,28 in patients carrying the COL4A3-G1334E mutation as compared with obese persons (Figure 8). Notably, BiP staining in the glomerulus of these patients is more evident, whereas tubular BiP expression is similar between patients and obese persons. Further experiments should be carried out in order to examine whether it is collagen accumulation per se that causes BiP upregulation in glomeruli of these patients or it is a result of other downstream mutational effects. Still, our data agree with previous studies depicting an increase in ER stress proteins in kidney biopsy specimens from patients with other nephropathies, such as diabetic nephropathy,29 FSGS, and membranous nephropathy.30,31
UPR Phenotype in a Mouse Carrying the Col4A3-G1332E Mutation
Our data demonstrated (for the first time, to our knowledge), UPR activation in a relevant in vivo mouse model, the knockin Col4a3-G1332E mouse. These mice developed severe AS-like GBM pathology as early as at 3 months of age, based on electron microscopy of renal biopsy specimens. The histologic phenotype became more evident at the 7-month time point, while the mice also exhibited hematuria and proteinuria (not shown). UPR activation was evident by significant upregulation of the UPR markers BiP, p-PERK, and p-eIF2α in the mutant mice both at the mRNA and the protein level, as well as nearly significant increase of XBP1 splicing in mutant mice (Figure 11). Contrary to the in vitro overexpression results, in these mice we did not observe CHOP upregulation at the 3-month time point. However, the in vivo data best resembled the COL4A3 knockdown experiments with upregulation of the markers p-PERK and p-eIF2a and no change in the proapoptotic marker CHOP. This finding is interesting because it seems that the stress exerted by the partial knockdown of one chain seems to resemble best the in vivo situation.
Follow-up studies using more and older mice could determine whether collagen IV mutations could evoke the proapoptotic branch of the UPR pathway at a later time point. Still, the significant eIF2a activation observed in these mice could mean translational attenuation, which could still be detrimental in disrupting the highly coordinated events in the podocyte’s specialized secretory capacity, leading to defective GBMs. Tsang et al. showed similar results in which transgenic mice expressing mutant collagen X in chondrocytes activated the UPR pathway without inducing cell death but caused altered chondrocyte differentiation and function causing chondrodysplasia.32 It is interesting that in the in vitro system, overexpression of collagen chains results in significant CHOP upregulation and unaltered p-eIF2a, whereas in the Col4a3-G1332E–carrying mice the opposite is observed and is more similar to the knockdown system. This discrepancy could be due to overexpression of the mutant chains in the cellular system, resulting in severe stress and initiating the proapoptotic branch of the UPR, or it might reflect the various homeostatic mechanisms present in the in vivo situation that shifts the UPR pathway toward the adaptive cytoprotective site. Nonetheless, this appears to be the first time that UPR upregulation is evident in a knock-in Col4a3 mouse. Thus, it would be interesting to examine the current established models of AS33–36 for a similar UPR activation effect. The existing knockout models are null for the Col4a3 or the Col4a5 chain, meaning that the α3α4α5 collagen protomer is compromised. On the basis of the existing knowledge of collagen biochemistry and protomer formation, it is reasonable to assume that knocking down or entirely knocking out one α chain leaves the other two WT chains in excess and vulnerable to degradation, perhaps as a result of the quality control applied by the ER machinery and UPR activation.
Interestingly, it was previously shown that ectopic expression of COL4A3 and COL4A4 in the lens of transgenic mice results in cataract,37 while it is accompanied by UPR activation and ER expansion. This transgene expression had adverse effects on lens fiber cell differentiation and eventually induced cell death in a group of transgenic fiber cells.37
To our knowledge, this study is the first to generate results in a podocyte culture model and in human kidney biopsy specimens, as well as results in knockin mice carrying the Col4a3-G1332E mutation that provide evidence of UPR activation. Consequently, on the basis of these data we can hypothesize that UPR activation might be part of the overall AS/TBMN phenotype at the intracellular level. We are also justified in hypothesizing that different amino acid substitutions may activate the UPR pathway differentially and thereby lead to disparate phenotypes.
In conclusion, we herewith showed experimental data based on cultured human podocytes and human renal biopsy specimens as well as preliminary data from a knockin mouse model for a Col4a3 mutation that collectively implicate the UPR pathway in the disease process. To date the exact pathogenic mechanism resulting from COL4 mutations in patients with AS or TBMN is poorly characterized. ER stress and UPR activation make up a central pathogenic mechanism in various conditions, including several connective tissue disorders associated with the expression of mutant extracellular matrix proteins.14 ER stress in renal pathophysiology is a relatively new area of research.38–40 Therefore, recognizing its contributory role to the deleterious consequences of collagen IV trafficking defects would greatly improve AS/TBMN patient prognosis and would pave ways for development of novel therapeutics, such as the use of molecular chaperones to improve or ameliorate disease symptoms. Finally, the clinical heterogeneity observed in patients with AS/TBMN could be associated with a varying pattern in UPR activation. Because the level of UPR activation is linked directly to the duration and severity of the ER experienced by the cell, this is something that could vary between individuals carrying different mutations, leading to diverse clinical outcomes. Therefore, the pursuit for modifier genes could include the search for UPR related genes.
Concise Methods
Plasmid Vectors
A plasmid vector containing the full-length human COL4Α3 cDNA subcloned into the pCMV6-AC-HA was purchased from Origene. The vector was confirmed for expression of C-terminal fusion of the Collagen α3 with the hemagglutinin (HA) tag. The COL4A3-G1334E point mutation was introduced in the WT COL4A3 by PCR-based site-directed mutagenesis (QuikChange Site-Directed Mutagenesis, Stratagene, La Jolla, CA). Mutagenic primers were as follows: G1334E-A3-For: 5′-CCATTGGACCTCCAGAACCAATTGGGCCAAAAGG-3′; G1334E-A3-Rev: 5′-CCTTTTGGCCCAATTGGTTCTGGAGGTCCAATGG-3′. Correct incorporation of the mutation as well as elimination of any nonspecific changes was confirmed by both restriction digest and DNA sequencing. Mutation G1334E was chosen because of its known pathogenic effect; it is endemic in Cyprus and has been found in a cohort of 169 TBMN patients as a strong founder effect.19
Cell Culture and Transfections
The AB8/13 undifferentiated podocyte cells41 were incubated at 33°C at 5% CO2 and cultured in RPMI medium, supplemented with 10% FBS (Invitrogen, Carlsbad, CA), 1% of 100 U/ml penicillin/streptomycin (Invitrogen), and 1% insulin-transferrin-selenium (Invitrogen). When needed, differentiated podocytes were obtained by culturing the cells at 37°C for 14 days. At 70% confluence, cells were transiently transfected with the vectors containing the collagen cDNAs, WT or mutant, using lipofectamine 2000 according to the manufacturer’s instructions. Forty-eight hours after transfection, cellular lysates and cellular medium were collected for experiments. Importantly, 12 hours before extraction, cells were treated with 50 μg/ml ascorbic acid (Sigma-Aldrich, St. Louis, MO) in serum-free medium. Transfection efficiency was assessed by immunofluorescence using an anti-HA antibody in a transfection control sample; transfection efficiency was typically about 60%. In all experiments, the empty pCMV6-AC-HA plasmid was used as a control to determine transfection toxicity. Collagen construct expression was similar in all transfected cells as assessed by qPCR on the collagen mRNA levels, at each single experiment, using a reverse primer on the engineered C-terminal tag. Primers were as follows: COL4A3-For: CCAGCTGGATCAGATGGAT; HA-FLAG-Rev: AGCGTAATCTGGAACATCGTATGGGTA. COL4A3 mRNA expression after transfection strongly increased (>30-fold) compared with nontransfected AB8/13 cells as determined by qPCR (data not shown).
RNA Preparation and Microarray Experimental Design
To study the genetic regulation of COL4A3 overexpression in the podocyte, 24 hours after transfection, total RNA was extracted from podocytes transfected only with the pcDNA6-HA vector or vectors expressing WT COL4A3 or the mutant COL4A3-G1334E chain. Before cDNA synthesis, the quality of total RNA was checked by a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) using an RNA 6000 LabChip kit (Agilent Technologies). All samples were of good quality, and 100 ng RNA was used to synthesize biotin-labeled target cRNA using the GeneChip 3′ IVT Express Kit (Affymetrix, Santa Clara, CA). Two hundred micrograms of the labeled and fragmented RNA from V1 (vector only), WT (WT COL4A3), and G (COL4A3-G1334E) were hybridized for 16 hours to the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array, which represents approximately 39,000 of the best-characterized human genes. The hybridization step was followed by staining and washes using the GeneChip Hybridization, Wash and Stain Kit from Affymetrix as well as the GeneChip Fluicids Station 450 robot (Affymetrix). The arrays were scanned at 5-μm resolution using the Affymetrix GeneChip Scanner 3000 7G. Quality control was performed using the Affymetrix expression Console. The raw microarray data were background corrected, quantile normalized, and log2 transformed using the SAS JMP7 Genomics, version 4, package (SAS Institute, Cary, NC). Differential mRNA expression was analyzed by one-way ANOVA. A P value of 0.05 with false discovery rate <10% was considered as the cutoff level of significance.
Pathway Analysis, Gene Ontology, and Clustering
Pathway analysis and gene ontology for the significantly deregulated genes was performed using the KEGG (http://www.genome.jp/kegg/pathway.html) and The Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov/) databases. A P value of 0.05 using the Fisher exact test with Bonferroni correction was set as a cutoff. We used two-way hierarchical clustering and K-means clustering with Euclidean distance as a metric to cluster the most deregulated genes. Clustering was performed using Genesis software, version 1.7.6 (Graz University of Technology).
qPCR
Twenty-four hours after transfection, total RNA was extracted from cells using an RNA extraction kit (Macherey-Nagel, Düren, Germany). The integrity of the RNA was assessed via gel electrophoresis, and the concentration was measured spectrophotometrically (Nanodrop Technologies, Montchanin, DE). One microgram of total RNA from all samples was reverse transcribed simultaneously using an oligo-dT (dT23VN) primer and the ProtoScript First Strand cDNA Synthesis Kit (New England Biolabs, United Kingdom). The qPCR amplifications were performed on a LightCycler system (Roche Applied Science) using the LightCycler FastStart DNA Master SYBR Green I kit (Roche, Germany) in a reaction volume of 20 μl. Relative quantification analysis was carried out on LightCycler software, version 4.1. The primer sets for the genes whose differential expression was analyzed by qPCR are shown in Table 1. Differences in starting material were compensated by normalization to the endogenous reference genes GAPDH and L19.
Table 1.
Sets of qPCR oligonucleotides synthesized to measure expression of the mRNA levels of these genes.
| Genes | Primer Sequence (5′ to 3′) |
|---|---|
| Human genes | |
| BiP_F | TGTTCAACCAATTATCAGCAAACTC |
| BiP_R | TTCTGCTGTATCCTCTTCACCAGT |
| CHOP_F | AGAACCAGGAAACGGAAACAGA |
| CHOP_R | TCTCCTTCATGCGCTGCTTT |
| Calnexin_F | TCCGCCTCTCTCTTTACTGC |
| Calnexin_R | GCAACCACTTCCCTTCCAT |
| Calreticulin_F | GCAAATTCGTCCTCAGTTCTGG |
| Calreticulin_R | CCATGCATGTCCTTCTGGTC |
| Gadd34_F | ATGTATGGTGAGCGAGAGGC |
| Gadd34_R | GCAGTGTCCTTATCAGAAGGC |
| EDEM_F | CAAGTGTGGGTACGCCACG |
| EDEM_R | AAAGAAGCTCTCCATCCGGTC |
| sXBP-1_F | CTGAGTCCGCAGCAGGTGCAG |
| sXBP-1_R | CCAGAACATCTCCCCATGGAT |
| GAPDH_F | TTGGTATCGTGGAAGGACTCA |
| GAPDH_R | TGTCATCATATTTGGCAGGTTT |
| L19_F | GCGGAAGGGTACAGCCAAT |
| L19_R | GCAGCCGGCGCAAA |
| Mouse genes | |
| COL4A3_F | CCGAGCCAGTCCATTTATAGAAT |
| COL4A3_R | CAGCGAAGCCAGCCAGAA |
| BiP_F | TCATCGGACGCACTTGGAA |
| BiP_R | CAACCACCTTGAATGGCAAGA |
| CHOP_F | GTCCCTAGCTTGGCTGACAGA |
| CHOP_R | TGGAGAGCGAGGGCTTTG |
| sXbp1_F | CTGAGTCCGCAGCAGGTGCAG |
| sXbp1_R | CCAGAACATCTCCCCATGGAT |
| GAPDH_F | GCATGGCCTTCCGTGTTCCTA |
| GAPDH_R | CCTGCTTCACCACCTTCT |
| Oligos for Xbp1 splicing assay | |
| hXbp1_F | GGAGTTAAGACAGCGCTTGG |
| hXbp1_R | ACTGGGTCCAAGTTGTCCAG |
| mXbp1_F | GAACCAGGAGTTAAGAACACG |
| mXbp1_R | AGGCAACAGTGTCAGAGTCC |
All supplied by M.W.G., Ebersberg, Germany.
Immunoblotting
Forty-eight hours after transfection, cellular lysates or cellular medium were collected for experiments. Cells were lysed in equal volumes of preheated 2× SDS loading buffer (SDS 25 mM, Tris-HCl with pH of 6.8, 20% glycerol, 2% SDS, 2% b-mercaptoethanol, and bromophenol blue) and homogenized via sonication. The transblots were probed with a primary antibody (anti-HA; Santa Cruz Biotechnology), targeted to the fused HA-tag epitope engineered in the carboxyl terminus of the collagen chain. The collagen chains were seen at around 200 kDa. Antibodies used against the ER-stress proteins were anti-BiP, anti-PERK, anticalnexin (Cell Signaling Technology, Danvers, MA) and anti-CHOP, and anti–p-eIF2a (Santa Cruz Biotechnology), followed by peroxidase-labeled secondary antibodies—either goat antimouse or donkey antirabbit (Santa Cruz Biotechnology). Proteins were detected using the Enhanced ChemiLuminescence Plus Blotting Detection system (Amersham Biosciences, Buckinghamshire, United Kingdom) and were visualized by autoradiography on photographic film (Kodak X-OMAT; Eastman Kodak, Rochester, NY).
All transblots were reprobed with anti–β-tubulin antibody (Santa Cruz Biotechnology) to prove equal amounts of protein were loaded on the membrane. Band density was defined the ImageJ Software (http://imagej.nih.gov/ij).
Semiquantitative Analysis and XBP1 Splicing
AB8/13 cells were transfected with COL4A3-WT or COL4A3-G1334E constructs in equal amounts as confirmed by qPCR and Western blots (not shown). Twenty-four hours post-transfection total RNA was isolated and cDNA was synthesized as described above. For every RNA sample, a control reaction without reverse transcription was performed to exclude genomic DNA contamination. cDNA was further used as a template for PCR with XBP or L19 RNA primers (primers shown in Table 1). PCR products were then analyzed on a 3% agarose gel.
Experiments with siRNA
A total of 400,000 podocytes were seeded on a 6-cm culture dish. Differentiated podocytes were transfected with siRNA COL4A3 (Santa Cruz Biotechnology) using the lipofectant RNAiMax (Invitrogen) Briefly, 7.5 μl of the lipofectant was mixed with 80 pmol of siRNA in 1 ml of Opti MEM. The mix was added to 4 ml of complete media without antibiotics to the dishes. Cells were incubated overnight at 37°C and 5% CO2. The following day, the same concentrations of lipofectant and siRNA in 1 ml of Opti-MEM Were again added to the dishes. Cells were harvested 72 hours after the second transfection.
Immunocytochemistry
Forty-eight hours after transfection, cells on coverslips were fixed with 4% paraformaldehyde in PBS for 10 minutes and permeabilized for an additional 10 minutes with 0.5% Triton X-100 in PBS. All cells were quenched with 50 mM ammonium chloride in PBS for 10 minutes, blocked with 2% BSA, 2% FCS, 0.2% fish skin gelatin in PBS (blocking mix) for 30 minutes, and incubated with primary and then secondary antibodies in PBS, containing 5% blocking mix, either overnight at 4°C or for 1 hour at room temperature. Cells were incubated with Hoechst solution (DAKO, Denmark) at a 1:1000 dilution for 1 minute for nucleus staining and were subsequently mounted with fluorescence mounting medium (DAKO).
Fluorescence Microscopy
Immunofluorescent preparations were analyzed on a Zeiss Axiovert 200M inverted fluorescence microscope equipped with Zeiss Axiovision 4.2 software. Digital images were recorded and composed using Adobe Photoshop 5.0 and Illustrator 10 for Macintosh. Quantification and co-localization were determined using ImageJ software.
Immunohistochemistry
For human kidney staining experiments, kidney sections were cut and mounted on slides coated with suitable tissue adhesive. Then sections were deparaffinized using xylene and rehydrated through washes in graded alcohols. Endogenous peroxidase was neutralized using 0.5% vol/vol hydrogen peroxide/methanol for 10 minutes. Slides were washed with water. Retrieval was carried out using 0.01 M citrate retrieval solution (pH, 6.0). Sections were subsequently washed with TBS and blocked for 10 minutes using diluted normal serum (Novocastra Protein Block; Leica Microsystems). Sections were incubated with primary antibody anti-BiP (Cell Signaling Technology) diluted 1:200 in antibody diluent solution (DAKO) overnight at 4°C. Slides were then washed and incubated with DAKO REAL EnVision, HRP Rabbit/Mouse (ENV). Subsequently, slides were incubated with a suitable peroxidase substrate, washed thoroughly in running tap water, and counterstained with hematoxylin, before being dehydrated and mounted. Patient tissue was used after bioethics approval and informed consent. The patients who carry mutation G1334E belong to families that have been described before.4
Generation of Col4a3-G1332E Knock-In Mice
All animal experiments were carried out in compliance with Cyprus law for protection of animals and were approved by the local authorities. Details on targeting construct generation, production, and genotyping of mice are described in the supplemental materials (Supplemental Figure 2). Comprehensive phenotypic details of this line will be documented elsewhere (M.P. and C.D., unpublished data). Animals were hosted in the Cyprus Institute of Neurology and Genetics animal house under specific pathogen-free conditions.
Electron Microscopy of Mouse Kidney Sections
For electron microscopy studies, left kidney sections were processed as usual and examined under a transmission electron microscope (JEM2100HR; JEOL, Inc., Tokyo, Japan) equipped with an ES500W Erlangshen CCD camera (Gatan GmbH, Munchen, Germany). GBM thickness was measured in open capillary loops using Digital-Micrograph software (Gatan).
Glomeruli Isolation
Glomeruli were isolated following the protocol by Takemoto et al.42 Briefly, mice were anesthetized by an intraperitoneal injection of Avertin (2,2,2-tribromoethyl and tertiary amyl alcohol; 17 μl/g mouse) and perfused with 8×107 Dynabeads through the heart. The kidneys were removed, minced, and digested in collagenase. The digested tissue was gently pressed through a 100-μm cell strainer and the cell suspension was then centrifuged at 200×g for 5 minutes. The supernatant was discarded and the cell pellet was resuspended in HBSS. Glomeruli containing Dynabeads were gathered by a magnetic particle concentrator. Glomeruli total RNA was isolated using a commercially available kit (Macherey-Nagel), according to the manufacturer’s instructions.
Statistical Analyses
All statistical analyses with the exception of the microarrays were performed using GraphPad Prism, version 5, statistical software. Data are expressed as mean±SEM; the Mann-Whitney U test was performed for pairwise comparisons. For comparisons involving more than two groups, nonparametric analysis of relative contrasts was performed with Tukey post hoc analysis. No prior transformation of the data was performed. For microarray analysis we used the SAS JMP7 Genomics, version 4, package (SAS Institute). Differential mRNA expression was analyzed by ANOVA. A P value < 0.05 with a false discovery rate < 10% was considered as the cutoff level of significance. For pathway analysis and gene ontology enrichment, a P value of 0.05 using the Fisher exact test with Bonferonni correction was set as a cutoff.
Disclosures
None.
Supplementary Material
Acknowledgments
The authors thank Dr. S. Malas and his student, E. Panayiotou, for assistance with the animal procedures. Part of this work was presented orally during the 8th Meeting of the COST Action BM0702, Kidney and Urine Proteomics, March 29–April 1, 2012, Sounion, Athens, Greece, during the 25th European Renal Cell Study Group meeting in Oxford, United Kingdom, March 21–24, 2013, and as a poster during the 50th Congress of the European Renal Association-European Dialysis and Transplant Association, May 18–21, 2013.
The work was supported mainly by a grant from the Cyprus Research Promotion Foundation, HEALTH/ΒΙΟS/0308(ΒΕ)/17 and NEW INFRASTRUCTURE/STRATEGIC/0308/24 (cofunding by the EU Structural Funds) to C.D.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2012121217/-/DCSupplemental.
References
- 1.Haas M: Alport syndrome and thin glomerular basement membrane nephropathy: a practical approach to diagnosis. Arch Pathol Lab Med 133: 224–232., 2009 [DOI] [PubMed] [Google Scholar]
- 2.Thorner PS: Alport syndrome and thin basement membrane nephropathy. Nephron Clin Pract 106: c82–c88, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Temme J, Peters F, Lange K, Pirson Y, Heidet L, Torra R, Grunfeld JP, Weber M, Licht C, Müller GA, Gross O: Incidence of renal failure and nephroprotection by RAAS inhibition in heterozygous carriers of X-chromosomal and autosomal recessive Alport mutations. Kidney Int 81: 779–783, 2012 [DOI] [PubMed] [Google Scholar]
- 4.Voskarides K, Damianou L, Neocleous V, Zouvani I, Christodoulidou S, Hadjiconstantinou V, Ioannou K, Athanasiou Y, Patsias C, Alexopoulos E, Pierides A, Kyriacou K, Deltas C: COL4A3/COL4A4 mutations producing focal segmental glomerulosclerosis and renal failure in thin basement membrane nephropathy. J Am Soc Nephrol 18: 3004–3016, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Parkin JD, San Antonio JD, Pedchenko V, Hudson B, Jensen ST, Savige J: Mapping structural landmarks, ligand binding sites, and missense mutations to the collagen IV heterotrimers predicts major functional domains, novel interactions, and variation in phenotypes in inherited diseases affecting basement membranes. Hum Mutat 32: 127–143, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miner JH: The glomerular basement membrane. Exp Cell Res 318: 973–978, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi T, Kakihara T, Uchiyama M: Mutational analysis of type IV collagen alpha5 chain, with respect to heterotrimer formation. Biochem Biophys Res Commun 366: 60–65, 2008 [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi T, Uchiyama M: Characterization of assembly of recombinant type IV collagen alpha3, alpha4, and alpha5 chains in transfected cell strains. Kidney Int 64: 1986–1996, 2003 [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi T, Uchiyama M: Mutant-type alpha5(IV) collagen in a mild form of Alport syndrome has residual ability to form a heterotrimer. Pediatr Nephrol 25: 1169–1172, 2010 [DOI] [PubMed] [Google Scholar]
- 10.Abrahamson DR, Hudson BG, Stroganova L, Borza DB, St John PL: Cellular origins of type IV collagen networks in developing glomeruli. J Am Soc Nephrol 20: 1471–1479, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heidet L, Cai Y, Guicharnaud L, Antignac C, Gubler MC: Glomerular expression of type IV collagen chains in normal and X-linked Alport syndrome kidneys. Am J Pathol 156: 1901–1910, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boot-Handford RP, Briggs MD: The unfolded protein response and its relevance to connective tissue diseases. Cell Tissue Res 339: 197–211, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshida H: ER stress and diseases. FEBS J 274: 630–658, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Rajpar MH, McDermott B, Kung L, Eardley R, Knowles L, Heeran M, Thornton DJ, Wilson R, Bateman JF, Poulsom R, Arvan P, Kadler KE, Briggs MD, Boot-Handford RP: Targeted induction of endoplasmic reticulum stress induces cartilage pathology. PLoS Genet 5: e1000691, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierides A, Voskarides K, Athanasiou Y, Ioannou K, Damianou L, Arsali M, Zavros M, Pierides M, Vargemezis V, Patsias C, Zouvani I, Elia A, Kyriacou K, Deltas C: Clinico-pathological correlations in 127 patients in 11 large pedigrees, segregating one of three heterozygous mutations in the COL4A3/ COL4A4 genes associated with familial haematuria and significant late progression to proteinuria and chronic kidney disease from focal segmental glomerulosclerosis. Nephrol Dial Transplant 24: 2721–2729, 2009 [DOI] [PubMed]
- 16.Voskarides K, Pierides A, Deltas C: COL4A3/COL4A4 mutations link familial hematuria and focal segmental glomerulosclerosis. Glomerular epithelium destruction via basement membrane thinning? Connect Tissue Res 49: 283–288, 2008 [DOI] [PubMed] [Google Scholar]
- 17.Zoll WL, Horton LE, Komar AA, Hensold JO, Merrick WC: Characterization of mammalian eIF2A and identification of the yeast homolog. J Biol Chem 277: 37079–37087, 2002 [DOI] [PubMed] [Google Scholar]
- 18.Oslowski CM, Urano F: Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol 490: 71–92, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cawley K, Deegan S, Samali A, Gupta S: Assays for detecting the unfolded protein response. Methods Enzymol 490: 31–51, 2011 [DOI] [PubMed] [Google Scholar]
- 20.Deltas C, Pierides A, Voskarides K: The role of molecular genetics in diagnosing familial hematuria(s). Pediatr Nephrol 27: 1221–1231, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayne R, Wiedemann H, Irwin MH, Sanderson RD, Fitch JM, Linsenmayer TF, Kühn K: Monoclonal antibodies against chicken type IV and V collagens: Electron microscopic mapping of the epitopes after rotary shadowing. J Cell Biol 98: 1637–1644, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trüeb B, Gröbli B, Spiess M, Odermatt BF, Winterhalter KH: Basement membrane (type IV) collagen is a heteropolymer. J Biol Chem 257: 5239–5245, 1982 [PubMed] [Google Scholar]
- 23.Fatemi SH: The role of secretory granules in the transport of basement membrane components: Radioautographic studies of rat parietal yolk sac employing 3H-proline as a precursor of type IV collagen. Connect Tissue Res 16: 1–14, 1987 [DOI] [PubMed] [Google Scholar]
- 24.Leinonen A, Netzer KO, Boutaud A, Gunwar S, Hudson BG: Goodpasture antigen: Expression of the full-length alpha3(IV) chain of collagen IV and localization of epitopes exclusively to the noncollagenous domain. Kidney Int 55: 926–935, 1999 [DOI] [PubMed] [Google Scholar]
- 25.Fukuda K, Hori H, Utani A, Burbelo PD, Yamada Y: Formation of recombinant triple-helical [alpha 1(IV)]2 alpha 2(IV) collagen molecules in CHO cells. Biochem Biophys Res Commun 231: 178–182, 1997 [DOI] [PubMed] [Google Scholar]
- 26.Jeanne M, Labelle-Dumais C, Jorgensen J, Kauffman WB, Mancini GM, Favor J, Valant V, Greenberg SM, Rosand J, Gould DB: COL4A2 mutations impair COL4A1 and COL4A2 secretion and cause hemorrhagic stroke. Am J Hum Genet 90: 91–101, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee AS: The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods 35: 373–381, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D: Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2: 326–332, 2000 [DOI] [PubMed] [Google Scholar]
- 29.Lindenmeyer MT, Rastaldi MP, Ikehata M, Neusser MA, Kretzler M, Cohen CD, Schlöndorff D: Proteinuria and hyperglycemia induce endoplasmic reticulum stress. J Am Soc Nephrol 19: 2225–2236, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bek MF, Bayer M, Müller B, Greiber S, Lang D, Schwab A, August C, Springer E, Rohrbach R, Huber TB, Benzing T, Pavenstädt H: Expression and function of C/EBP homology protein (GADD153) in podocytes. Am J Pathol 168: 20–32, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markan S, Kohli HS, Joshi K, Minz RW, Sud K, Ahuja M, Anand S, Khullar M: Up regulation of the GRP-78 and GADD-153 and down regulation of Bcl-2 proteins in primary glomerular diseases: A possible involvement of the ER stress pathway in glomerulonephritis. Mol Cell Biochem 324: 131–138, 2009 [DOI] [PubMed] [Google Scholar]
- 32.Tsang KY, Chan D, Cheslett D, Chan WC, So CL, Melhado IG, Chan TW, Kwan KM, Hunziker EB, Yamada Y, Bateman JF, Cheung KM, Cheah KS: Surviving endoplasmic reticulum stress is coupled to altered chondrocyte differentiation and function. PLoS Biol 5: e44, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rheault MN, Kren SM, Thielen BK, Mesa HA, Crosson JT, Thomas W, Sado Y, Kashtan CE, Segal Y: Mouse model of X-linked Alport syndrome. J Am Soc Nephrol 15: 1466–1474, 2004 [DOI] [PubMed] [Google Scholar]
- 34.Lu W, Phillips CL, Killen PD, Hlaing T, Harrison WR, Elder FF, Miner JH, Overbeek PA, Meisler MH: Insertional mutation of the collagen genes Col4a3 and Col4a4 in a mouse model of Alport syndrome. Genomics 61: 113–124, 1999 [DOI] [PubMed] [Google Scholar]
- 35.Cosgrove D, Meehan DT, Grunkemeyer JA, Kornak JM, Sayers R, Hunter WJ, Samuelson GC: Collagen COL4A3 knockout: A mouse model for autosomal Alport syndrome. Genes Dev 10: 2981–2992, 1996 [DOI] [PubMed] [Google Scholar]
- 36.Beirowski B, Weber M, Gross O: Chronic renal failure and shortened lifespan in COL4A3+/- mice: An animal model for thin basement membrane nephropathy. J Am Soc Nephrol 17: 1986–1994, 2006 [DOI] [PubMed] [Google Scholar]
- 37.Firtina Z, Danysh BP, Bai X, Gould DB, Kobayashi T, Duncan MK: Abnormal expression of collagen IV in lens activates unfolded protein response resulting in cataract. J Biol Chem 284: 35872–35884, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitamura M: Endoplasmic reticulum stress in the kidney. Clin Exp Nephrol 12: 317–325, 2008 [DOI] [PubMed] [Google Scholar]
- 39.Inagi R: Endoplasmic reticulum stress in the kidney as a novel mediator of kidney injury. Nephron, Exp Nephrol 112: e1–e9, 2009 [DOI] [PubMed] [Google Scholar]
- 40.Dickhout JG, Krepinsky JC: Endoplasmic reticulum stress and renal disease. Antioxid Redox Signal 11: 2341–2352, 2009 [DOI] [PubMed] [Google Scholar]
- 41.Saleem MA, O’Hare MJ, Reiser J, Coward RJ, Inward CD, Farren T, Xing CY, Ni L, Mathieson PW, Mundel P: A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol 13: 630–638, 2002 [DOI] [PubMed] [Google Scholar]
- 42.Takemoto M, Asker N, Gerhardt H, Lundkvist A, Johansson BR, Saito Y, Betsholtz C: A new method for large scale isolation of kidney glomeruli from mice. Am J Pathol 161: 799–805, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.