Abstract
Acute myeloid leukemia (AML) is a heterogeneous hematologic malignancy that originates from leukemia-initiating cells (LICs). The identification of common mechanisms underlying LIC development will be important in establishing broadly effective therapeutics for AML. Constitutive NF-κB pathway activation has been reported in different types of AML; however, the mechanism of NF-κB activation and its importance in leukemia progression are poorly understood. Here, we analyzed myeloid leukemia mouse models to assess NF-κB activity in AML LICs. We found that LICs, but not normal hematopoietic stem cells or non-LIC fractions within leukemia cells, exhibited constitutive NF-κB activity. This activity was maintained through autocrine TNF-α secretion, which formed an NF-κB/TNF-α positive feedback loop. LICs had increased levels of active proteasome machinery, which promoted the degradation of IκBα and further supported NF-κB activity. Pharmacological inhibition of the proteasome complex markedly suppressed leukemia progression in vivo. Conversely, enhanced activation of NF-κB signaling expanded LIC frequency within leukemia cell populations. We also demonstrated a strong correlation between NF-κB activity and TNF-α secretion in human AML samples. Our findings indicate that NF-κB/TNF-α signaling in LICs contributes to leukemia progression and provide a widely applicable approach for targeting LICs.
Introduction
Acute myeloid leukemia (AML) is a highly aggressive hematologic malignancy characterized by a relentless proliferation of immature myeloid blasts. Recent studies have demonstrated that the apparently uniform leukemia cell population is organized as a hierarchy that originates from leukemia-initiating cells (LICs) (1, 2). Although intensive chemotherapy is initially effective in most cases of AML, the surviving LIC clones repopulate the disease, leading to subsequent relapse and an ultimately dismal prognosis (3). Another problem is that AML is a heterogeneous disease with different cytogenetic and molecular abnormalities. This heterogeneity has increasingly been unveiled by recent work involving the screening of recurrent mutations seen in AML cells using high-throughput sequencing technology, which is useful for constructing individualized therapeutics (4, 5). At the same time, however, these findings indicate that it is difficult to develop a treatment strategy in addition to standard chemotherapy that is widely applicable to AML. Therefore, to establish effective treatments, it is important to identify the universally essential mechanisms involved in the LIC phenotype, irrespective of the cells’ diverse genetic abnormalities.
NF-κB is a transcription factor initially discovered in B cells (6). Although well known for its role in controlling various aspects of immune responses, the NF-κB pathway is now also recognized as an important regulator of cell survival, proliferation, and differentiation (7–9). Its constitutive activation has been reported in a variety of malignancies and mostly plays a cancer-promoting role (10–12). There is some evidence that this pathway activity is also seen in the AML CD34+CD38– fraction, which is considered to be enriched for LICs (13, 14). Given that NF-κB activity is not restricted to specific AML subtypes or genetic abnormalities, it is possible that the signaling is universally essential for myeloid leukemia progression, and a variety of agents have been reported to induce apoptosis in cultured leukemia cells via NF-κB pathway inhibition (15–19). The effect of specific inhibition of the NF-κB pathway on LICs in vivo, however, has not been sufficiently studied. Furthermore, the mechanism of this pathway’s activation remains to be elucidated. Although several gene mutations found in hematologic malignancies have been reported to be associated with enhanced NF-κB signaling (20–22), these findings do not fully explain why the activation of NF-κB is observed in a number of different types of leukemia. It is more intriguing, as well as reasonable, to consider that NF-κB activation arises from the signaling pathways that are commonly involved in LICs. Another limitation of the previous studies is that LIC-enriched populations in AML are highly heterogeneous among patients and are not necessarily confined to the CD34+CD38– fraction, as they are in normal HSCs. Therefore, it is problematic to strictly define LICs by their surface-marker antigens (23, 24).
To overcome these challenges, we used variable myeloid leukemia mouse models, in which LIC-enriched fractions were well characterized using a surface marker phenotype and revealed that NF-κB signaling is constitutively activated in LICs, but not in normal cells or non-LIC fractions within leukemic BM cells. We also elucidate the mechanism of NF-κB activation in LICs in each model and demonstrate that the inhibition of NF-κB or its upstream machinery in LICs markedly suppresses leukemia progression in vivo.
Results
The NF-κB pathway is activated in LICs of different types of myeloid leukemia models.
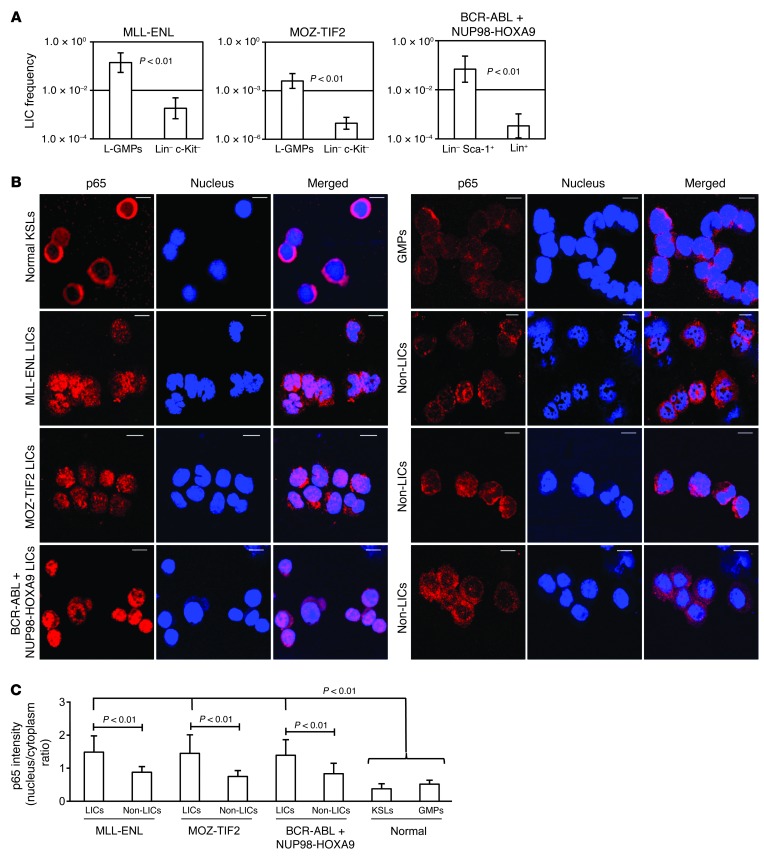
To extensively investigate NF-κB activity in LICs of different types of myeloid leukemia, we used three types of mouse models of myeloid leukemia induced by the retroviral transduction of granulocyte-monocyte progenitors (GMPs) with MLL-ENL and MOZ-TIF2 and the cotransduction of GMPs with BCR-ABL and NUP98-HOXA9 (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI68101DS1). LIC-enriched populations of these myeloid leukemia models have been investigated in previous studies: GMP-like leukemia cells (L-GMPs) in MLL-ENL and MOZ-TIF2 models and the lineage– Sca-1+ fraction in the BCR-ABL/NUP98-HOXA9 model (Supplemental Figure 2, A–C, and refs. 25–27). In order to obtain cell populations that would barely contain LICs, we also sorted lineage– c-Kit– cells in MLL-ENL and MOZ-TIF2 leukemic mice and lineage+ cells in a BCR-ABL/NUP98-HOXA9 model. There were striking differences in clonogenic potential (Supplemental Figure 3) and LIC frequencies, as determined by in vivo limiting dilution assays in the two populations of each model (Figure 1A and Supplemental Table 1). Therefore, we confirmed that LIC and non-LIC fractions can be clearly isolated through the surface antigen profiles of the three leukemia models. Next, we visualized the subcellular distribution of the major NF-κB subunit p65 in LICs, non-LICs, and normal cells by immunofluorescence staining and confocal microscopy. As shown in Figure 1B, prominent nuclear translocation of p65 was observed in the LICs of each model, while it was retained mostly in the cytoplasm in normal lineage– c-Kit+ Sca-1+ cells (KSLs), which are enriched for HSCs and GMPs. Interestingly, non-LICs also had relatively reduced p65 nuclear translocation signal compared with that in LICs in all three leukemia models. We quantified the nucleus/cytoplasm ratio of p65 staining intensity in these images, which also showed that the LICs in each model had significant nuclear localization compared with that observed in non-LICs, normal KSLs, and GMPs (Figure 1C).
Figure 1. NF-κB pathway is activated in LICs of different murine myeloid leukemia models.
(A) LIC frequency in the two fractions of each leukemia model as determined by limiting dilution assay. See Supplemental Table 1 for detailed transplantation results. (B) Immunofluorescence assessment for p65 nuclear translocation in KSLs, GMPs, LICs, and non-LICs in three leukemia models. Scale bars: 10 μm. (C) Quantification of p65 nuclear translocation assessed by the mean nucleus/cytoplasm intensity ratio. More than 50 cells were scored in each specimen, and the average intensity ratio with SD is shown.
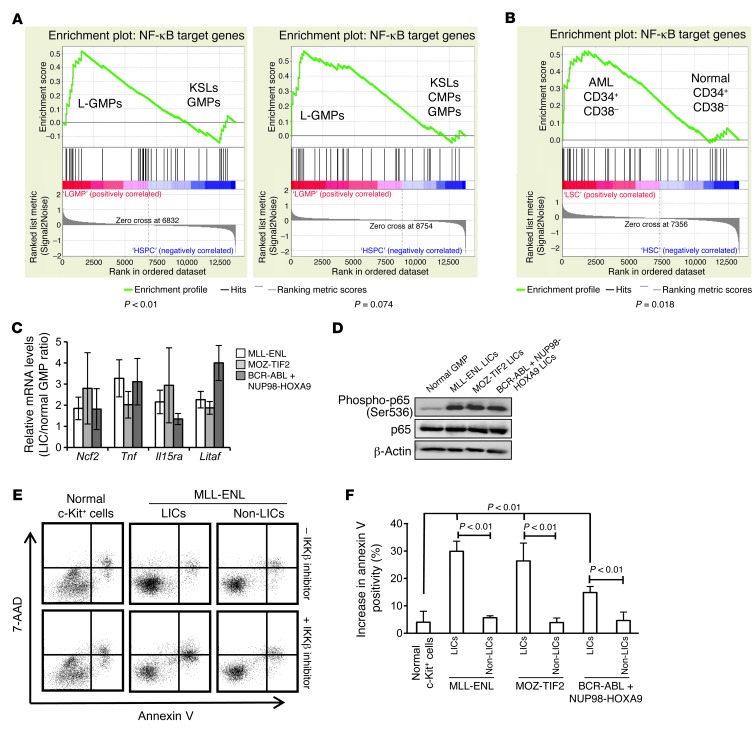
To further test NF-κB transcription activity in LICs, we investigated the expression profiles of a subset of genes regulated by the NF-κB pathway. We first used two sets of published gene expression microarray data, which compared the expression profiles of MOZ-TIF2 L-GMPs (26), MLL-AF9 L-GMPs, and HOXA9-MEIS1 L-GMPs (28) with those of normal hematopoietic stem or progenitor cells (HSPCs). The expression profiles of previously identified NF-κB target genes were assessed by gene set enrichment analysis (GSEA) (Supplemental Table 2 and ref. 29), which showed that L-GMPs had increased expression levels of NF-κB target genes compared with those in normal HSPCs in both sets of gene expression microarray data (Figure 2A). We also compared the expression profiles of the same gene set in CD34+CD38– human AML cells with those of the equivalent cell population in normal BM cells, which corresponded to the HSC fraction, and observed a similar tendency (Figure 2B and ref. 30). Then, we validated these results using quantitative real-time PCR by comparing the expression levels of several NF-κB target genes in LICs and non-LICs from our three mouse models with those in normal GMPs and found increased expression levels of most of the genes in different types of LICs, but no significant elevation of these levels in non-LICs (Figure 2C and Supplemental Figure 4). Furthermore, the level of p65 phosphorylation, which is important for enhancing its transcription activity, was significantly increased in LICs compared with the level observed in normal GMPs (Figure 2D). Consistent with these findings, LICs showed a more prominent increase in apoptosis than did normal cells or non-LICs when treated with sc-514, a selective inhibitor of IκB kinase β (IKKβ) (Figure 2, E and F, and ref. 31). Although LICs from BCR-ABL/NUP98-HOXA9–induced leukemia were rather resistant to sc-514 compared with cells from MLL-ENL– and MOZ-TIF2–induced leukemia, they still showed higher sensitivity than non-LICs. Collectively, these data fully support the hypothesis that the NF-κB pathway is constitutively activated in the LICs of different types of myeloid leukemia.
Figure 2. NF-κB transcription activity is increased in LICs.
(A) GSEA of NF-κB target genes in the published gene expression data comparing LICs in leukemia mouse models with normal HSPCs. Left panel: comparison of MOZ-TIF2 L-GMP with normal KSLs and GMPs (GSE24797). Right panel: comparison of MLL-AF9 and HOXA9-MEIS1 L-GMPs with normal KSLs, common myeloid progenitors (CMPs), and GMPs (GSE20377). (B) GSEA of NF-κB target genes in CD34+CD38– fractions in human AML versus healthy controls (GSE24006). (C) Quantitative real-time PCR analysis of a subset of NF-κB target genes in LICs of MLL-ENL, MOZ-TIF2, and BCR-ABL/NUP98-HOXA9 leukemia models relative to normal GMPs (n = 4). Error bars indicate SD. (D) Immunoblotting of total and phosphorylated p65 in normal GMPs and LICs in the three leukemia models. (E) Representative annexin V and 7-AAD profiles of normal c-Kit+ cells, L-GMPs, and Lin–c-Kit– cells in MLL-ENL leukemic mice after a 24-hour culture with or without 10 μM IKK inhibitor (sc-514). (F) Average percentage increase in apoptotic cells in LICs of the three leukemia models compared with that in non-LICs and normal c-Kit+ cells treated with 10 μM IKK inhibitor (sc-514) (n = 4 each). Error bars indicate SD.
LICs maintain their constitutive NF-κB activity via autocrine TNF-α signaling.
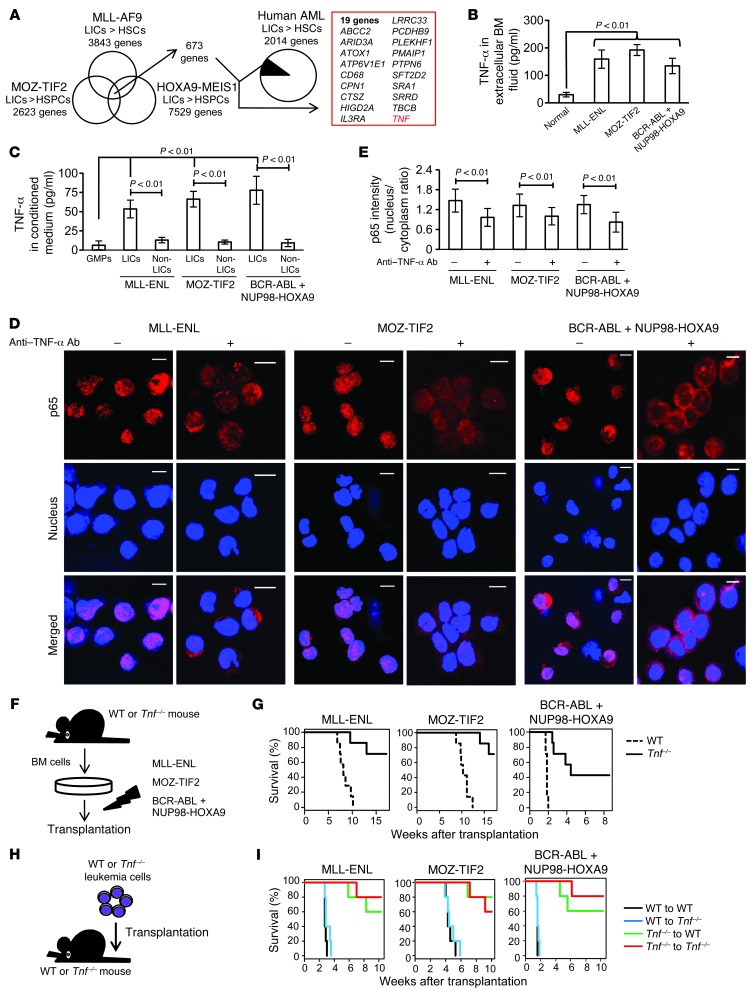
In the next step, we addressed the question of how LICs maintain constitutive NF-κB activity in different types of leukemia models. In order to investigate genes prevalently dysregulated in LICs, we analyzed the previously published microarray-based gene expression profiles comparing murine and human LICs with normal HSPCs (26, 28, 30). After narrowing down our analysis to the genes commonly upregulated in LICs in three different types of murine leukemia models, we further selected nineteen genes whose expression is elevated in human AML CD34+CD38– cells (Figure 3A). Among the nineteen genes with typically elevated expression levels in LICs, we focused on Tnf, because it is well known as an activator of NF-κB and as an NF-κB–regulated gene. For the purpose of directly evaluating TNF-α abundance in the BM of leukemic mice, we measured the concentration of TNF-α in the BM extracellular fluid and confirmed that it was conspicuously enriched in leukemic BM cells compared with normal BM cells (Figure 3B). We also examined the TNF-α concentration in culture media conditioned by LICs, non-LICs, and normal cells, respectively, to determine whether leukemia cells themselves have the ability to secrete TNF-α. We found that TNF-α secretion was distinctly elevated in LICs, while the normal GMP-conditioned media barely included TNF-α (Figure 3C). Although non-LICs also had TNF-α secretory ability, it was much lower that that of LICs. We therefore reasoned that LICs might maintain their NF-κB pathway activity via autocrine TNF-α signaling. To test this hypothesis, we cultured freshly isolated LICs in serum-free media with a TNF-α–neutralizing antibody or its isotype control and observed p65 subcellular distribution. While LICs treated with isotype control antibodies maintained p65 nuclear translocation even after serum-deprived culture, the p65 translocation signal we observed in three types of LICs was significantly attenuated when these cells were cultured with neutralizing antibodies against TNF-α (Figure 3D). The results were also confirmed by quantification of p65 intensity (Figure 3E). These data strongly suggest that different types of LICs have a similarly increased potential for TNF-α secretion, which maintains constitutive NF-κB activity in an autonomous fashion.
Figure 3. Autocrine TNF-α secretion maintains constitutive NF-κB activity and confers proliferative advantage in LICs.
(A) Thorough investigation of genes with elevated expression in murine and human LICs compared with that in normal HSPCs in the published gene expression data. (B) TNF-α ELISA in extracellular fluid of normal or leukemic BM (n = 4 each). Error bars indicate SD. (C) TNF-α secretory ability in LICs compared with that of non-LICs and normal GMPs assessed by ELISA in cultured media (n = 4 each). Error bars indicate SD. (D) Immunofluorescence assessment for p65 nuclear translocation in LICs in serum-free culture medium with neutralizing antibody against TNF-α or isotype control. Scale bars: 10 μm. (E) Quantification of p65 nuclear translocation of LICs treated with neutralizing antibody against TNF-α or isotype control assessed by the mean nucleus/cytoplasm intensity ratio. More than 50 cells were scored in each specimen, and the average intensity ratio with SD is shown. (F) Schematic representation of the experiments. BM cells derived from WT or Tnf-knockout mice were transduced with MLL-ENL, MOZ-TIF2, and BCR-ABL plus NUP98-HOXA9 and transplanted into sublethally irradiated mice. (G) Survival curves of mice in the experiments shown in F (n = 7 each). (H) Schematic representation of the experiments. WT or Tnf–/– leukemia cells were secondarily transplanted into WT or Tnf–/– recipient mice. (I) Survival curves of mice in the experiments shown in H (n = 5 each).
Autocrine TNF-α signaling promotes leukemia cell progression.
We were then interested in exploring the effect of autocrine TNF-α secretion on leukemia progression. BM cells derived from WT or Tnf-knockout mice were transplanted into sublethally irradiated WT recipient mice after transduction with MLL-ENL and MOZ-TIF2, and cotransduction with BCR-ABL and NUP98-HOXA9 (Figure 3F). Although several mice did develop leukemia with prolonged latency, Tnf-deficient cells were significantly (P < 0.01) impaired in their ability to initiate leukemia (Figure 3G). We confirmed that Tnf-deficient LICs show a distinct decrease in nuclear localization of p65 compared with the that in LICs derived from WT BM cells (Supplemental Figure 5, A and B). Next, we examined whether paracrine TNF-α from the BM microenvironment contributes to leukemia progression. When the established leukemia cells were secondarily transplanted into WT or Tnf-knockout recipient mice, Tnf-deficient leukemia cells failed to effectively establish AML in all three models (Figure 3, H and I). Interestingly, there was no significant difference in leukemogenicity among the recipient genotypes. These results indicate that autocrine TNF-α secretion is important for AML progression and that the contribution of paracrine effects derived from stromal cells is minimal.
The impact of specific NF-κB inhibition on leukemia progression.
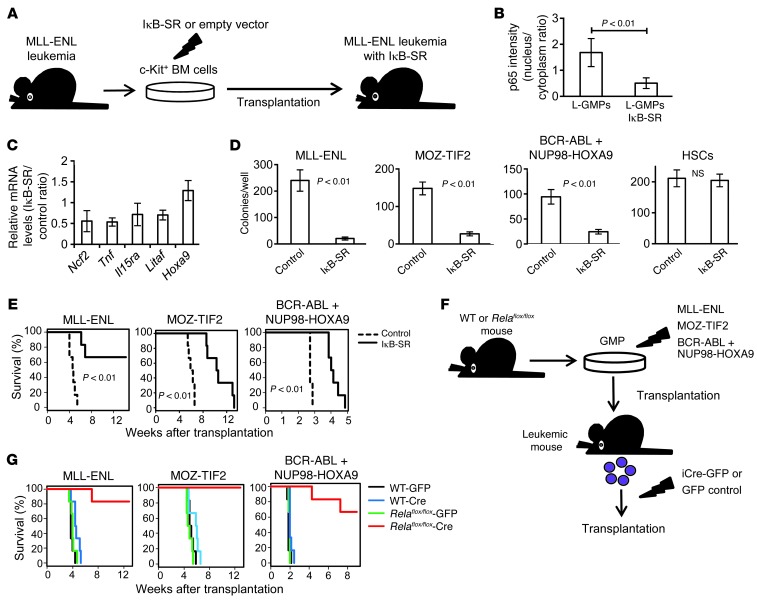
To investigate the influence of specific NF-κB pathway inhibition on leukemia progression in vivo, we transduced MLL-ENL leukemia cells with a retroviral vector expressing a dominant-negative form of IκBα (super repressor, referred to herein as IκB-SR) or with a control vector, transplanted them into recipient mice, and compared the characteristics of the repopulating cells (Figure 4A). Although the introduction of IκB-SR did not affect the morphology of MLL-ENL leukemia cells (Supplemental Figure 6A), p65 was almost completely sequestered in the cytoplasm of L-GMPs with IκB-SR (Figure 4B and Supplemental Figure 6B), and the expression levels of NF-κB target genes, including Tnf, were substantially decreased (Figure 4C). Considering that the blockage of autocrine TNF-α attenuated NF-κΒ signaling, we hypothesized that NF-κΒ activity and TNF-α secretion form a positive feedback loop in LICs. We therefore established MOZ-TIF2 and BCR-ABL/NUP98-HOXA9 leukemia cells with IκB-SR. The introduction of IκB-SR significantly decreased a proportion of the cells in the S and G2/M phases of the cell cycle and resulted in a substantial growth delay of those cells in liquid culture (Supplemental Figure 6, C and D). Moreover, leukemia cells with IκB-SR had a reduced colony-forming capacity, while the transduction of IκB-SR into normal HSCs had no significant influence on their colony-forming ability (Figure 4D). Finally, we transplanted leukemia cells with IκB-SR into sublethally irradiated mice and observed a remarkable delay in leukemia progression (Figure 4E). We also confirmed that the developed leukemia cells with IκB-SR had reduced nuclear translocation of p65 compared with that seen in control cells (Supplemental Figure 6E). In contrast, when normal BM cells were transduced with IκB-SR and transplanted into lethally irradiated mice, we observed no significant differences in the reconstitution capacity of the transplanted cells, nor did we find significant differences in peripheral blood cell counts or PBL surface-marker profiles, indicating that NF-κB pathway inhibition exerts a marginal influence on normal hematopoiesis (Supplemental Figure 7, A–C). Collectively, these findings clearly demonstrate that enhanced NF-κB activity in LICs plays a supportive role in leukemia progression and that NF-κB inhibition severely attenuates the proliferative ability of these cells.
Figure 4. Specific inhibition of NF-κB significantly inhibits leukemia progression in vivo.
(A) Schematic representation of the following experiments: c-Kit+ BM cells isolated from MLL-ENL leukemic mice were transduced with IκB-SR or control vector and transplanted into sublethally irradiated mice. (B) Quantification of p65 nuclear translocation assessed by the mean nucleus/cytoplasm intensity ratio by immunofluorescence staining. More than 50 cells were scored in each specimen, and the average intensity ratio with SD is shown. (C) Relative expression profiles of NF-κB target genes in MLL-ENL leukemia cells with or without IκB-SR. The change in Hoxa9 expression is shown as a control gene not regulated by NF-κB. Error bars indicate SD (n = 3 each). (D) CFC assay of leukemia cells and normal HSCS with or without IκB-SR. Cells were seeded at 2,000 cells per well in MLL-ENL or BCR-ABL/NUP98-HOXA9–induced leukemia cells, at 500 cells per well in MOZ-TIF2–induced leukemia cells, and at 1,000 cells per well in normal HSCs (n = 6 in each experiment). (E) Survival curves of mice transplanted with MLL-ENL, MOZ-TIF2, and BCR-ABL/NUP98-HOXA9 leukemia cells with or without IκB-SR (n = 6 each). (F) Schematic representation of the following experiments: WT or Relaflox/flox mice were transduced with MLL-ENL, MOZ-TIF2, or BCR-ABL plus NUP98-HOXA9 and transplanted into sublethally irradiated mice. The developed leukemia cells were transduced with iCre-IRES-GFP or control GFP, and GFP+ cells were secondarily transplanted into mice. (G) Survival curves of mice in the experiments shown in F (n = 6 each).
To further validate the importance of the NF-κB pathway in leukemia progression, we used BM cells from Relaflox/flox mice (32). We similarly established leukemia cells derived from Relaflox/flox BM cells. Then, the developed leukemia cells were infected with codon-improved Cre recombinase–IRES-GFP (iCre-IRES-GFP) or GFP empty vector, and GFP-positive cells were isolated and secondarily transplanted into sublethally irradiated mice (Figure 4F). Remarkably, most of the mice transplanted with Rela-deleted leukemia cells did not develop leukemia (Figure 4G). Compared with controls, several mice did develop leukemia after longer latencies, but they did not develop leukemia after tertiary transplantation (data not shown), indicating that the complete ablation of NF-κB drastically reduced leukemogenicity.
High proteasome activity in LICs yields differences in NF-κB activity between leukemia cell populations.
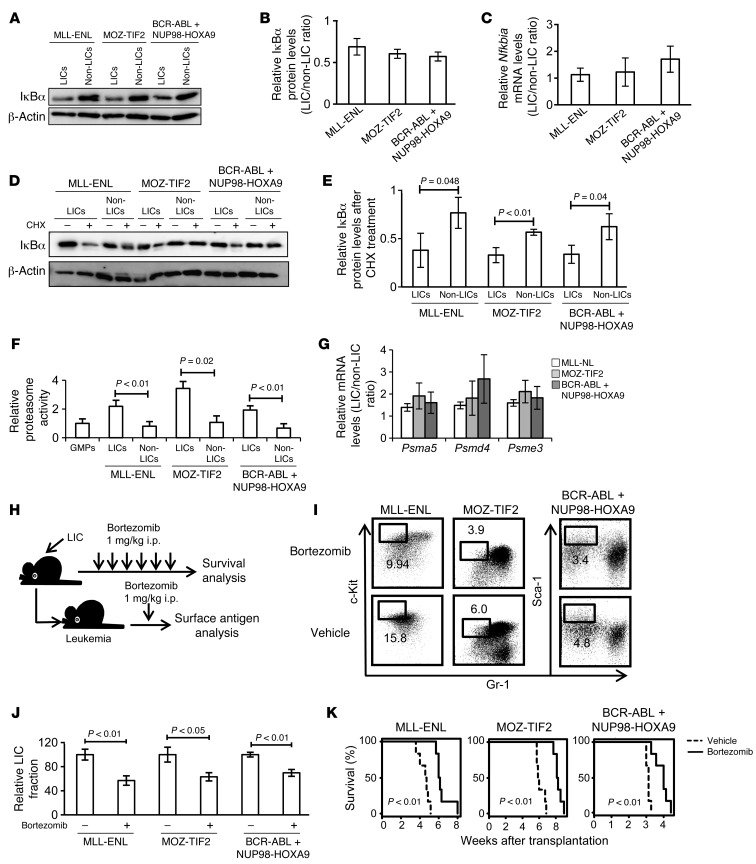
We next sought to elucidate the mechanisms underlying the differences in p65 nuclear translocation status between LICs and non-LICs. We confirmed that LICs had substantially lower IκBα protein levels compared with those of non-LICs in all three models (Figure 5, A and B). These results are very consistent with the p65 distribution status of LICs and non-LICs, considering that NF-κB is usually sequestered in the cytoplasm, bound to IκBα, and translocates to the nucleus, where IκBα is phosphorylated and degraded upon stimulation with a variety of agents such as TNF-α (33). We initially tested whether the expression of IκBα is downregulated in LICs at the transcription level and found that LICs had a tendency toward increased Nfkbia mRNA expression levels compared with non-LICs (Figure 5C). Moreover, when Nfkbia mRNA translation was inhibited by treatment with cycloheximide, the reduction in IκBα protein levels was more prominent in LICs than in non-LICs (Figure 5, D and E). These data indicate that the differences in IκBα levels are caused by the protein’s predominant degradation in LICs. Since both LICs and non-LICs are similarly exposed to high levels of TNF-α within leukemic BM cells, we considered that there would be differences in response to the stimulus and sequentially examined the downstream signals. We first hypothesized that there is a difference in TNF-α receptor expression levels between LICs and non-LICs that leads to greater TNF-α signal transmission in LICs. The expression patterns of TNF receptors I and II were, however, almost similar in LICs and non-LICs, although they varied between leukemia models (Supplemental Figure 8A). We next tested the phosphorylation capacity of IκB kinase (IKK) by examining the ratio of phosphorylated IκBα to total IκBα after treatment with the proteasome inhibitor MG132. Contrary to our expectation, a similar accumulation of the phosphorylated form of IκBα was seen in both LICs and non-LICs, implying that they had no significant difference in IKK activity (Supplemental Figure 8B). Another possibility is that the differences in IκBα protein levels are caused by predominant proteasome activity in LICs, because it is required for the degradation of phosphorylated IκBα. We measured 20S proteasome activity in LICs and non-LICs in each leukemia model by quantifying the fluorescence produced upon cleavage of the proteasome substrate SUC-LLVY-AMC and observed a 2- to 3-fold higher proteasome activity in LICs (Figure 5F). Furthermore, the expression of several genes encoding proteasome subunits was elevated in LICs compared with that in non-LICs (Figure 5G). Similarly, the published gene expression data on human AML samples revealed that CD34+CD38– cells had increased expression levels of proteasome subunit gene sets compared with those in CD34– cells (Supplemental Figure 9 and ref. 30). These findings suggest that enhanced proteasome activity in LICs leads to more efficient degradation of IκBα in response to TNF-α, thus resulting in elevated NF-κB activity. We then tested the effect of bortezomib, a well-known proteasome inhibitor, on LICs in vivo (Figure 5H). First, we treated mice with full-blown leukemia with a single injection of bortezomib and compared their BM surface-marker profiles with those of the vehicle-treated mice. Notably, bortezomib-treated mice showed a significant decrease in LIC-enriched populations in each type of leukemia (Figure 5, I and J). Finally, we treated mice with bortezomib after LIC transplantation and observed significant improvement in survival in those treated with bortezomib (Figure 5K). These results are very consistent with the selectively elevated proteasome activity we observed in LICs.
Figure 5. LICs have higher proteasome activity than non-LICs.
(A and B) Immunoblotting of IκBα in LICs and non-LICs (A). Protein levels were quantified with ImageJ software (B). Data representative of four experiments with SD are shown. (C) Relative mRNA expression of Nfkbia in LICs compared with that in non-LICs (n = 4 each). Error bars indicate SD. (D and E) Immunoblotting of IκBα in LICs and non-LICs. Cells were pretreated with MG132 for 1 hour and incubated for an additional hour with or without cycloheximide (CHX) (D). IκBα protein levels were quantified with ImageJ software, and the relative decrease in IκBα after cycloheximide treatment was calculated (n = 3 each). Error bars indicate SD (E). (F) Analysis of 20S proteasome activity quantified with fluorescence produced upon cleavage of the proteasome substrate SUC-LLVY-AMC (n = 4 each). Error bars indicate SD. (G) Relative mRNA expression of proteasome subunits in LICs compared with that in non-LICs (n = 4 each). Error bars indicate SD. (H) Schematic representation of the experiments. Each type of LIC was secondarily transplanted into mice. Bortezomib was injected twice weekly or injected once after incidence of leukemia. (I and J) Comparison of surface marker profiles in leukemic mice treated with bortezomib or vehicle. Representative FACS data (I) and relative percentages of Gr-1loc-Kithi fraction in MLL-ENL– or MOZ-TIF2–induced leukemic mice, and Gr-1loSca-1hi fraction in BCR-ABL/NUP98-HOXA9–induced leukemic mice are shown (n = 3 each) (J). Values of control mice were normalized to 100%. Error bars indicate SD. (K) Survival curves of mice in the experiments shown in H (n = 6 each).
Enforced activation of the NF-κB pathway increases LIC frequency in leukemic BM.
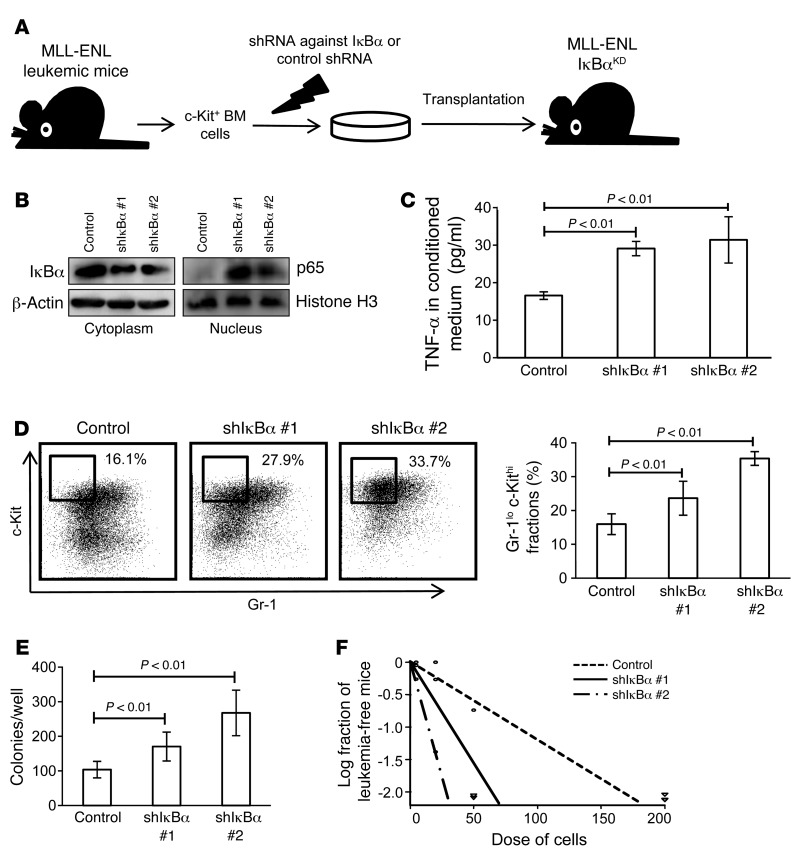
Given the supportive role of the NF-κB pathway in LIC proliferation as well as the differences in its activation status observed between LICs and non-LICs, we reasoned that the attenuation of NF-κB activity might be related to the transition from LICs to non-LICs. To test this hypothesis, we transduced MLL-ENL leukemia cells with a retrovirus encoding shRNA against IκBα and transplanted them into sublethally irradiated mice (Figure 6A). Because IκBα works as an inhibitor of NF-κB by holding it in the cytoplasm, its downregulation should function to enhance NF-κB activity, regardless of the basal proteasome activity. We first confirmed that MLL-ENL leukemia cells with shRNA-mediated knockdown of IκBα (MLL-ENL-IκBαKD) showed decreased IκBα protein levels in the cytoplasm and increased nuclear p65 protein levels, which would indicate that NF-κB signal was enhanced by the reduction of its cytoplasmic inhibitor (Figure 6B). In accordance with this finding, MLL-ENL-IκBαKD cells had a significantly greater ability to secrete TNF-α than did control cells, reflecting an activated NF-κB/TNF-α signaling loop (Figure 6C). We further investigated the phenotype of leukemic mice with MLL-ENL-IκBαKD. Interestingly, the BM of these MLL-ENL-IκBαKD mice showed a marked increase in immature Gr-1loc-Kithi cell populations (Figure 6D). Consistent with this change, we found that these leukemic cells had a greater CFC capacity (Figure 6E). Additionally, in order to investigate the frequency of LICs in BM mononuclear cells, we performed limiting dilution analysis by secondary transplantation of leukemia cells. Although the disease latency for leukemia development was not significantly different among the leukemia cells, MLL-ENL-IκBαKD leukemia cells had a marked abundance of LICs in the leukemic BM mononuclear cells compared with the control shRNA cells (Figure 6F and Supplemental Figure 10A). These data indicate that enforced NF-κB activation expands the LIC fraction in MLL-ENL leukemic BM cells. We also transduced normal BM cells with shRNAs against IκBα and transplanted them into lethally irradiated mice to test whether NF-κB activation by itself can induce leukemia or myeloproliferative-like disease. Over the 4-month follow-up period, the mice exhibited no significant change in peripheral blood values, indicating that NF-κB signal alone is not sufficient for leukemogenesis (Supplemental Figure 10B).
Figure 6. Forcible maintenance of NF-κB activity in leukemia cells enhances LIC frequency.
(A) Schematic representation of the experiments. c-Kit+ BM cells isolated from MLL-ENL leukemic mice were transduced with shRNA against IκBα or control shRNA and transplanted into sublethally irradiated mice. (B) Immunoblotting of cytoplasmic IκBα and nuclear p65 in BM mononuclear cells from MLL-ENL-IκBαKD mice compared with those from control leukemic mice. (C) TNF-α secretory ability of MLL-ENL- IκBαKD leukemia cells compared with that of control leukemia cells (n = 4 each). Error bars indicate SD. (D) Surface marker profiles of MLL-ENL leukemic mice with or without knockdown of IκBα. Representative FACS plots and mean percentages of Gr-1loc-Kithi fractions (n = 6 each). (E) CFC assay of MLL-ENL leukemia cells with or without knockdown of IκBα (n = 6). Cells were seeded at 500 cells per well. Error bars indicate SD. (F) LIC frequency in BM mononuclear cells derived from MLL-ENL-IκBαKD leukemic mice compared with those from control mice as determined by limiting dilution transplantation assay.
Significant correlation between NF-κB and TNF-α is observed in human AML LICs.
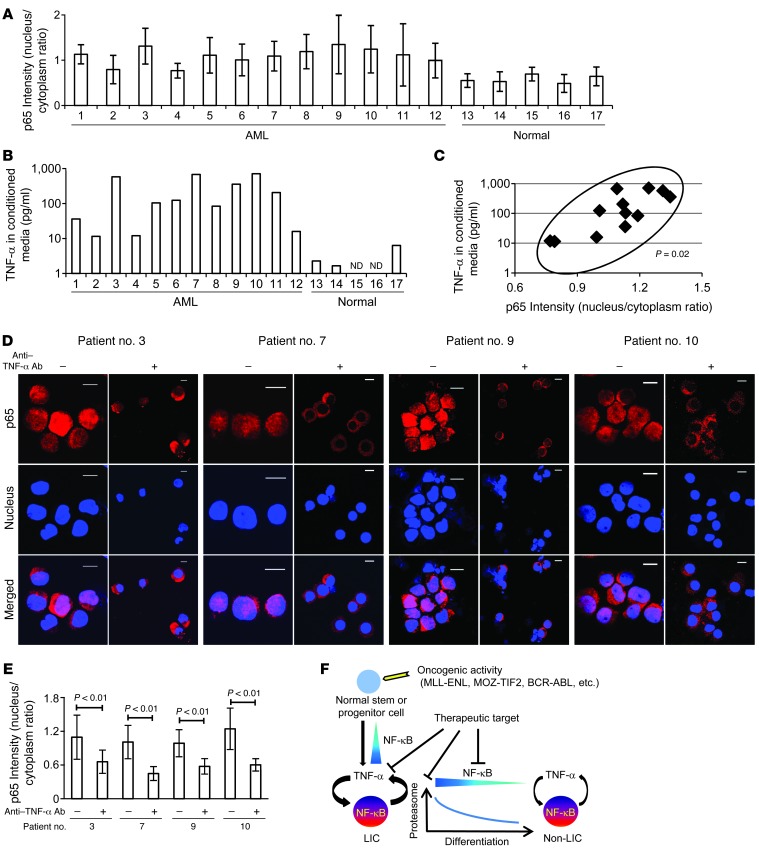
Finally, we investigated NF-κB/TNF-α positive feedback signaling in human AML LICs. We analyzed CD34+CD38– cells derived from 12 patients with previously untreated or relapsed AML and the same cell population from 5 normal BM specimens (Table 1) and evaluated their NF-κB signal intensity. We also quantified the concentration of TNF-α in the culture media conditioned by CD34+CD38– cells from each patient in order to measure the TNF-α secretory ability of these cells. As expected, our data from both of these analyses showed a wide variation among patients, one that might reflect a heterogeneous distribution and frequency of the LIC fraction in human AML cells, as was previously described (23). LICs in most of the patients did, however, show increased p65 nuclear translocation and TNF-α secretory potential compared with normal HSCs (Figure 7, A and B, and Supplemental Figure 11). We plotted these two parameters for each patient to compare between patients. Interestingly, a significant positive correlation was demonstrated statistically (P = 0.02), as LICS with enhanced p65 nuclear translocation showed a tendency toward abundant TNF-α secretion (Figure 7C). We also compared p65 intensity between LICs and non-LICs in 2 patients (patients 1 and 3) and found that p65 nuclear translocation was predominant in LICs, which is also consistent with the data obtained in murine AML cells (Supplemental Figure 11). Moreover, we cultured LICs with or without neutralizing antibodies against TNF-α and assessed p65 nuclear translocation to determine the effect of autocrine TNF-α on NF-κB activity. When incubated in the presence of TNF-α–neutralizing antibodies, nuclear translocation of p65 was significantly suppressed in LICs (Figure 7, D and E). These results support our hypothesis that a positive feedback loop exists between NF-κB and TNF-α in human AML LICs.
Table 1.
Clinical characteristics of the 12 patients with AML and the 5 patients with normal BM findings
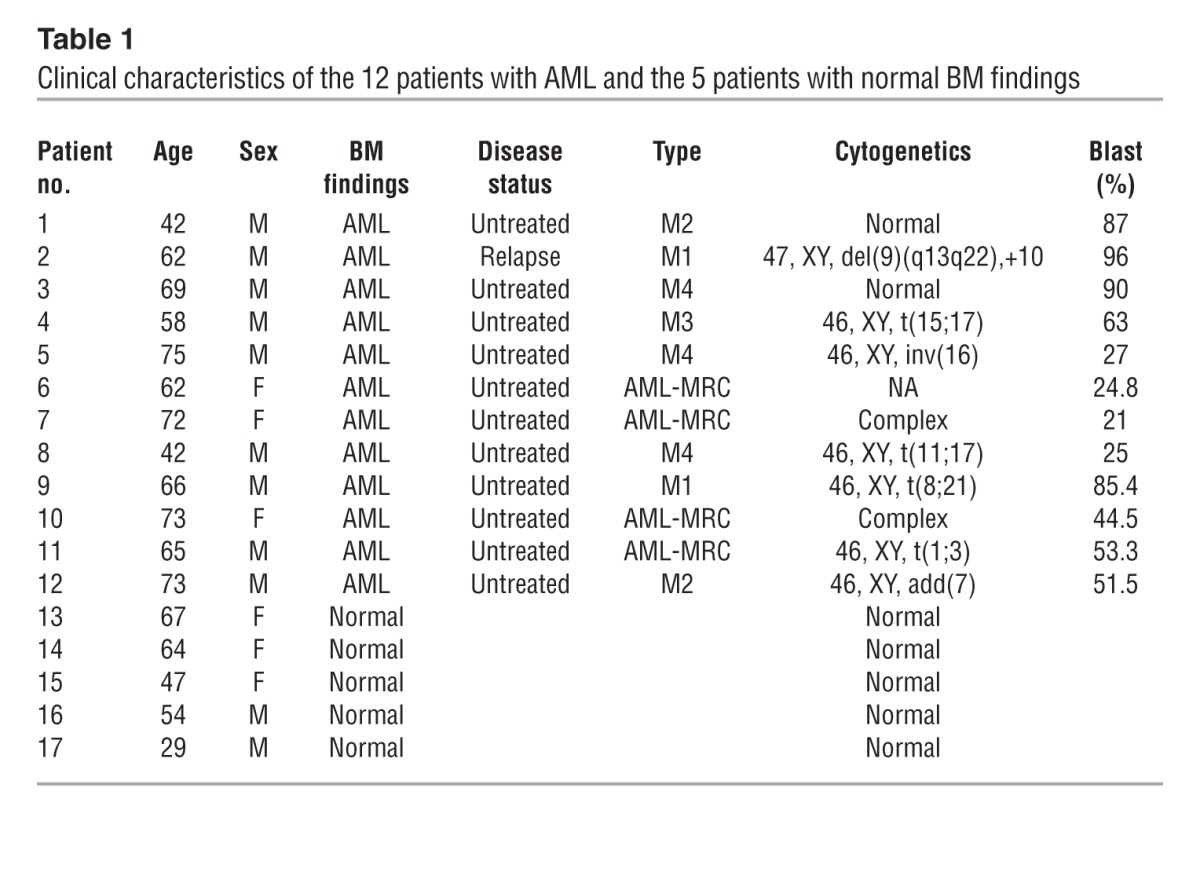
Figure 7. NF-κB/TNF-α positive feedback loop is activated in human AML LICs.
(A) Quantification of p65 nuclear translocation assessed by the mean nucleus/cytoplasm intensity ratio by immunofluorescence staining. The CD34+CD38– fractions isolated from AML or normal BM were analyzed. More than 50 cells were scored in each specimen, and the average intensity ratio with SD is shown. (B) TNF-α concentration of culture media conditioned by human AML LICs and normal HSCs measured by ELISA. ND, not detected. (C) Correlation between p65 nuclear translocation intensity ratio and TNF-α secretory ability of human AML LICs. (D) Immunofluorescence assessment of p65 nuclear translocation in LICs purified from 4 patients after serum-free culture with neutralizing antibody against TNF-α or isotype control. Scale bars: 10 μm. (E) Quantification of p65 nuclear translocation of LICs with or without neutralizing antibody against TNF-α assessed by the mean nucleus/cytoplasm intensity ratio. (F) Proposed model showing the role of NF-κB signaling in LICs. Positive feedback loop involving NF-κB/TNF-α promotes the maintenance and proliferation of LICs. The signaling is supported by active proteasome machinery, which declines with LIC differentiation.
Discussion
In the present study, we provide evidence that LICs, but not normal HSPCs or non-LIC fractions within leukemic BM, exhibit constitutive NF-κB pathway activity in different types of myeloid leukemia models. Moreover, we identified the underlying mechanism involved in the maintenance of this pathway activity, which had yet to be elucidated. We found that autocrine TNF-α secretion, with the support of enhanced proteasome activity, contributed to a constitutive activation of the NF-κB pathway in LICs. Although we observed different sensitivities to the inhibition of these signaling cascades according to the type of leukemia, these cascades play an important role in LIC proliferation, especially considering that the complete ablation of Tnf or Rela distinctly suppressed leukemia progression in vivo. These findings, which we validated in human AML LICs, could translate into improved AML treatment strategies.
The strong connection between inflammation and cancer has been increasingly discussed, and the NF-κB pathway is now recognized as a major regulator bridging the two pathological conditions in different types of malignancies. In most of these malignancies, aberrant activation of the NF-κB pathway derives from inflammatory microenvironments that are mainly created by proinflammatory immune cells such as tumor-infiltrating macrophages, neutrophils, and lymphocytes (34, 35). In this study, however, LICs retained their p65 nuclear translocation even after serum-free culture, suggesting that the constitutive NF-κB activity of LICs is maintained in an autonomous fashion. Through our investigation of gene expression profiles in LICs and normal HSCs, we found that LICs had distinctly elevated TNF-α expression levels that contributed to the maintenance of NF-κB activation in LICs. Conversely, the introduction of IκB-SR markedly suppressed TNF-α expression levels, indicating that NF-κB activity and TNF-α secretion create a positive feedback loop in LICs. Moreover, our hypothesis is strongly supported by our findings that a positive correlation exists between NF-κB and TNF-α secretory activities in human AML CD34+CD38– cells and that inhibition of autocrine TNF-α signaling attenuates p65 nuclear translocation. The role of TNF-α in the process of tumor promotion has recently been demonstrated in various types of solid tumors (36–39). It has also been reported that TNF-α is required for clonal evolution of myeloid malignancies (40). On the other hand, there has been controversy over the effect of TNF-α on leukemia cells when it was exogenously administered (41, 42). However, these previous studies did not address the critical question of whether endogenously secreted TNF-α is required for the maintenance of established leukemia cells, which is a crucially important aspect when considering therapeutic applications. We clearly reveal that the autonomously secreted TNF-α had beneficial effects on LIC proliferation through NF-κB activation, while the contribution of paracrine TNF-α secretion from BM microenvironments was minimal. Another important aspect of cytokine secretion by LICs that was not investigated in the present study is whether this secretion can exert some influence on BM stromal cells. Since the importance of bidirectional crosstalk between leukemia and niche cells through a variety of cytokines has increasingly been recognized (43), TNF-α secreted from LICs might also modulate the function of BM stromal cells, which could also have an impact on leukemia progression. Unveiling the role of TNF-α as a paracrine mediator would further extend the therapeutic options for AML.
Few studies have compared the NF-κB activity of different fractions within leukemia cells, and the mechanism underlying the difference in this activity has not been analyzed (44). We focused on proteasome activity as the essential machinery supporting NF-κB activity in LICs. Although high proteasome activity has been reported in various types of cancers (45, 46), its actual role in the malignant phenotype remained to be elucidated. In this study, we found that proteasome activity was especially high in LICs, which contributed to selective NF-κB activity in LICs via the efficient degradation of IκBα. Conversely, the inefficient NF-κB nuclear translocation we observed in non-LICs, despite TNF-α–enriched leukemic BM cells, could be explained by the low proteasome activity in these cells. Therefore, we postulate that both an activating stimulus such as TNF-α and high proteasome activity are required for efficient NF-κB signaling (Figure 7F). Both of these conditions are present exclusively in LICs, which acquire selective NF-κB activation. We also found that the expression levels of proteasome subunit genes were elevated in LICs compared with those in non-LICs, genes that could be involved in regulating proteasome function. Because we observed similar expression patterns in LICs and non-LICs in human AML cells, an elevated expression level of proteosome subunit genes might be one of the common characteristics of the LIC phenotype. Further studies will be needed to elucidate the regulatory mechanism of the proteasome gene families.
Our findings provide several advantages when considering their application to the clinical care setting. First, an activated NF-κB/TNF-α feedback loop was seen in AML LICs that had different genetic abnormalities. Although the therapeutic strategy of targeting aberrant molecules based on genetic abnormalities such as FLT3-ITD is promising, its application is limited to a particular group of patients. In contrast, inhibition of the NF-κB signal in addition to standard chemotherapy would show beneficial effects in most AML patients. Second, because there was a strong positive correlation between the NF-κB signal and TNF-α secretion, therapeutic efficacy could easily be inferred from the abundance of TNF-α instead of from evaluation of the activation status of NF-κB. Third, the NF-κB/TNF-α signal and enhanced proteasome activity are selectively seen in LICs, but not in normal HSCs. A recent study has shown that complete ablation of p65 in hematopoietic cells attenuates the long-term capacity for hematopoietic reconstitution (47). However, our data from the experiments in which we introduced IκB-SR into normal BM cells show that partial repression of NF-κB activity exerted minimal influence on normal hematopoiesis, while it markedly inhibited leukemia progression. These results indicate that there is a therapeutic window during which LICs can selectively be killed by NF-κB inhibition without seriously affecting normal hematopoiesis. Alternatively, there is some evidence that TNF-α has suppressive effects on normal HSCs (48, 49). The opposing role of TNF-α in LICs and HSCs is additionally beneficial, since anti–TNF-α therapy contributes to the recovery of normal hematopoiesis and attenuates LIC proliferation. Now that the TNF-α antagonist etanercept is widely used in inflammatory diseases such as rheumatoid arthritis, this drug might be a promising candidate for treating patients with AML.
In summary, the present study shows that blocking the NF-κB pathway offers a promising therapeutic approach for targeting LICs in various types of myeloid leukemia, without disturbing normal hematopoiesis. We further determined that autocrine TNF-α signaling and enhanced proteasome activity are crucial for maintaining constitutive NF-κB activity in LICs, findings that may also provide a new therapeutic opportunity.
Methods
Animals.
C57BL/6 mice and BALB/c mice were purchased from Japan SLC, Inc. Tnf-knockout mice on a BALB/c background were established as described previously (50). Rela-floxed mice on a C57BL/6 background were provided by H. Algül and R.M. Schmid (32). BALB/c mice were used as the controls in the experiments using Tnf-knockout mice, and C57BL/6 mice were used in the other experiments.
Retrovirus production and BM transplantation assays.
To obtain retrovirus supernatants, platinum-E (Plat-E) packaging cells were transiently transfected with each retrovirus vector, and the viral supernatants were collected 48 hours after transfection and used immediately for infection. To establish each myeloid leukemia mouse model, we used pMSCV-neo-MLL-ENL; pMSCV-MLL-ENL–internal ribosome entry site–EGFP (IRES-EGFP); pGCDNsam-MLL-ENL-IRES-Kusabira-Orange; pGCDNsam-MOZ-TIF2-IRES-EGFP; pGCDNsam-MOZ-TIF2-IRES-Kusabira-Orange; pGCDNsam-BCR-ABL-IRES-EGFP; pGCDNsam-BCR-ABL-IRES-Kusabira-Orange; and pMSCV-neo-NUP98-HOXA9. GMPs isolated from the BM of 8- to 10-week-old mice were transduced with the respective vectors and injected into sublethally irradiated (7.5 Gy) recipient mice. For experiments involving the generation of leukemia cells with IκB-SR, MLL-ENL leukemia cells were transduced with pBabe-GFP or pBabe-GFP-IκB-SR. MOZ-TIF2, and BCR-ABL/NUP98-HOXA9 leukemia cells were transduced with pGCDNsam-Kusabira-Orange or pGCDNsam-IκB-SR-IRES-Kusabira-Orange. For experiments involving the deletion of p65 in Rela-floxed mice, leukemia cells were established using Kusabira-Orange–containing retroviral vectors. The developed leukemia cells were transduced with pGCDNsam-EGFP or pGCDNsam-iCre-EGFP and transplanted into sublethally irradiated mice.
In vivo limiting dilution assays.
Varying numbers of cells from different populations were transplanted into sublethally irradiated mice and monitored for disease development (see Supplemental Table 1 for the injected cell numbers).
Immunofluorescence and quantification of p65 nuclear translocation.
A total of 1 × 104 to 5 × 104 cells were cytospun onto glass slides. The cells were fixed with 3.7% formaldehyde in PBS for 30 minutes, permeabilized by treatment with 0.2% Triton X in PBS for 10 minutes, and blocked with 1% BSA in PBS for 60 minutes. Then, the slides were incubated with rabbit anti–p65 polyclonal antibody (sc-372; 1:100 dilution; Santa Cruz Biotechnology Inc.) overnight at 4°C, followed by incubation with Alexa Fluor 555 goat anti-mouse IgG (1:250 dilution; Invitrogen) and TO-PRO3 (1:1,000 dilution; Invitrogen) for 90 minutes. For immunofluorescence staining of Kusabira-Orange+ leukemia cells, Alexa Fluor 647 goat anti-mouse IgG (1:250 dilution; Invitrogen) was used as a secondary antibody, and the nucleus was stained with DAPI. After the cells were washed, they were treated with ProLong Gold Antifade Reagent (Invitrogen). Images were acquired using an Olympus FluoView FV10i confocal microscope with a ×60 objective oil immersion lens. The mean intensity of p65 in the nucleus and cytoplasm of each cell was measured within a region of interest (ROI) placed within the nucleus and cytoplasm. Similarly, the background intensity was quantified within an ROI placed outside the cells. All the measurements were performed using FluoView software. The background-subtracted intensity ratio of nucleus/cytoplasm was calculated in more than 50 cells in each specimen, and the average intensity with SD is presented.
Flow cytometry.
Isolation of each fraction from normal or leukemic BM cells was performed using a FACSAria II (BD) cell sorter. For isolation of GMPs and KSLs, biotinylated antibodies against Gr-1 (RB6-8C5), CD11b (M1/70), B220 (RA-3-6B2), CD3 (145-2C11), CD4 (GK1.5), CD8 (53-6.7), and TER119 were used for lineage staining. A PerCP-Cy5.5–labeled streptavidin antibody was used for secondary staining, together with APC–anti–c-Kit (2B8), PE-Cy7–anti–Sca-1 (E13-161.7). FITC–anti-CD34 (RAM34), and PE–anti-CD16/32b antibodies (clone 93). The following antibodies were used for isolation of L-GMPs from GFP-containing leukemia cells: APC-Cy7–anti–streptavidin, PE-Cy5–anti–c-Kit (2B8), PE-Cy7–anti–Sca-1 (E13-161.7), Alexa Fluor 647–anti–CD34 (RAM34), and PE–anti-CD16/32b (clone 93). APC-antistreptavidin and PE-Cy7–anti–Sca-1 antibodies (E13-161.7) were used for sorting LICs and non-LICs in the BCR-ABL plus NUP98-HOXA9 leukemia model. See Supplemental Figures 1 and 2 for detailed FACS plots. For analysis of TNF receptor expression in leukemia cells, biotinylated antibodies against TNF receptor I or II (55R-170) and an APC-Cy7–antistreptavidin antibody were used. Analysis was performed using FlowJo software (Tree Star Inc.).
Real-time quantitative PCR.
Real-time quantitative PCR was carried out on the LightCycler480 system (Roche) using SYBR green reagents according to the manufacturer’s instructions. The results were normalized to Gapdh levels. Relative expression levels were calculated using the 2-ΔΔCt method (51). The following primers were used for real-time PCR experiments: Gapdh forward, TGGCCTCCAAGGAGTAAGAA, and reverse, GGTCTGGGATGGAAATTGTG; Ncf2 forward, CCAGAAGACCTGGAATTTGTG, and reverse, AAATGCCAACTTTCCCTTTACA; Tnf forward, TCTTCTCATTCCTGCTTGTGG, and reverse, GGTCTGGGCCATAGAACTGA; Il15ra forward, TAAGCGGAAAGCTGGAACAT, and reverse, TGAGGTCACCTTTGGTGTCA; Litaf forward, CTCCAGGACCTTACCAAGCA, and reverse, AGGTGGATTCATTCCCTTCC; Hoxa9 forward, GGTGCCTGCTGCAGTGTAT, and reverse, GTTCCAGCCAGGAGCGCATAT; Psma5 forward, CGAGTACGACAGGGGTGTG, and reverse, TGGATGCCAATGGCTGTAG; Psmd4 forward, GTACATGCGGAACGGAGACT, and reverse, TGTGGTCAGCACCTCACAGT; Psme3 forward, TTTCAGAGAGCGGATCACAA, and reverse, GGTCATGGATATTTAGAATTGGTTC.
siRNA interference.
Specific shRNAs targeting murine Ikba mRNA were designed and cloned into pSIREN-RetroQ-ZaGreen vectors. Control shRNA is a nonfunctional construct provided by Clontech. The target sequences, from 5′ to 3′, were: CCGAGACTTTCGAGGAAAT (shIκBα number 1), and AGCTGACCCTGGAAAATCT (shIκBα number. 2).
Immunoblotting.
Membranes were probed with the following antibodies: anti-IκBα (Cell Signaling Technology), anti–phospho-IκBα (Ser32) (Cell Signaling Technology), anti-p65 (Santa Cruz Biotechnology Inc.), anti–phospho-p65 (Ser536) (Cell Signaling Technology), anti–β-actin (Cell Signaling Technology), and anti–histone H3 (Cell Signaling Technology). Protein levels were quantified with ImageJ software (NIH). To obtain nuclear and cytoplasmic extracts, an Active Motif Nuclear Extract Kit was used according to the manufacturer’s instructions. Cycloheximide treatment assay was performed as described previously, with modification (52). Cells were pretreated with MG132 (20 μM) for 1 hour to initially inhibit the proteasomal degradation of IκBα. Cells were washed twice with medium, then cultured with or without 10 μg/ml of cycloheximide for an additional hour and harvested.
CFC assays.
In each experiment, cells were plated onto MethoCult GF M3434 medium (STEMCELL Technologies). Colony numbers in each dish were scored on day 7.
Measurement of TNF-α levels in BM extracellular fluid and conditioned media.
BM extracellular fluid was obtained by flushing bilateral femurs and tibia of individual mice with 400 μl PBS. The supernatant was collected after centrifugation. To obtain conditioned media, 0.3–1.0 × 106 murine leukemia cells or normal GMPs were cultured in RPMI medium containing 10% FBS and 10 ng/ml IL-3. After a 48-hour incubation, the culture supernatants were collected. The concentration of TNF-α was measured using a murine TNF-α ELISA kit (Gen-Probe Diaclone) according to the manufacturer’s instructions. Similarly, 0.5 × 104 to 2.0 × 104 human AML or normal CD34+CD38– cells were cultured for 48 hours in RPMI medium containing 10% FBS and 100 ng/ml SCF, IL-3, and thrombopoietin. The concentration of TNF-α in the harvested supernatants was measured with a human TNF-α Quantikine ELISA kit (R&D Systems).
20S proteasome activity.
A 20S proteasome activity assay kit (Cayman Chemical) was used to analyze proteasome activity. A total of 5 × 104 freshly isolated normal GMPs, LICs, and non-LICs in each model were assayed according to the manufacturer’s protocol. As a control, the proteasome activity of each cell was also assayed after the specific proteasome inhibitor epigallocatechin gallate was added. Fluorescence was measured with a Wallac ARVO V (PerkinElmer), and the proteasome activity of each cell type was calculated by subtracting the respective control value.
Bortezomib treatment studies.
For in vivo treatment experiments, LICs of each leukemia model were injected into sublethally irradiated mice: 1 × 103 cells in the MLL-ENL or BCR-ABL/NUP98-HOXA9 models, and 1 × 104 cells in the MOZ-TIF2 model. Bortezomib was administrated i.p. at doses of 1.0 mg/kg twice weekly for 3 weeks. Treatment was started 1 week after transplantation in the MLL-ENL or BCR-ABL/NUP98-HOXA9 models, and 2 weeks after transplantation in the MOZ-TIF2 model. For experiments analyzing changes in LIC populations, bortezomib was administrated i.p. at doses of 1.0 mg/kg into fully developed leukemic mice. GFP+ BM cells were collected 24 hours after injection, and surface marker profiles were analyzed.
Analysis of microarray data.
We analyzed publicly available gene expression microarray data on murine and human samples from the Gene Expression Omnibus (GEO) database (GEO GSE24797, GSE20377, and GSE24006). A set of CEL files were downloaded from GEO and normalized using the JustRMA function from the Affy package 1.22.1 in Bioconductor. To compare expression profiles of the NF-κB target genes, normalized data were tested for GSEA using previously described NF-κB target gene sets (29), and a nominal P value was calculated. For screening of genes with elevated expression levels in LICs compared with those in normal HSPCs, the expression values of individual genes were compared between groups. Genes significantly elevated in LICs from all three leukemia models as determined by an unpaired Student’s t test (P < 0.05) were selected, among which genes also elevated in human AML LICs (Student’s t test set at P < 0.01) were ultimately selected.
Statistics.
Statistical significance of differences between groups was assessed with a 2-tailed unpaired Student’s t test. Differences were considered statistically significant at a P value of less than 0.05. LIC frequency was calculated by Poisson statistics. In leukemia cell transplantation experiments, the overall survival of mice in BM transplantation assays is depicted by a Kaplan-Meier curve. Survival between groups was compared using the log-rank test. To measure the correlation between NF-κB intensity and TNF-α secretion in human AML samples, the Spearman’s rank correlation coefficient was used.
Study approval.
A total of 12 BM cells derived from patients with AML were obtained from the Department of Hematology and Oncology of the University of Tokyo Hospital. Five BM cells from patients diagnosed with lymphoid neoplasia without BM invasion were used as normal controls. The study was approved by the ethics committee of the University of Tokyo, and written informed consent was obtained from all patients whose samples were collected. All animal experiments were approved by the University of Tokyo Ethics Committee for Animal Experiments.
Supplementary Material
Acknowledgments
We thank T. Kitamura for the Plat-E packaging cells; H. Nakauchi and M. Onodera for the pGCDNsam-IRES-EGFP retroviral vector; R. Ono and T. Nosaka for the MLL-ENL cDNA; I. Kitabayashi for the MOZ-TIF2 cDNA; W. Hahn for the pBabe-GFP and pBabe-GFP-IκB-SR; and H. Algül and R.M. Schmid for providing the Rela-floxed mice. This work was supported by a Grant-in-Aid for Scientific Research A (KAKENHI 12020240) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2014;124(2):528–542. doi:10.1172/JCI68101.
References
- 1.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 2.Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa F, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25(11):1315–1321. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
- 4.Marcucci G, Haferlach T, Döhner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J Clin Oncol. 2011;29(5):475–486. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 5.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sen R, Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-κB by a posttranslational mechanism. Cell. 1986;47(6):921–928. doi: 10.1016/0092-8674(86)90807-X. [DOI] [PubMed] [Google Scholar]
- 7.La Rosa FA, Pierce JW, Sonenshein GE. Differential regulation of the c-myc oncogene promoter by the NF-κB rel family of transcription factors. Mol Cell Biol. 1994;14(2):1039–1044. doi: 10.1128/mcb.14.2.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttridge DC, Albanese C, Reuther JY, Pestell RG, Baldwin AS., Jr NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol. 1999;19(8):5785–5799. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duckett CS. Apoptosis and NF-κB: the FADD connection. J Clin Invest. 2002;109(5):579–580. doi: 10.1172/JCI15197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karin M, Greten FR. NF-κB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5(10):749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 11.Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441(7092):431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 12.Pikarsky E, et al. NF-κB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431(7007):461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 13.Guzman ML, et al. Nuclear factor-κB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood. 2001;98(8):2301–2307. doi: 10.1182/blood.V98.8.2301. [DOI] [PubMed] [Google Scholar]
- 14.Guzman ML, et al. Preferential induction of apoptosis for primary human leukemic stem cells. Proc Natl Acad Sci U S A. 2002;99(25):16220–16225. doi: 10.1073/pnas.252462599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frelin C, et al. Targeting NF-κB activation via pharmacologic inhibition of IKK2-induced apoptosis of human acute myeloid leukemia cells. Blood. 2005;105(2):804–811. doi: 10.1182/blood-2004-04-1463. [DOI] [PubMed] [Google Scholar]
- 16.Carvalho G, et al. Inhibition of NEMO, the regulatory subunit of the IKK complex, induces apoptosis in high-risk myelodysplastic syndrome and acute myeloid leukemia. Oncogene. 2007;26(16):2299–2307. doi: 10.1038/sj.onc.1210043. [DOI] [PubMed] [Google Scholar]
- 17.Guzman ML, et al. An orally bioavailable parthenolide analog selectively eradicates acute myelogenous leukemia stem and progenitor cells. Blood. 2007;110(13):4427–4435. doi: 10.1182/blood-2007-05-090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkins C, et al. Nuclear factor-κB as a potential therapeutic target for the novel cytotoxic agent LC-1 in acute myeloid leukaemia. Br J Haematol. 2008;143(5):661–671. doi: 10.1111/j.1365-2141.2008.07392.x. [DOI] [PubMed] [Google Scholar]
- 19.Jin Y, et al. Antineoplastic mechanism of niclosamide in acute myelogenous leukemia stem cells: inactivation of the NF-κB pathway and generation of reactive oxygen species. Cancer Res. 2010;70(6):2516–2527. doi: 10.1158/0008-5472.CAN-09-3950. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi S, et al. Over-expression of Flt3 induces NF-κB pathway and increases the expression of IL-6. Leuk Res. 2005;29(8):893–899. doi: 10.1016/j.leukres.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Liu S, et al. Sp1/NFκB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell. 2010;17(4):333–347. doi: 10.1016/j.ccr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagawa M, et al. AML1/RUNX1 functions as a cytoplasmic attenuator of NF-κB signaling in the repression of myeloid tumors. Blood. 2011;118(25):6626–6637. doi: 10.1182/blood-2010-12-326710. [DOI] [PubMed] [Google Scholar]
- 23.Eppert K, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17(9):1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 24.Sarry JE, et al. Human acute myelogenous leukemia stem cells are rare and heterogenous when assayed in NOD/SCID/IL2Rγc-deficient mice. J Clin Invest. 2011;121(1):384–395. doi: 10.1172/JCI41495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu T, et al. Functional characterization of meningioma 1 as collaborating oncogene in acute leukemia. Leukemia. 2010;24(3):601–612. doi: 10.1038/leu.2009.272. [DOI] [PubMed] [Google Scholar]
- 26.Kvinlaug BT, et al. Common and overlapping oncogenic pathways contribute to the evolution of acute myeloid leukemias. Cancer Res. 2011;71(12):4117–4129. doi: 10.1158/0008-5472.CAN-11-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neering SJ, et al. Leukemia stem cells in a genetically defined murine model of blast-crisis CML. Blood. 2007;110(7):2578–2585. doi: 10.1182/blood-2007-02-073031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, et al. The Wnt/β-catenin pathway is required for the development ofleukemia stem cells in AML. Science. 2010;327(5973):1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinz M, et al. Nuclear factor κB-dependent gene expression profiling of Hodgkin’s disease tumor cells, pathogenetic significance, and link to constitutive signal transducer and activator of transcription 5a activity. J Exp Med. 2002;196(5):605–617. doi: 10.1084/jem.20020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 2010;304(24):2706–2715. doi: 10.1001/jama.2010.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kishore N, et al. A selective IKK-2 inhibitor blocks NF-κB-dependent gene expression in interleukin-1 β-stimulated synovial fibroblasts. J Biol Chem. 2003;278(35):32861–32871. doi: 10.1074/jbc.M211439200. [DOI] [PubMed] [Google Scholar]
- 32.Algül H, et al. Pancreas-specific RelA/p65 truncation increases susceptibility of acini to inflammation-associated cell death following cerulein pancreatitis. J Clin Invest. 2007;117(6):1490–1501. doi: 10.1172/JCI29882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beg AA, Finco TS, Nantermet PV, Baldwin AS., Jr Tumor necrosis factor and interleukin-1 lead to phosphorylation and loss of IκBα: a mechanism for NF-κB activation. Mol Cell Biol. 1993;13(6):3301–3310. doi: 10.1128/mcb.13.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeNardo DG, Coussens LM. Inflammation and breast cancer. Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9(4):212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLean MH, et al. The inflammatory microenvironment in colorectal neoplasia. PLoS One. 2011;6(1):e15366. doi: 10.1371/journal.pone.0015366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charles KA, et al. The tumor-promoting actions of TNF-α involve TNFR1 and IL-17 in ovarian cancer in mice and humans. J Clin Invest. 2009;119(10):3011–3023. doi: 10.1172/JCI39065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore RJ, et al. Mice deficient in tumor necrosis factor-α are resistant to skin carcinogenesis. Nat Med. 1999;5(7):828–831. doi: 10.1038/10552. [DOI] [PubMed] [Google Scholar]
- 38.Popivanova BK, et al. Blocking TNF-α in mice reduces colorectal carcinogenesis associated with chronic colitis. J Clin Invest. 2008;118(2):560–570. doi: 10.1172/JCI32453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egberts JH, et al. Anti-tumor necrosis factor therapy inhibits pancreatic tumor growth and metastasis. Cancer Res. 2008;68(5):1443–1450. doi: 10.1158/0008-5472.CAN-07-5704. [DOI] [PubMed] [Google Scholar]
- 40.Li J, et al. TNF-α induces leukemic clonal evolution ex vivo in Fanconi anemia group C murine stem cells. J Clin Invest. 2007;117(11):3283–3295. doi: 10.1172/JCI31772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoang T, Levy B, Ouetto N, Hainan A, Rodriguez-Cimadevilla JC. Tumor necrosis factor α stimulates the growth of the clonogenic cells of acute myeloblastic leukemia in synergy with granulocyte-macrophage colonystimulating factor. J Exp Med. 1989;170(1):15–26. doi: 10.1084/jem.170.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khoury E, et al. Tumor necrosis factor alpha (TNF α) downregulates c-kit proto-oncogene product expression in normal and acute myeloid leukemia CD34+ cells via p55 TNF alpha receptors. . Blood. 1994;84(8):2506–2514. [PubMed] [Google Scholar]
- 43.Zhang B, et al. Altered microenvironmental regulation of leukemic and normal stem cells in chronic myelogenous leukemia. Cancer Cell. 2012;21(4):577–592. doi: 10.1016/j.ccr.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerbauy DM, Lesnikov V, Abbasi N, Seal S, Scott B, Deeg HJ. NF-κB and FLIP in arsenic trioxide (ATO)-induced apoptosis in myelodysplasticsyndromes (MDSs). Blood. 2005;106(12):3917–3925. doi: 10.1182/blood-2005-04-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5(5):417–421. doi: 10.1016/S1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 46.Chen L, Madura K. Increased proteasome activity, ubiquitin-conjugating enzymes, and eEF1A translation factor detected in breast cancer tissue. Cancer Res. 2005;65(13):5599–5606. doi: 10.1158/0008-5472.CAN-05-0201. [DOI] [PubMed] [Google Scholar]
- 47.Stein SJ, Baldwin AS. Deletion of the NF-κB subunit p65/RelA in the hematopoietic compartment leads to defects in hematopoietic stem cell function. Blood. 2013;121(25):5015–5024. doi: 10.1182/blood-2013-02-486142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iversen PO, Wiig H. Tumor necrosis factor α and adiponectin in bone marrow interstitial fluid from patients with acute myeloidleukemia inhibit normal hematopoiesis. Clin Cancer Res. 2005;11(19 pt 1):6793–6799. doi: 10.1158/1078-0432.CCR-05-1033. [DOI] [PubMed] [Google Scholar]
- 49.Pronk CJ, Veiby OP, Bryder D, Jacobsen SE. Tumor necrosis factor restricts hematopoietic stem cell activity in mice: involvement of 2 distinct receptors. J Exp Med. 2011;208(8):1563–1570. doi: 10.1084/jem.20110752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taniguchi T, Takata M, Ikeda A, Momotani E, Sekikawa K. Failure of germinal center formation and impairment of response to endotoxin in tumor necrosis factor alpha-deficient mice. Lab Invest. 1997;77(6):647–658. [PubMed] [Google Scholar]
- 51.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the Comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 52.Jain AK, Bloom DA, Jaiswal AK. Nuclear import and export signals in control of Nrf2. J Biol Chem. 2005;280(32):29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.