Abstract
The accumulation of autophagosomes in postischemic kidneys may be renoprotective, but whether this accumulation results from the induction of autophagy or from obstruction within the autophagic process is unknown. Utilizing the differential pH sensitivities of red fluorescent protein (RFP; pKa 4.5) and enhanced green fluorescent protein (EGFP; pKa 5.9), we generated CAG-RFP-EGFP-LC3 mice to distinguish early autophagic vacuoles from autolysosomes. In vitro and in vivo studies confirmed that in response to nutrient deprivation, renal epithelial cells in CAG-RFP-EGFP-LC3 mice produce autophagic vacuoles expressing RFP and EGFP puncta. EGFP fluorescence diminished substantially in the acidic environment of the autolysosomes, whereas bright RFP signals remained. Under normal conditions, nephrons expressed few EGFP and RFP puncta, but ischemia-reperfusion injury (IRI) led to dynamic changes in the proximal tubules, with increased numbers of RFP and EGFP puncta that peaked at 1 day after IRI. The number of EGFP puncta returned to control levels at 3 days after IRI, whereas the high levels of RFP puncta persisted, indicating autophagy initiation at day 1 and autophagosome clearance during renal recovery at day 3. Notably, proliferation decreased in cells containing RFP puncta, suggesting that autophagic cells are less likely to divide for tubular repair. Furthermore, 87% of proximal tubular cells with activated mechanistic target of rapamycin (mTOR), which prevents autophagy, contained no RFP puncta. Conversely, inhibition of mTOR complex 1 induced RFP and EGFP expression and decreased cell proliferation. In summary, our results highlight the dynamic regulation of autophagy in postischemic kidneys and suggest a role of mTOR in autophagy resolution during renal repair.
Autophagy is a lysosomal degradation pathway that is essential for cellular stress adaptation and normal homeostasis.1–3 It involves a series of membrane rearrangements to form autophagosomes, which are double-membraned vacuoles that contain cytoplasmic contents and organelles. Fusion of autophagosomes with lysosomes results in the formation of autolysosomes in which captured materials are degraded for removal of damaged organelles and recycling of nutrients within the cells. Autophagy has been recognized as a protective mechanism after renal ischemia-reperfusion injury (IRI).4–8 Increased levels of autophagy have been reported in the postischemic kidneys by accumulation of autophagosomes under electron microscopy or increased Atg proteins by immunoblot analysis.4,7,8 However, electron microscopy can only provide static information and does not distinguish whether the accumulation of autophagosomes is due to the induction of autophagy or a blockage in downstream processes of autophagy. Immunoblot analysis of Atg proteins detects autophagy in a heterogeneous and asynchronous cell population and does not reflect autophagy in individual compartments of the kidney. New tools are needed to study autophagic flux, which will provide a more accurate assessment of autophagic activity in individual cells of the organ.
LC3 protein is the mammalian homology of Atg8 in yeasts and is essential for autophagy to occur. LC3 is cleaved to LC3-I immediately after its synthesis. LC3-I is an ubiquitin-like protein that can be conjugated to phosphatidylethanolamine and possibly phosphatidylserine. The lipidated forms are referred to LC3-II, which is present in all autophagic vacuoles. LC3-II is the most widely used Atg protein to quantify autophagic levels by immunoblot analysis.3 Methods for immunostaining of LC3-II have been recently developed.9 However, it is not always possible to detect low levels of endogenous LC3-II. Transfection of cells with plasmids expressing enhanced green fluorescent protein (EGFP) fused with LC3 enables the visualization of EGFP puncta in autophagic vacuoles. Transgenic mice expressing EGFP-LC3 fusion protein under the cytomegalovirus immediate-early enhancer and chicken β-actin (CAG) promoter have been generated.10 The mice are useful to study autophagy induction in many organs, including the kidney, based on the appearance of EGFP puncta identified with fluorescence microscopy.5 However, EGFP fluorescence is quenched in the acidic environment of autolysosomes, and thus loses its ability to track the autophagic process.
We generated a new strain of autophagy reporter mice that express a tandem red fluorescent protein (RFP)-EGFP-LC3 fusion protein ubiquitously under the CAG promoter to advance our understanding of the dynamics of autophagy. Specifically, the inclusion of a relatively acid-insensitive RFP (pKa 4.5) and acid-sensitive EGFP (pKa 5.9) is expected to result in the quenching of EGFP but persistence of RFP signals in the low pH environment of autolysosomes (pH=4–5), whereas both EGFP and RFP fluorescence will be maintained before autophagosomes fuse with lysosomes.11,12 The mice were used as novel tools to study the dynamics of renal epithelial autophagy under stress conditions such as starvation and ischemic injury.
Results
Renal Epithelial Cells of CAG-RFP-EGFP-LC3 Mice Respond to Starvation with Distinct Fluorescence Puncta
Transgenic mice expressing RFP-EGFP-LC3 fusion protein were morphologically indistinguishable from their wild-type littermates. The mouse line that expressed the fusion protein at a similar level to the endogenous LC3 protein was selected for our studies.
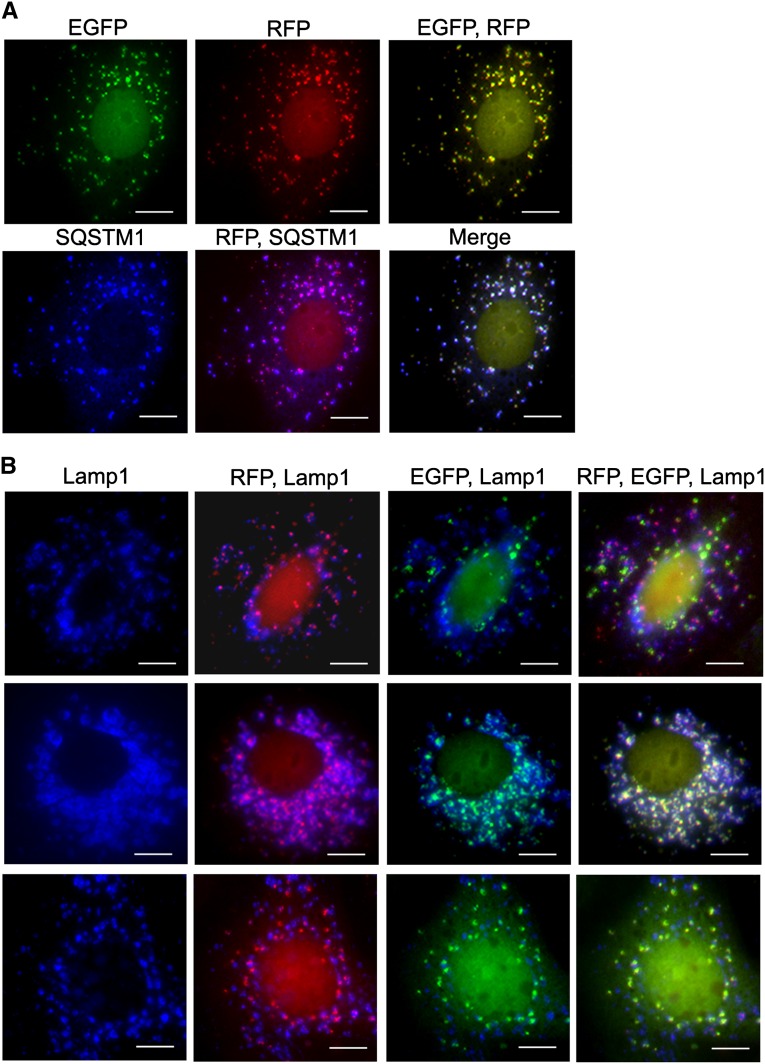
First, we isolated cells from the kidneys for primary cultures and detected few EGFP and RFP puncta in cells grown in nutrient-abundant medium. However, incubation of the cells with Earle’s basic salt solution (EBSS) that contained no glucose or amino acid for 2 hours resulted in a time-dependent appearance of bright EGFP and RFP puncta (Figure 1A). Immunostaining showed the presence of tight junction protein ZO-1 and the epithelial cadherin (E-cadherin), indicating that cells that responded to autophagic stimulation were tubular epithelial cells (Figure 1, B and C). Because EGFP could form weak dimers and self-aggregation of EGFP has been reported,13 we tested whether this could occur in renal epithelial cells. Immunocytochemistry with an antibody to sequestosome 1 (SQSTM1/p62), which was a polyubiquitin-binding protein that directly interacted with LC3 on the isolation membrane and incorporated into the autophogosome,3 showed that >94% of puncta that emitted EGFP and RFP also contained with SQSTM1/p62, suggesting that fluorescence puncta represented autophagic vacuoles rather than random aggregates (Figure 2A).
Figure 1.
EGFP and RFP puncta are easily detected in response to starvation. (A) Primary cultures isolated from kidneys of CAG-RFP-EGFP-LC3 mice show increasing numbers of EGFP or RFP puncta in response to autophagy stimulation with glucose and amino acid deprivation when incubated in EBSS medium for 0–120 minutes. (B and C) Cells respond to autophagic stimulation with the appearance of RFP puncta and have renal epithelial cell characteristics with the expression of tight junction protein ZO-1 (blue) (B) and E-cadherin (blue) (C). Control cells are cultured in nutrient-abundant condition. Scale bar, 10 µm.
Figure 2.
EGFP and RFP puncta represent autophagic vacuoles and display differential pH sensitivity. (A) EGFP and RFP fluorescent puncta are colocalized with SQSTM1, thus representing autophagic vacuoles. (B) RFP puncta that do not emit EGFP are costained with Lamp1, indicating their localization to the autolysosomes. Approximately 14% of RFP puncta in the autolysosomes coemit weak EGFP signals (top), indicating that the majority of EGFP loses its fluorescence in the autolysosomes. Treatment of the cells with chloroquine to increase lysosomal pH results in an increased number of EGFP puncta in the autolysosomes, supporting the pH sensitivity of the EGFP (middle). Immunostaining of FITC-labeled anti-EGFP reveals that all RFP-containing protein emits FITC signals, indicating the presence of EGFP in the fusion protein (bottom). Scale bar, 10 µm.
All EGFP signals were colocalized with RFP as the fusion protein was expressed. However, 35% of the RFP puncta did not emit EGFP. Because EGFP fluorescence is sensitive to acidic pH whereas RFP signals are not, we costained the cells with lysosomal marker Lamp1 to distinguish fluorescence puncta in the autolysosomes from those in the phagophores and autophagosomes. The results showed that RFP puncta that did not emit EGFP were costained with Lamp1, suggesting that they were localized to the autolysosomes. Among all RFP puncta that were localized to the autolysosomes, 14% coemitted weak EGFP signals, indicating that the majority of the fusion protein lost its native EGFP fluorescence in the autolysosomes (Figure 2B, top panel).
Next, we treated the cells with chloroquine to increase the lysosomal pH and detected a 5.8-fold increase in the number of EGFP puncta in the autolysosome, further supporting the pH sensitivity of the EGFP (Figure 2B, middle panel). Immunostaining with FITC-labeled anti-EGFP antibody revealed that all RFP-containing protein emitted FITC signals (Figure 2B, bottom panel), thus confirming the presence of EGFP in the fusion protein. Because reappearance of green fluorescent protein (GFP) fluorescence has been reported after fixation with 4% paraformaldehyde (PFA) (pH 7.4) in some cell types,3 we compared the percentage of yellow puncta (both EGFP- and RFP-positive) over total RFP puncta and found no statistically significant difference in nonfixed cells (68%) and cells promptly fixed with PFA (65%), indicating that renal epithelial cells of CAG-RFP-EGFP-LC3 mice can be used to study autophagy in real time or fixed for further immunostaining.
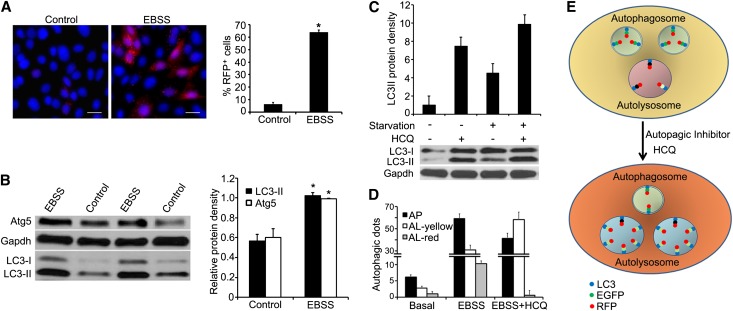
Next, we counted the number of cells that contained RFP puncta and determined whether RFP puncta corresponded to Atg5 and endogenous LC3-II protein levels. Under nutrient-abundant conditions, 6.4%±1.5% cells contained ≥3 RFP puncta. However, 63.9%±2.0% cells contained ≥3 RFP puncta when deprived of nutrients via incubation with EBSS for 2 hours (Figure 3A). Immunoblot analyses of Atg5 and endogenous LC3-II showed 1.6- and 2.0-fold increases in the protein levels, respectively (Figure 3B). These results indicate that the abundance of fluorescence puncta parallels the increase in endogenous Atg proteins that are commonly used to measure the degree of autophagy.3
Figure 3.
RFP puncta corresponds to Atg5 and endogenous LC3-II protein levels. (A) Glucose and amino acid deprivation of primary cultures of renal epithelia cells by incubating cells with EBSS for 2 hours results in a 10-fold increase in cells containing ≥3 RFP puncta. (B) Immunoblot analysis shows parallel increases in Atg5 and endogenous LC3-II protein levels in cells with nutrient deprivation. Cells grown under nutrient-abundant conditions are used as controls. Values are the mean±SEM; n=3. *P<0.05. (C) Immunoblot analysis shows that chloroquine inhibits LC3-II degradation under basal and starvation conditions. (D) The addition of chloroquine to cells incubated with EBSS results in a decrease in the number of autophagosomes (yellow dots without Lamp1). However, within the autolysosomes that are Lamp1 positive, there is an increase in yellow dots (yellow) and a reciprocal decrease in red dots (red), reflecting the recovery of EGFP in the presence of chloroquine. (E) The illustration demonstrates the number and localization of fluorescence dots in cells treated with chloroquine. AL, autolysosome; AP, autophagosome; HCQ, chloroquine. Scale bar, 10 µm.
In CAG-RFP-EGFP-LC3 mice, RFP-EGFP-LC3 is driven by an ubiquitous CAG promoter. To confirm that the number and localization of fluorescent puncta reflect autophagic activity and flux in response to autophagic stimulation and inhibition, we treated cells with chloroquine to attenuate autolysosomal degradation. As expected, cells responded to chloroquine treatment with an accumulation of endogenous LC3-II protein under basal and starvation conditions (Figure 3C). We then examined fluorescent puncta that were colocalized with and without Lamp1. Incubation of cells with EBSS led to an increased number of both autophagosomes (yellow dots without Lamp 1) and autolysosomes (red and yellow dots with Lamp 1) compared with those under basal conditions. The addition of chloroquine to cells incubated with EBSS resulted in suppression of autophagic activity with reduced autophagosome numbers. However, within the autolysosomes, there was an increase in yellow dots and a reciprocal decrease in red dots (Figure 3D), reflecting the recovery of EGFP fluorescence. These results further support the usefulness of using fluorescence dots to study epithelial autophagy.
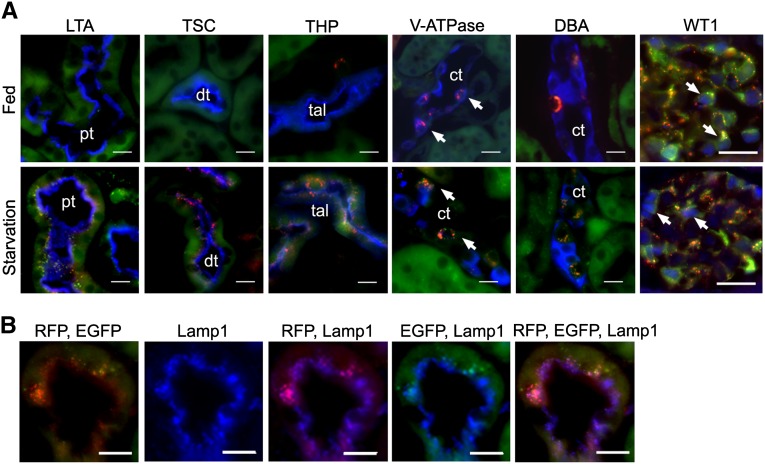
We then subjected the mice to starvation, which has been shown to be a powerful stimulus for autophagy.14 Under ad libitum feeding conditions, epithelial cells of nephron segments exhibited occasional EGFP and few RFP puncta. However, in the collecting ducts, abundant RFP puncta were detected in intercalated cells and some RFP puncta also coemitted EGFP fluorescence. The high level of autophagy in intercalated cells could reflect a high turnover rate in membrane vesicles in this cell type.15 In the glomerulus, podocytes also exhibited high levels of autophagy, which were consistent with the finding in CAG-EGFP mice.10 After 32 hours of starvation, the number of cells that contained RFP and EGFP puncta increased in all nephron segments, the collecting ducts, and the podocytes (Figure 4A). Similarly to epithelial cells in culture, the majority of RFP puncta that coemitted EGFP were not stained for Lamp1 and represented phagophores and autophagosomes, whereas the RFP stand-alone puncta were costained for Lamp1 and thus represented autolysosomes (Figure 4B).
Figure 4.
Mice respond to starvation with visible EGFP and RFP puncta in kidney tubules and glomeruli. (A) Under normal conditions, epithelial cells of nephron segments exhibit occasional EGFP and few RFP puncta. Intercalated cells (arrows, stained with anti–V-ATPase, blue), but not principal cells (stained with DBA, blue), of the collecting ducts show abundant RFP and some EGFP puncta (top panel). Starvation for 32 hours results in increased numbers of EGFP and RFP puncta in epithelial cells of all nephron segments and collecting ducts (bottom panel). Markers of nephron segments (blue) are LTA, anti-TSC, and anti-THP to identify the proximal tubules, distal tubules, and thick ascending limbs, respectively. Glomerular podocytes labeled with WT1 (blue nuclear staining, arrows) show abundant RFP and EGFP signals under basal and starvation conditions. (B) Immunostaining of Lamp1 shows that the majority of RFP puncta that coemit EGFP are not stained for Lamp1, thus representing phagophores and autophagosomes. In comparison, RFP stand-alone puncta are costained for Lamp1, thus representing autolysosomes. ct, collecting duct; DBA, Dolichos biflorus agglutinin; dt, distal tubule; LTA, Lotus tetragonolobus agglutinin; pt, proximal tubule; tal, thick ascending limb; THP, Tamm-Horsfall protein; TSC, thiazide-sensitive NaCl cotransporter. Scale bar, 20 µm.
Taken together, these results indicate that renal epithelial cells of CAG-RFP-EGFP-LC3 mice can respond to autophagic stimuli with easily detectable EGFP and RFP punta that represent autophagic vacuoles. Although EGFP fluorescence can be lost in the acidic environment of the autolysosomes, RFP puncta persisted in all stages of autophagic vacuoles. Additional use of lysosomal marker Lamp1 can distinguish autolysosomes from phagophores and autophagosomes, which allows us to study the dynamics of autophagy.
CAG-RFP-EGFP-LC3 Mice Reveal the Dynamics of Autophagy in Postischemic Kidneys
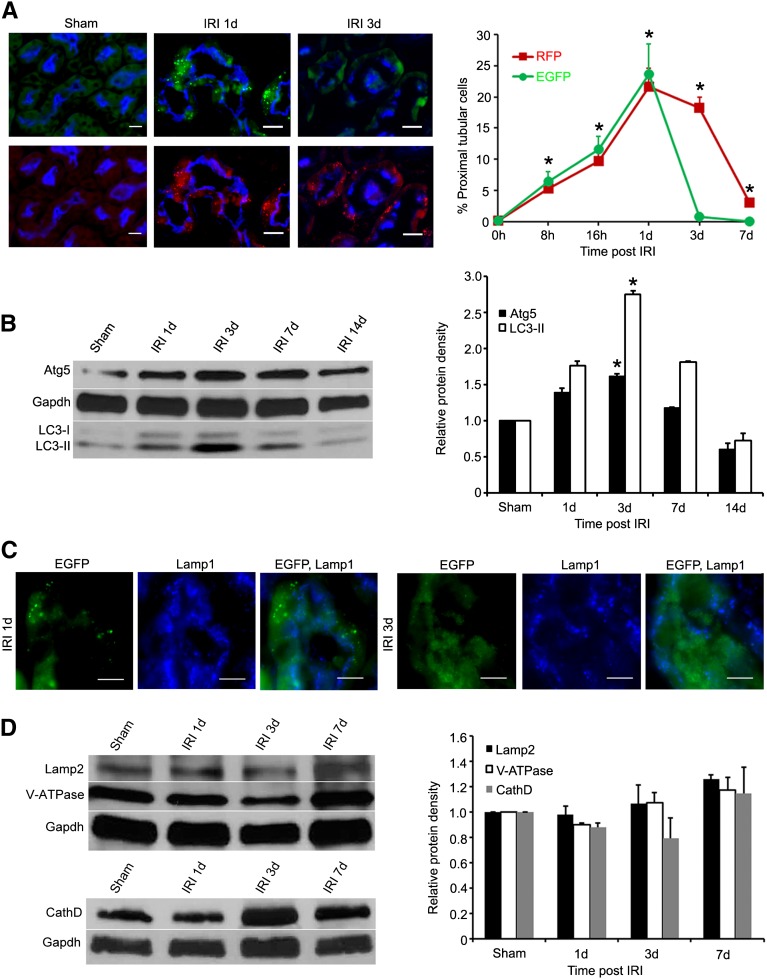
To understand the dynamics and the flux of autophagy in the kidneys with IRI, we used the CAG-RFP-EGFP-LC3 mice to examine RFP puncta in the proximal tubules where the most significant stress and injury occurred.16 In sham-operated mice, 0.24%±0.16% of proximal tubular cells contained RFP puncta. There was no increase in RFP puncta immediately after the vascular clamp release or 4 hours after IRI. At 8 hours after IRI, 5.4%±1.2% of proximal tubular cells showed bright RFP puncta. The number of cells with RFP puncta increased to 9.7%±1.8%, 21.6%±3.0%, and 18%±1.8% at 16 hours and 1 and 3 days after IRI, respectively, and returned to near control level at 7 days (Figure 5A). The percentage of RFP-containing proximal tubular cells mirrors the autophagy protein levels of LC3-II and Atg5 collected from the cortex and the outer strip of the outer medulla (Figure 5B), thus supporting the usefulness of RFP puncta in evaluating tubular epithelial autophagy after IRI.
Figure 5.
Dynamic changes of autophagy in proximal tubules after IRI. (A) The percentage of proximal tubules containing EGFP or RFP puncta peaks at 1 day and returns to close to baseline at 7 days after IRI. Proximal tubules are stained with LTA (blue). (B) Immunoblot analysis shows increased protein levels of Atg5 and endogenous LC3-II in the kidneys after IRI. Values are the mean±SEM; n=3. *P<0.05. (C) The vast majority of EGFP puncta are not stained for Lamp1 (blue) at 1 and 3 days after IRI, indicating that EGFP puncta represent early autophagic vacuoles. (D) The levels of lysosomal proteins Lamp2, V-ATPase, and cathepsin D do not change significantly in postischemic kidneys. LTA, Lotus tetragonolobus agglutinin. Values are the mean±SEM; n=3. Scale bar, 20 µm.
Bright EGFP puncta were also detectable in proximal tubular cells after IRI. To identify the intracellular localization of EGFP puncta, immunostaining of Lamp1 was performed in kidneys harvested at 1 and 3 days after IRI. The vast majority of EGFP was not stained for Lamp1, suggesting its localization to early autophagic vacuoles (i.e., phagophores and autophagosomes) (Figure 5C). Quantitative analysis of autolysosomes, shown by the presence of both Lamp1 and RFP, demonstrated that 7% at 1 day and 8% at 3 days after IRI also emitted weak EGFP, indicating that most EGFP was quenched in the lysosomal environment and there was no difference in the degree of quenching at 1 or 3 days after IRI. These results suggested that, similarly to epithelial cells in culture, differential pH sensitivity of EGFP and RFP could be used to study autophagic processes in postischemic kidneys.
We therefore compared the percentage of proximal tubular cells that contained EGFP or RFP puncta. At 1 day after IRI, 23.3% of proximal tubular cells contained EGFP puncta, which is not significantly different from those that contained RFP puncta (21.6%). However, at 3 days, proximal tubular cells that contained EGFP puncta reduced to 0.9% compared with the 18% that still contained RFP puncta. Both EGFP and RFP fluorescence returned to near control level at 7 days (Figure 5A, line graph). Because most EGFP puncta represent phagophores and autophagososomes, these results suggested that at 1 day after IRI, there was a higher rate of formation of early autophagic vacuoles and/or reduced fusion of autophagosomes with lysosomes. As kidneys were recovering from injury at 3 days,16 formation of early autophagic vacuoles decreased and autophagosomes proceeded to fuse with lysomes for clearance. Our results also indicated that the presence of RFP puncta at 3 days could be used to track cells with recent autophagy.
To test whether more cells containing EGFP puncta at 1 day was due to reduced degradation as a consequence of low levels of lysosomal proteins or enzyme activities, we performed immunoblot analyses of a lysosomal marker Lamp2 and lysosomal enzymes V-ATPase and cathepsin D. The results showed no significant differences in protein levels in the kidneys 1–7 days after IRI (Figure 5D). To examine whether there was a reduced V-ATPase activity that could cause higher pH and less quenching of EGFP fluorescence at different time points after IRI, we focused on cells with autophagic vacuoles and quantified the ratio of EGFP and RFP puncta that were localized to the autolysosomes. The results showed that the ratio of EGFP over RFP puncta was not significantly different at 1 and 3 days (0.20 versus 0.15), suggesting a similar lysosomal V-ATPase activity to maintain lysosomal pH. We therefore concluded that there was no significant change in lysosomal degradation in the postischemic kidneys.
Resolution of Autophagy Is Accompanied by Activation of Mechanistic Target of Rapamycin
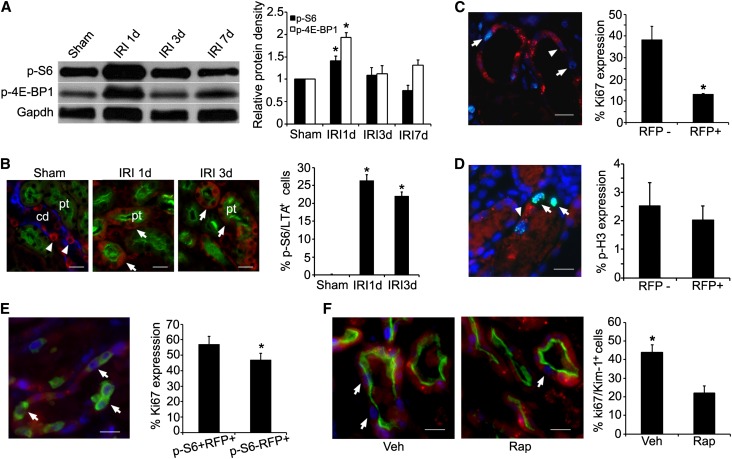
To understand the molecular events leading to the resolution of epithelial autophagy, we examined the pathway of mechanistic target of rapamycin (mTOR) (previously referred to as mammalian target of rapamycin), which was shown to inhibit autophagy in many types of cells.17,18 Immunoblot analyses showed an increase in the phosphorylation of mTOR substrates S6 kinase and 4E-BP1 proteins in the postischemic kidneys, indicating mTOR activation (Figure 6A). Phospho-S6 kinase (p-S6) that was normally expressed in the distal nephron and collecting ducts but not detectable in intact proximal tubules could now be easily detected by immunostaining after IRI. At 3 days, 24.0%±0.3% of proximal tubular cells expressed p-S6 (Figure 6B). Interestingly, 81.7%±3.8% of the p-S6–expressing cells contained no RFP puncta (not shown). Inhibition of mTOR complex 1 (mTORC1) activity by treating the mice with rapamycin resulted in a significantly increased number of proximal tubular cells to contain RFP puncta at 3 days (36.0%±4.1% in rapamycin-treated mice versus 24.0%±4.1% in vehicle-treated mice, n=3, P<0.05). Quantification of early autophagic vacuoles represented by the presence of EGFP puncta showed a 5-fold increase in EGFP-containing proximal tubular cells, indicating increased initiation of autophagy when mTORC1 was inhibited.
Figure 6.
mTOR activation accompanied with autophagy resolution and renal repair. (A) Immunoblot analysis shows increased levels of p-S6 kinase and p-4E-BP1 protein expression in postischemic kidneys. (B) Immunostaining shows that p-S6 protein (red, arrows) is easily detected in proximal tubular epithelial cells (stained green with LTA) after IRI. In sham-operated mice, proximal tubules (green with LTA staining) show no p-S6 expression. However, p-S6 (red, arrowheads) is expressed in collecting ducts (blue with DBA staining). (C) Ki67 expression (blue) is reduced in autophagic cells that contain RFP puncta (arrowhead) compared with nonautophagic cells that contain no RFP puncta (arrows). (D) No significant differences in phospho-histone H3 expression (cyan) are detected in autophagic cells that contain RFP puncta (arrowhead) compared with nonautophagic cells (arrows) that contain no RFP puncta. Nuclei are stained with 4′,6-diamidino-2-phenylindole. (E) Activation of p-S6 kinase (green) in autophagic cells results in increased cell proliferation with the expression of Ki67 (blue, arrows). (F) Rapamycin injection results in a 2-fold decrease in Ki67 expression (blue, arrows) in injured proximal tubular cells that express kidney injury molecule 1 (green). Values are the mean±SEM; n=3. *P<0.05. cd, collecting duct; DBA, Dolichos biflorus agglutinin; LTA, Lotus tetragonolobus agglutinin; pt, proximal tubule; rap, rapamycin; veh, vehicle. Scale bar, 20 µm.
To examine whether resolution of autophagy is accompanied by tubular repair, we quantified Ki67 expression that identified proliferating cells in the late G1 to M phases19 in the outer stripe of the outer medulla where the S3 segments of proximal tubules were localized (Figure 6C). At 3 days after IRI, Ki67 expression was significantly higher in cells that did not contain RFP puncta (38.3%±3.5%) compared with cells that contained RFP puncta (13.0%±0.3%). Because cell cycle arrest has been reported in autophagic cells,20 we examined autophagic and nonautophagic cells for the expression of phospho-histone H3 (Figure 6D), which identified cells in the late G2 and M phase.21 The results showed no difference in the expression of phospho-histone H3 (2.03% versus 2.54%, P>0.05), suggesting that the lower rate of proliferation in autophagic cells was likely due to reduced cell cycle entry and/or slow progression in the early phase of the cell cycle. Next, we examined whether activation of mTOR in autophagic cells could stimulate proliferation by comparing Ki67 expression in RFP-containing cells in which p-S6 was activated or inactivated. The results showed a significantly higher proliferation rate when p-S6 was activated (Figure 6E). Finally, treating mice with rapamycin that prevented the phosphorylation of S6 kinase resulted in a 2-fold decrease in Ki67 expression in injured proximal tubules that expressed kidney injury molecule 1 (Figure 6F), further supporting the role of mTOR activation for autophagy resolution and tubular repair.
Discussion
In renal tubular cells of CAG-RFP-EGFP-LC3 mice, the EGFP and RFP puncta outside the autolysosome were distinctly bright, which offered the advantage of easy distinction from autofluorescence that might be present in the kidney. We perfused the kidneys with PFA at the time the mice were euthanized to fix the protein immediately and to eliminate any possible autophagy induction during sample preparations. Although using 4% PFA solution (pH=7.4) could potentially raise the lysosomal pH, resulting in less quenching of EGFP fluorescence in the autolysosomes in some cell types, our results suggested that this was not the case in tubular epithelial cells. When the same method was used for all samples, we found that a significantly lower number of proximal tubular cells contained EGFP puncta at 3 days (0.9%) compared with 1 day (23%) after IRI, arguing against the nonspecific pH effect of PFA. Because levels of lysosomal proteins including V-ATPase were similar at 1–7 days after IRI, a lower number of EGFP puncta at 1 day was unlikely due to more rapid degradation. Indeed, 14% of EGFP in the autolysosome (pH = 4–5) continued to emit weak green fluorescence. Because the EGFP signal is dependent on the enzymatic degradation and the speed at which the acidic pH of the lysosome quenches the fluorescence,22 the presence of EGFP signals in autolysosomes limits the utility of using the yellow dots to identify early autophagic vacuoles (before the fusion with lysosomes) and red dots to identify autolysosomes. Additional immunostaining of Lamp1 with the RFP fluorescence could help to distinguish autolysosomes (containing Lamp1) from early autophagic vacuoles (containing no Lamp1). A more acid-sensitive fluorescent protein such as mWasabi, which has a pKa at 6.5 and is predicted to have a more complete loss of fluorescence in the autolysosomes,12 may be used to generate future mouse tools to track autophagic flux more directly.
RFP protein, but not EGFP protein, is reported to be accumulated in lysosomes of mouse embryonic fibroblasts transfected with plasmids expressing recombinant GFP or GFP-like proteins.23 This raises the question of whether RFP-positive dots may be lysosomes, but not autolysosomes, in mice expressing RFP-EGFP-LC3 fusion protein, although the inclusion of LC3 in the fusion protein could possibly facilitate the incorporation of the protein into autophagic vacuoles rather than lysosomes. We examined cells stained with FITC-labeled anti-EGFP antibody and showed that all RFP puncta emitted FITC signals. Because EGFP protein is susceptible to degradation when directly exposed to low pH and lysosomal contents,23 our results suggest that these RFP puncta are most likely not inside the lysosome in renal epithelia cells.
Autophagy is essential for energy reutilization under stress and nutrient deprivation.2 In renal proximal tubules, autophagy was not increased during the acute ischemic period or 4 hours after reperfusion, but was increased at 8 hours and reached peak at 1–3 days after IRI. This finding suggests that, in addition to metabolic adaptation, autophagy may play a role in the clearance of damaged organelles and in the remodeling of tubular cells during renal repair. Under physiologic conditions, autophagy is required to maintain epithelial structural and functional integrity. Deletion of Atg5 in tubular epithelial cells resulted in a time-dependent accumulation of concentric membrane bodies, deformed mitochondria, and p62- and ubiquitin-positive inclusion bodies.7,8 Mice exhibited elevated basal levels of creatinine, indicating renal functional impairment. When subjected to IRI, mice with Atg5 deletion had more apoptosis in proximal tubules and a higher level of urinary neutrophil gelatinase-associated lipocalin, a biomarker for renal injury.24 The mice also showed persistent elevation of serum urea nitrogen and creatinine for up to 16 days after IRI.8 Because autophagy was more pronounced at 1–3 days after IRI when tubular repair is most active,16 it is conceivable that mice with autophagy defects could also have defects in tubular repair. Future studies using autophagy inhibitors or inducible systems to delete Atg protein during the reparative phase could test this possibility and provide insights into the role of autophagy in tubular quality control and regeneration.
Taking advantage of being able to identify the individual autophagic cells, we examined the initiation and resolution of autophagy and studied the molecular events associated with this dynamic process in the same cell. mTOR is an energy sensor and its activity is reduced with nutrient and growth factor deficiency.25 mTORC1 is activated upon energy restoration and/or in the presence of growth factors. In many cell types, activation of mTORC1 is sufficient to suppress autophagy by preventing the formation of Atg complexes.17 mTOR phosphorylates and inhibits the kinases ULK1/2, catalytic subunits of a protein kinase complex that initiates the autophagic cascade.26,27 Similar to reports by Lieberthal et al.,28,29 we showed activation of mTORC1 in the proximal tubules of the postischemic kidneys. At 3 days after IRI, cells with mTORC1 activation had fewer RFP puncta. This raises the possibility that activation of mTOR could prevent autophagy initiation or lead to autophagy resolution by accelerating the fusion of auphgosomes with lysosomes or enhancing the degradation of autophagosomes contents. The fact that lysososomal protein levels remained unchanged after IRI did not support the possibility of more rapid degradation. On the other hand, treatment of mice with rapamycin to inhibit mTORC1 activity resulted in higher numbers of EGFP puncta, which represented the early autophagic vacuoles, and supported the possibility of suppression of early autophagic processing.
We showed that activation of mTOR resulted in increased cell proliferation and inhibition of mTOR with rapamycin led to a 2-fold reduction in proliferation. These results are consistent with the study showing that rats receiving rapamycin treatment had a lower rate of tubular proliferation and delayed recovery of renal function after ischemic injury.28 In mice with cisplatin-induced AKI, however, rapamycin treatment led to a modest but statistically significant reduction in BUN and creatinine and a better renal morphology. The beneficial effect was related to the increase in LC3-II levels at 2 days after cisplatin injection, suggesting that early activation of autophagy during the injury phase protected against chemical toxicity. During the repairing phase, we demonstrated that autophagic cells had reduced Ki67 expression and therefore were less likely to function as precursors for cell division required for tubular repair. We proposed that mTOR activation provides a feedback mechanism to release cells from autophagy and to stimulate cell cycle entry. At this time, signals permitting the transition from cell catabolism to cell growth are not clear, but could include known mTOR activators such as insulin-like growth factor 1 and hepatocyte growth factor, which have been shown to be upregulated in the postischemic kidneys.30–32 Interestingly, data obtained from cell-free system and cultured HEK-293T cells showed that amino acids in the lysosomes activate Rag GTPases, which recruit mTORC1 to the lysosomal surface for its activation.33 Another study in normal rat kidney cells indicated that nutrients generated by starvation-induced autophagy activated mTOR, leading to autophagy termination. Inhibition of protein degradation with lysosomal enzyme inhibitors abolished mTOR activation.34 Future studies will test whether nutrients generated by autophagy in the postischemic kidneys function in a similar fashion to activate mTOR, resulting in the switch from catabolism for cell survival to proliferation for tubular repair.
Concise Methods
Generation of CAG-RFP-GFP-LC3 Mice
To generate the CAG-RFP-GFP-LC3 transgenic construct, we first amplified the RFP-GFP-LC3 sequence from ptfLC3 (PMID: 17534139). To facilitate protein purification, a Strep-tag sequence (WSHPQFEK) was added in front of RFP. The PCR product with XhoI at 5′ and NotI at 3′ was inserted to pCR4TA (Invitrogen) and sequenced from both ends (Genewiz). The fragment was then cloned into pCAGEN vector (PMID: 1660837) between XhoI and NotI sites. The transgene was under the control of a CAG promoter. The CAG-RFP-GFP-LC3 construct was then linearized by SpeI and HindIII digestion. The 4.2 kb fragment was purified and submitted for pronuclear injection into mice with C57/B6 background by the Transgenic Core facility at the University of Texas Southwestern Medical Center. Genotyping was performed with specific primers for a product of 208 bp in size (forward: CATGGACGAGCTGTACAAGT; reverse: CACCGTGATCAGGTACAAGGA).
Renal IRI in Mice
Methods for renal IRI were previously described.16 Briefly, female CAG-RFP-GFP-LC3 mice (aged 6–8 weeks) were anesthetized with isoflurane, and renal IRI was created by clamping the left renal pedicles for 45 minutes followed by clamp release to allow reperfusion to the kidney. The right kidney was left intact. Sham-operated mice underwent the identical procedure except that clamping of the renal pedicles was omitted. For immunohistochemistry, the mice were perfused with 4% PFA via the left ventricle, and kidney cryosections at 6-µm thickness were used. For immunoblot analysis, the renal cortex and the outer strips of the outer medulla were snap-frozen in liquid nitrogen for total cellular protein isolation. To inhibit mTORC1 activity, mice were given daily intraperitoneal injection with rapamycin (LC Laboratories) at 5 mg/kg of body weight 1 day before and 2 days after renal ischemic injury. All experiments involving animals were performed with the approval of the Institutional Animal Care and Research Advisory Committee at Columbia University and the University of Texas Southwestern Medical Center at Dallas.
Renal Tubular Epithelial Cell Culture and Treatment
Methods for primary renal tubular epithelial cells culture were previously described.35 Briefly, kidneys were collected from 3-week-old female CAG-RFP-GFP-LC3 mice after perfusion with 30 ml of sterile PBS followed by 3 ml of 0.2% collagenase (Sigma-Aldrich). Kidneys were cut into 1- to 2-mm3 pieces and digested with collagenase for 10 minutes at 37°C with gentle shaking. The digestion mixture was filtered through a 40-µm cell strainer (Biosciences), washed, and plated into 10-cm cell culture dishes in DMEM/F12 medium supplemented with insulin (8.3×10−7 M), selenium (6.8×10−9 M), transferrin (6.2×10−8 M), triiodothyronine (2×10−9 M), and 2% FCS (Gemini Bio-Product). Medium was changed 24 hours after initial plating. Confluent cells were transferred into chamber slides or petri dishes and incubated with the above culture medium or EBSS (Sigma-Aldrich) for 2 hours to deprive glucose and amino acids. Some cells grown on chamber slides were incubated with chloroquine diphosphate salt (Sigma-Aldrich) at 40 mg/ml for 2 hours during starvation with EBSS before fixation with 4% PFA for further analysis.
Immunohistochemistry and Image Analyses
Immunohistochemistry was performed using established methods.36 The following antibodies at the indicated dilutions were used: Lamp1 (1:200; Abcam), kidney injury molecule 1 (1:800; R&D Systems), phospho-S6 ribosomal protein (Ser-235/236, 1: 200; Cell Signaling Technology), V-ATPase B1/2 (1:500; Santa Cruz Biotechnology), thiazide-sensitive NaCl cotransporter (1:200; generous gift from Mark Knepper at the National Institutes of Health [NIH]), Tamm-Horsfall protein (1:200; Biomedical Technologies, Inc.), Ki67 (1:200; Leica Biosystems), phospho-histone H3 (1:1000; Sigma-Aldrich), SQSTM1or p62 (1:200; Abnova), ZO-1 (1:1000; Zymed Laboratories), E-cadherin (1:100; Invitrogen), WT1 (1:50; Santa Cruz Biotechnology), and FITC-conjugated anti-GFP (1:200; Rockland). Secondary antibodies were conjugated to Cy5 (1:100; Molecular Probes) or Alexa Fluor 488 for all the primary antibodies. Tissue sections were incubated with the primary antibody at 4°C overnight after incubation with blocking solution (10% goat serum and 0.3% BSA in PBS) and then the appropriate secondary antibody for 1 hour at room temperature. Biotin-conjugated Lotus tetragonolobus agglutinin (1:800; Vector Laboratories) was used to stain proximal tubules and fluorescence was detected with Daylight 649-conjugated streptavidin (1:400; Vector Laboratories). The slides were mounted with VECTASHIELD and examined using a Zeiss Observer Z1 fluorescence microscope. Images were acquired using a digital camera and analyzed with AxioVision Rel. 4.8.2 software (Carl Zeiss). Some images were obtained by laser scanning confocal microscopy (Zeiss LSM 510 META). Images were analyzed using the Zeiss LSM 510 software package.
Immunoblot Analyses
Preparation of cellular extracts and immunoblot analysis were performed as previously described.36 For protein extraction, 2% SDS buffer was used and kidney lysates were kept at room temperature for 20 minutes before centrifugation at 13,000 rpm for 5 minutes. Membranes were probed with antibodies to LC3 (1:700; Novus Biologicals), Atg5 (1:2000; Millipore), Lamp2 (1:5000; Abcam), cathepsin D (1:5000; Santa Cruz Biotechnology), V-ATPase B1/2 (1:5000; Santa Cruz Biotechnology), phospho-4E-BP1 (1:2000; Cell Signaling Technology), and phospho-S6 ribosomal protein (Ser-235/236, 1:2000; Cell Signaling Technology). The membrane was incubated with the appropriate secondary antibody conjugated with horseradish peroxidase and detected by chemiluminescence. Samples were examined with an antibody to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:1000; Santa Cruz Biotechnology) for loading controls. Protein intensities were quantified using NIH ImageJ 1.47 software and normalized to the intensities of the corresponding Gaphd as loading controls.
Quantification and Statistical Analyses
To quantify fluorescence signals in primary cultures of renal tubular epithelial cells, images at magnifications of ×400 and ×630 were randomly selected and at least a total of 500 cells were counted in each experiment. Cells that contained ≥3 RFP or EGFP puncta were selected and counted as autophagic cells.37 To quantify immunostaining in the kidney, 10 fields from the outer stripe of the outer medulla were randomly selected. The number of animals used is indicated in each experiment. Each immunostaining was conducted in duplicate. Data are presented as the mean±SEM. The t test was used to compare the differences between the two groups. A two-tailed P value of <0.05 was considered to be statistically significant.
Disclosures
None.
Acknowledgments
The authors thank Dr. Jonathan Barasch for helpful discussions, Cristina Fuente Mora and Catherine Ha for technical assistance, and Toni Marrow for administrative assistance.
This work was supported by grants from the NIH (R01DK083411) and March of Dimes to F.L., as well as grants from the NIH (HL-080144, HL-0980842, and HL-100401), the Cancer Prevention Research Institute of Texas (RP110486P3), the American Heart Association–Jon Holden DeHaan Foundation (0970518N), and the Fondation Leducq (11CVD04) to J.A.H. Z.V.W. was supported by a postdoctoral fellowship from the American Heart Association (10POST4320009). F.L. is a Samberg Scholar in Children’s Health at Morgan Stanley Children’s Hospital of the New York Presbyterian Hospital System.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Klionsky DJ: Autophagy: From phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 8: 931–937, 2007 [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G: Autophagy in the pathogenesis of disease. Cell 132: 27–42, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L,: Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8: 445–544, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang M, Liu K, Luo J, Dong Z: Autophagy is a renoprotective mechanism during in vitro hypoxia and in vivo ischemia-reperfusion injury. Am J Pathol 176: 1181–1192, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki C, Isaka Y, Takabatake Y, Tanaka H, Koike M, Shibata M, Uchiyama Y, Takahara S, Imai E: Participation of autophagy in renal ischemia/reperfusion injury. Biochem Biophys Res Commun 368: 100–106, 2008 [DOI] [PubMed] [Google Scholar]
- 6.Wu HH, Hsiao TY, Chien CT, Lai MK: Ischemic conditioning by short periods of reperfusion attenuates renal ischemia/reperfusion induced apoptosis and autophagy in the rat. J Biomed Sci 16: 19, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura T, Takabatake Y, Takahashi A, Kaimori JY, Matsui I, Namba T, Kitamura H, Niimura F, Matsusaka T, Soga T, Rakugi H, Isaka Y: Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J Am Soc Nephrol 22: 902–913, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S, Hartleben B, Kretz O, Wiech T, Igarashi P, Mizushima N, Walz G, Huber TB: Autophagy plays a critical role in kidney tubule maintenance, aging and ischemia-reperfusion injury. Autophagy 8: 826–837, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Ladoire S, Chaba K, Martins I, Sukkurwala AQ, Adjemian S, Michaud M, Poirier-Colame V, Andreiuolo F, Galluzzi L, White E, Rosenfeldt M, Ryan KM, Zitvogel L, Kroemer G: Immunohistochemical detection of cytoplasmic LC3 puncta in human cancer specimens. Autophagy 8: 1175–1184, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y: In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15: 1101–1111, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura S, Noda T, Yoshimori T: Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3: 452–460, 2007 [DOI] [PubMed] [Google Scholar]
- 12.Zhou C, Zhong W, Zhou J, Sheng F, Fang Z, Wei Y, Chen Y, Deng X, Xia B, Lin J: Monitoring autophagic flux by an improved tandem fluorescent-tagged LC3 (mTagRFP-mWasabi-LC3) reveals that high-dose rapamycin impairs autophagic flux in cancer cells. Autophagy 8: 1215–1226, 2012 [DOI] [PubMed] [Google Scholar]
- 13.Kuma A, Matsui M, Mizushima N: LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: Caution in the interpretation of LC3 localization. Autophagy 3: 323–328, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N: The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036, 2004 [DOI] [PubMed] [Google Scholar]
- 15.Schwartz GJ, Barasch J, Al-Awqati Q: Plasticity of functional epithelial polarity. Nature 318: 368–371, 1985 [DOI] [PubMed] [Google Scholar]
- 16.Lin F, Moran A, Igarashi P: Intrarenal cells, not bone marrow-derived cells, are the major source for regeneration in postischemic kidney. J Clin Invest 115: 1756–1764, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung CH, Ro SH, Cao J, Otto NM, Kim DH: mTOR regulation of autophagy. FEBS Lett 584: 1287–1295, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lieberthal W, Levine JS: The role of the mammalian target of rapamycin (mTOR) in renal disease. J Am Soc Nephrol 20: 2493–2502, 2009 [DOI] [PubMed] [Google Scholar]
- 19.Yu CC, Woods AL, Levison DA: The assessment of cellular proliferation by immunohistochemistry: A review of currently available methods and their applications. Histochem J 24: 121–131, 1992 [DOI] [PubMed] [Google Scholar]
- 20.Tasdemir E, Maiuri MC, Tajeddine N, Vitale I, Criollo A, Vicencio JM, Hickman JA, Geneste O, Kroemer G: Cell cycle-dependent induction of autophagy, mitophagy and reticulophagy. Cell Cycle 6: 2263–2267, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis CD, Sassone-Corsi P: Mitotic phosphorylation of histone H3: Spatio-temporal regulation by mammalian Aurora kinases. Mol Cell Biol 22: 874–885, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizushima N, Yoshimori T, Levine B: Methods in mammalian autophagy research. Cell 140: 313–326, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katayama H, Yamamoto A, Mizushima N, Yoshimori T, Miyawaki A: GFP-like proteins stably accumulate in lysosomes. Cell Struct Funct 33: 1–12, 2008 [DOI] [PubMed] [Google Scholar]
- 24.Paragas N, Qiu A, Zhang Q, Samstein B, Deng SX, Schmidt-Ott KM, Viltard M, Yu W, Forster CS, Gong G, Liu Y, Kulkarni R, Mori K, Kalandadze A, Ratner AJ, Devarajan P, Landry DW, D’Agati V, Lin CS, Barasch J: The Ngal reporter mouse detects the response of the kidney to injury in real time. Nat Med 17: 216–222, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laplante M, Sabatini DM: mTOR signaling in growth control and disease. Cell 149: 274–293, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganley IG, Lam H, Wang J, Ding X, Chen S, Jiang X: ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284: 12297–12305, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N: Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20: 1981–1991, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieberthal W, Fuhro R, Andry C, Patel V, Levine JS: Rapamycin delays but does not prevent recovery from acute renal failure: Role of acquired tubular resistance. Transplantation 82: 17–22, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Lieberthal W, Fuhro R, Andry CC, Rennke H, Abernathy VE, Koh JS, Valeri R, Levine JS: Rapamycin impairs recovery from acute renal failure: Role of cell-cycle arrest and apoptosis of tubular cells. Am J Physiol Renal Physiol 281: F693–F706, 2001 [DOI] [PubMed] [Google Scholar]
- 30.Igawa T, Matsumoto K, Kanda S, Saito Y, Nakamura T: Hepatocyte growth factor may function as a renotropic factor for regeneration in rats with acute renal injury. Am J Physiol 265: F61–F69, 1993 [DOI] [PubMed] [Google Scholar]
- 31.Kanda H, Tajima H, Lee GH, Nomura K, Ohtake K, Matsumoto K, Nakamura T, Kitagawa T: Hepatocyte growth factor transforms immortalized mouse liver epithelial cells. Oncogene 8: 3047–3053, 1993 [PubMed] [Google Scholar]
- 32.Supavekin S, Zhang W, Kucherlapati R, Kaskel FJ, Moore LC, Devarajan P: Differential gene expression following early renal ischemia/reperfusion. Kidney Int 63: 1714–1724, 2003 [DOI] [PubMed] [Google Scholar]
- 33.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM: mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334: 678–683, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, Hailey DW, Oorschot V, Klumperman J, Baehrecke EH, Lenardo MJ: Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465: 942–946, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, Truong P, Igarashi P, Lin F: Renal and bone marrow cells fuse after renal ischemic injury. J Am Soc Nephrol 18: 3067–3077, 2007 [DOI] [PubMed] [Google Scholar]
- 36.Li L, Zepeda-Orozco D, Black R, Lin F: Autophagy is a component of epithelial cell fate in obstructive uropathy. Am J Pathol 176: 1767–1778, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sengupta A, Molkentin JD, Yutzey KE: FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem 284: 28319–28331, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]