Abstract
Molecular mechanisms leading to myocardial injury during warm or cold ischemia are insufficiently understood. Although proteasomes are thought to contribute to myocardial ischemia-reperfusion injury, their roles during the ischemic period remain elusive. Because donor hearts are commonly exposed to prolonged global cold ischemia prior to cardiac transplantation, we evaluated the role and regulation of the proteasome during cold ischemic storage of rat hearts in context of the myocardial ATP content. When measured at the actual tissue ATP concentration, cardiac proteasome peptidase activity increased by 225% as ATP declined during cold ischemic storage of hearts in University of Wisconsin (UW) solution for up to 48h. Addition of the specific proteasome inhibitor epoxomicin to the UW solution inhibited proteasome activity in the cardiac extracts, significantly reduced edema formation and preserved the ultrastructural integrity of the cardiomyocyte. Utilizing purified 20S/26S proteasome enzyme preparations, we demonstrate that this activation can be attributed to a subset of 26S proteasomes which are stable at ATP concentrations far below physiological levels, that ATP negatively regulates its activity and that maximal activation occurs at ATP concentrations in the low mol/L range. These data suggest that proteasome activation is a pathophysiologically relevant mechanism of cold ischemic myocardial injury. A subset of 26S proteasomes appears to be a cell destructive protease that is activated as ATP levels decline. Proteasome inhibition during cold ischemia preserves the ultrastructural integrity of the cardiomyocyte.
Keywords: Proteasome, ATP, ischemia, hypothermia, injury, heart
Introduction
Hypothermic storage of a donor organ is a commonly used method to protect it from ischemic injury. However, in cardiac transplantation, cold ischemic storage of human hearts is limited to 4–6h and the risk of primary graft failure and death rises dramatically as ischemic time increases[1]. A significant amount of research has been focused on reperfusion injury in post-ischemic hearts. However, much less is known on the molecular mechanisms leading to organ injury within the ischemic period.
Recent observations implied a role of proteasomes in various cardiac pathologies, including myocardial warm and cold ischemia-reperfusion injury[2–4]. During reperfusion of post ischemic hearts partial inactivation of the proteasome has been reported and was suggested to contribute to impaired removal of oxidized proteins[4–7]. In addition, previous findings suggested that proteasome dysfunction may already occur in the cold ischemic heart prior to reperfusion[4].
ATP/Mg2+ is known to regulate 26S activity, its assembly and stability[8–11]. Although myocardial ischemia is associated with depletion of the tissue ATP content, the possible role and regulation of proteasomes during cardiac ischemia have not been evaluated in context of the actual tissue ATP content. Therefore, the purpose of this study was to evaluate the regulation of the cardiac proteasome during cold ischemia (CI) under conditions that resemble the cardiac ATP content and to assess whether proteasomes contribute to cold ischemic myocardial injury.
Material and Methods
Animal protocol
All procedures were approved by the Institutional Animal Care and Use Committee. Male Lewis rats (200–250g, Harlan) were anesthetized (1%–2% inhaled isoflurane; Baxter) and hearts were harvested as described[4]. In brief, the abdomen was opened and heparin (1000U) was injected into the inferior vena cava. A midline thoracotomy was performed, the heart dissected and flushed with normal saline. For cardioplegic arrest, the vena cavae were transsected to vent the heart and cold (4°C) University of Wisconson solution (UW, 10mL, ViaSpan, Duramed) were injected into the ascending aorta. Hearts were stored in UW ± 50 μmol/L epoxomicin (Boston Biochem) at 4°C for up to 48h. Biopsies from the left ventricle were taken for histopathological and ultrastructural evaluation and determination of wet-weight dry-weight (W/D) ratios. The remaining tissue was snap frozen and stored at −80°C until processing.
W/D ratios were determined gravimetrically, as described[12].
ATP concentrations ([ATP])
Hearts were homogenized in 1% TCA, centrifuged (2000g, 10min, 4°C) and supernatants collected. Supernatants were diluted 1:10(volume/volume) in 50mmol/L Tris-acetate, 2mmol/L EDTA, pH7.75, and ATP determined using a bioluminescence assay (Invitrogen), as described[11].
Preparation of heart extracts
Frozen hearts were homogenized in 1/10 phosphate buffered saline, pH7.4(1:5 weight/volume), centrifuged (20,000g, 4°C, 30min) and supernatants (= extracts) aliquoted[4].
Proteasomes
Highly purified 20S/26S enzyme preparations derived from human erythrocytes were obtained from Biomol. The 26S consist of 20S which are singly and doubly capped with 19S regulator complexes in a molar ratio of 1:1.5[11].
Peptidase activities
Peptidase activities were measured employing the fluorogenic peptide substrates N-Suc-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (chymotryptic-like, CT-L), Bz-Val-Gly-Arg-7-amino-4-methylcoumarin (tryptic-like, T-L) and Z-Leu-Leu-Glu-AMC (caspase-like, Casp-L) (all from Biomol), as described[4, 11]. Reaction mixtures contained 1mmol/L DTE, 0–5mmol/L ATP, 0–5mmol/L MgCl2, 10mmol/L Tris/HCl, pH7.5, 100 μmol/L peptide substrate and 1mg/mL cardiac extracts or 4 μg/mL 20S/26S proteasomes. To differentiate the proteasome from other peptidase activities in cardiac extracts, the epoxomicin sensitive proportions were determined by addition of 7 μmol/L epoxomicin to the incubation mixtures[4, 13]. Proteasome peptidase activity was calculated as total peptidase activity minus peptidase activity in the presence of epoxomicin. All enzyme assays were performed immediately after preparation of the cardiac extracts to prevent from proteasome inactivation by freeze-thawing. Enzyme time-progression curves showed linearity for 40min for all activities.
Proteasome ELISA
20S/26S ELISAs were performed as described[11]. For the 26S ELISA, all buffers contained ATP/Mg2+ as described in the results section.
Western blotting
Western blotting with anti-ubiquitin (Sigma) and densitometric quantification of the chemiluminescence signals were performed as described[4]. Anti-glyceraldehyde 3-phosphate dehydrogenase (Anti-GAPDH; 1:4000; Applied Biosciences) in combination with HRP labeled anti-mouse (1:5000; GE Healthcare) were used as control for the protein transfer to the blotting membranes.
Analytical gel filtration was performed on a Superose6-column (GE Healthcare) in 25mmol/L Tris/HCl, pH7.5, 100mmol/L NaCl, 50 μmol/L ATP, 5mmol/L Mg2+ at 5°C using the DuoFLow-chromatography system (Bio-Rad). The flow rate was 0.25mL/min and fractions of 0.5mL were collected. The column was calibrated using proteins of known molecular mass.
Light microscopy
Biopsies from the left ventricle were fixed in 10% formalin and embedded in paraffin. Sections (thickness: 5μm) were stained with hematoxylin and eosin (H&E) and Masson’s trichrome stain. Slides were examined under light microscopy (magnification ×40, ×100, ×200, ×400) by a pathologist (M.M.P.) who was blinded as to the treatment of the hearts.
Transmission electron microscopy (TEM)
Biopsies of the left ventricle were fixed with 4% glutaraldehyde, postfixed in 1%OsO4, 0.1mol/L cacodylate buffer, and embedded as monolayers in Embed 812. Ultrathin sections (90nm) were stained with uranyl acetate and lead citrate and analyzed by TEM (Hitachi H-600, Pleasanton, CA) by a pathologist (M.M.P.) who was blinded as to the treatment of the hearts.
Statistics
Data are expressed as mean±SEM. Student’s t-test and one-way ANOVA with Tukey post-hoc correction for multiple comparisons were calculated with the SPSS-program (Chicago, IL). Regression analyses were calculated with the GraphPad-Prism-program (GraphPad-Software, San Diego, CA). A two-tailed p<0.05 was considered significant.
Results
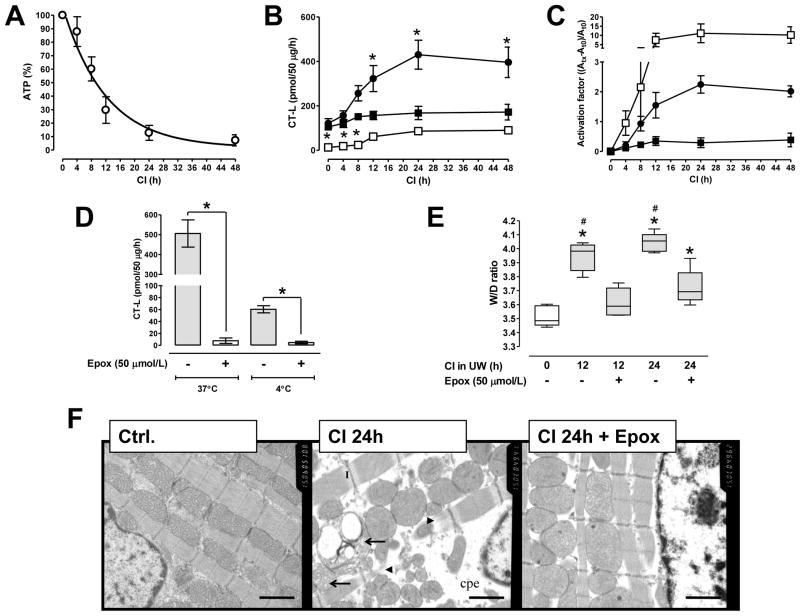
The myocardial ATP content in heart extracts during CI decreased with t1/2 of 8.3±2h (Fig. 1A). When measured at the physiologiocal myocardial [ATP,5mmol/L][14] and 37°C, proteasome CT-L activity increased during CI by 30–40%. When measured at the actual tissue [ATP], this activity increased by 225% within 24h of CI and remained constant until 48h (Fig. 1B/C). As compared with measurements at 37°C, proteasome CT-L activity in non-ischemic heart extracts was 8-fold lower at 4°C (pmol/50 μg/h: 105±14 at 37°C vs. 13±4 at 4°C;p<0.05). When measured at 4°C, proteasome activity increased within 12h of cold ischemic storage by 750% and remained constant thereafter (Fig. 1B/C).
Fig. 1.
A: ATP content in hearts during CI in UW solution, in % of non-ischemic hearts (n=5/time point). Data were analyzed by non-linear regression analysis (r2=0.81). B: Proteasome CT-L activity in hearts from A. Activity was measured at 5mmol/L ATP and 37°C(■), at the actual tissue ATP content (as determined in A: CI0h: 5mmol/L(=100%), CI4h: 4.25mmol/L(=85%), CI8h: 3mmol/L(=60%), CI12h: 1.5mmol/L(=30%), CI24 h: 0.75mmol/L(=15%) and CI48h: 0.5mmol/L(=10%)) and 37°C(●) or at 4°C(□). All incubation mixtures contained 5mmol/L Mg2+. *: p<0.05 vs. non-ischemic hearts measured at 5mmol/L ATP, 37°C. C: To assess proteasome peptidase activity activation at each condition (same symbols as in B), the activation factor was calculated as (Atx−At0)/At0 in which At0 is the activity in extracts from non-ischemic hearts (=0min CI) and Atx the activity after a given period of CI.
D: CT-L proteasome activity in extracts from hearts after 24h of CI in UW supplemented with (open bars) or without (grey bars) epoxomicin. n=5/group. Enzyme activities were measured at the actual tissue ATP content and at 37°C or 4°C, as indicated in the graph. *: p<0.05 for the comparison between hearts stored with and without epoxomicin. E: W/D ratios of hearts during CI in UW with (+) or without (−) epoxomicin, n=5/group. Boxes extend from the 25th to 75th percentile, the horizontal line shows the median. Error bars show the range of data (min/max). *: p<0.05 vs. non-ischemic hearts (open box). #: p<0.05 vs. hearts stored for the same duration in the presence of epoxomicin. F: Representative photomicrographs showing transmission electron microscopy (15000x) in a non-ischemic heart after cardioplegic arrest (ctrl.), in the heart after cardioplegic arrest and 24h of CI in UW (CI24h) and after CI in UW supplemented with 50 μmol/L of epoxomicin (CI24h+Epox). cpe: cytoplasmatic and perinuclear edema. I: I-bands. Arrows: ballooned and partly disrupted mitochondria. Arrowheads: disrupted myofibrils. Scale bars − 1 μm.
After CI in UW supplemented with epoxomicin, proteasome peptidase activity in the heart extracts was inhibited by more than 90% when measured at 37°C and 4°C (Fig. 1D). [ATP] were comparable after CI in UW with (200±37nmol ATP/g wet weight) and without (238±25nmol ATP/g wet weight) epoxomicin.
W/D ratios increased from 3.51±0.07 in non-ischemic hearts to 3.94±0.04 and 4.04±0.07 after 12h and 24h of CI, respectively (p<0.001 for both time points). This increase was significantly attenuated with epoxomicin (W/D ratios: 3.61±0.05 and 3.72±0.12 after 12h and 24h, respectively; p<0.01 both time points) (Fig. 1E).
Normal hearts and hearts after 24h of CI with or without epoxomicin were indistinguishable when evaluated by light microscopy (Fig. S1A/B). TEM demonstrated profound impairment of the myocardial ultrastructure after 24h of CI, as reflected by pronounced perinuclear and cytoplasmatic edema, ballooned and partly disrupted mitochondria with flocculent densities, prominent I-bands, disrupted myofibrills and clumped and marginated chromatin of the nucleus (Fig. 1F and Fig. S1C, center panels). All of these ultrastuctural changes were attenuated with epoxomicin (right panels).
A representative image from Western blotting experiments with extracts from a non-ischemic heart and hearts after 24h of CI in UW with and without epoxomicin is shown in Fig. S2A. Because the chemiluminescence signals for free ubiquitin were much stronger than the signals for ubiquitin-protein conjugates (>15kDa; Fig. S2A–left panel), the latter were visualized with a longer exposure time (Fig. S2A–right panel). The signal corresponding to free ubiquitin was not affected by CI (lanes 2–4). Ubiquitin-protein conjugates decreased in intensity after CI (lanes 6/7). Supplementation of UW with epoxomicin slightly increased the signals for ubiquitin-protein conjugates (lane 8). Quantification of the chemiluminescence signals from four independent experiments confirmed these observations (Fig. S2B).
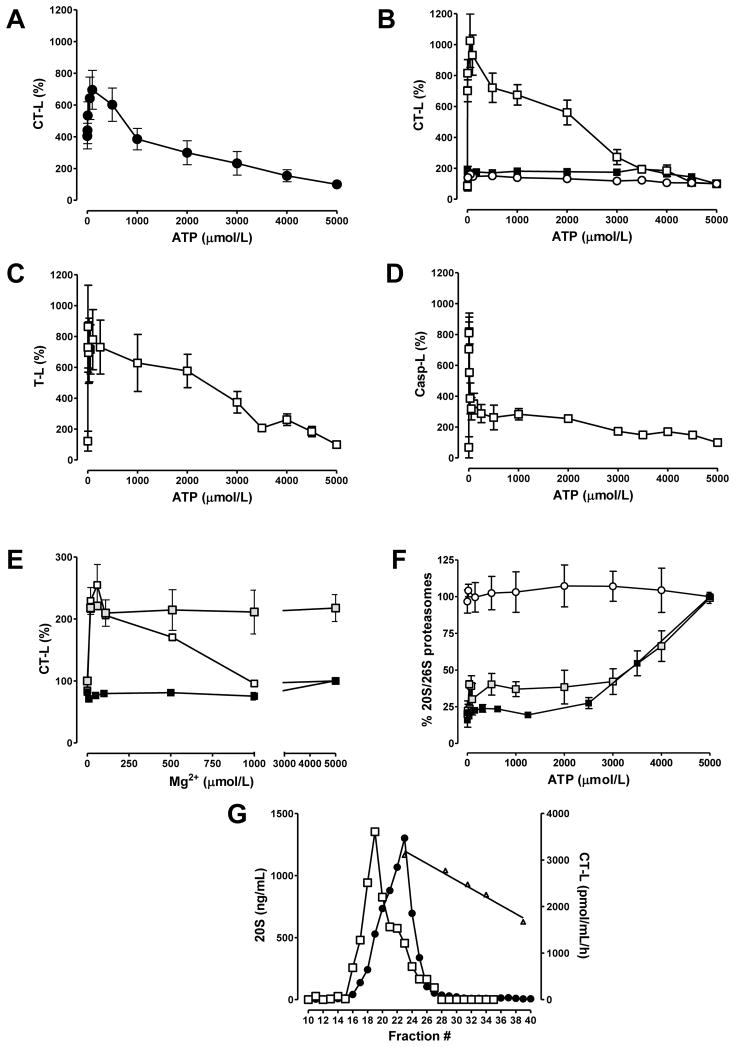
We further evaluated the effect of ATP on CT-L proteasome activity in cardiac extracts from non-ischemic hearts which were harvested without cardioplegia (Fig. 2A). The endogenous [ATP] in these cardiac extracts was 0.51±0.1nmol/g wet weight. This was 20-fold lower than the [ATP] in extracts from hearts that were harvested after cardioplegic arrest with UW (10.6±5nmol/g wet weight, n=5), suggesting that most of the endogenous ATP was consumed during tissue harvest. Based on the volume of extracts that were used in the incubation mixtures, the assay [ATP] in the absence of exogenous ATP was 10–20nmol/L. As compared with measurements at a physiological myocardial [ATP], proteasome CT-L activity increased the further the assay [ATP] was reduced. Maximal activation was reached at 100 μmol/L of exogenous ATP (695±122% of the activity at 5mmol/L ATP(=100%)).
Fig. 2.
A–D: Activities are expressed as % of activity measured at the normal myocardial ATP concentration (5mmol/L=100%); n=5. Mg2+ was constant (5mmol/L) in all assays. A: ATP dependency of the proteasome CT-L activity in extracts from non-ischemic hearts. Hearts were harvested without cardioplegic arrest; n=5. B–D: ATP dependency of proteasome CT-L (B), T-L (C) and Casp-L (D) activity in highly purified proteasome preparations. □:26S; ■:20S; ○:26S, activity measurements in the presence of 5mmol/L EDTA. Activity measurements at 0mmol/L ATP were performed in the presence of 5mmol/L EDTA without addition of ATP to the incubation mixtures (the total ATP concentration in these incubation mixtures containing 26S was 8 μmol/L and derived from the stock 26S solution). E: Mg2+ dependency of the CT-L proteasome activity. □:26S, 2mmol/L ATP;  :26S, 20 μmol/L ATP; ■: 20S, 2mmol/L ATP. Data are % of activity measured at 5mmol/L Mg2+, 2mmol/L ATP. Activity measurements at 0mmol/L Mg2+ were performed in the presence of 5mmol/L EDTA. F: Measurement of 20S and 26S content by ELISA. 160–500 ng of 20S or 26S were loaded onto the ELISA plate at the given ATP concentrations and 0 or 5mmol/L Mg2+, and incubated for 2h. For all subsequent steps, ATP/Mg2+ was kept constant at 5mmol/L. Data are expressed as % of proteasome content when assayed at 5mmol/L ATP and Mg2+; n=5–7. ○:20S measured with the 20S ELISA. ■:26S measured with the 26S ELISA; 0mmol/L Mg2+.
:26S, 20 μmol/L ATP; ■: 20S, 2mmol/L ATP. Data are % of activity measured at 5mmol/L Mg2+, 2mmol/L ATP. Activity measurements at 0mmol/L Mg2+ were performed in the presence of 5mmol/L EDTA. F: Measurement of 20S and 26S content by ELISA. 160–500 ng of 20S or 26S were loaded onto the ELISA plate at the given ATP concentrations and 0 or 5mmol/L Mg2+, and incubated for 2h. For all subsequent steps, ATP/Mg2+ was kept constant at 5mmol/L. Data are expressed as % of proteasome content when assayed at 5mmol/L ATP and Mg2+; n=5–7. ○:20S measured with the 20S ELISA. ■:26S measured with the 26S ELISA; 0mmol/L Mg2+.  :26S measured with the 26S ELISA; 5mmol/L Mg2+. G: Gel filtration of 26S on Superose6. The running buffer contained 50 μmol/L ATP, 5mmol/L Mg2+. Fractions were analyzed for 20S content by ELISA (●) and proteasome CT-L activity (□; incubation mixtures contained 50 μmol/L ATP and 5mmol/L Mg2+). △: elution positions of molecular mass standards. The line shows the standard curve derived from linear regression analysis (r2=0.98).
:26S measured with the 26S ELISA; 5mmol/L Mg2+. G: Gel filtration of 26S on Superose6. The running buffer contained 50 μmol/L ATP, 5mmol/L Mg2+. Fractions were analyzed for 20S content by ELISA (●) and proteasome CT-L activity (□; incubation mixtures contained 50 μmol/L ATP and 5mmol/L Mg2+). △: elution positions of molecular mass standards. The line shows the standard curve derived from linear regression analysis (r2=0.98).
To confirm the phenomenon of proteasome activation by low [ATP], we studied the effect of ATP on enzyme activities in highly purified 26S and 20S preparations. Measurements of the ATP dependency of CT-L (Fig. 2B), T-L (Fig. 2C) and Casp-L (Fig. 2D) 26S activities resulted in a similar activation by low [ATP] with peak activities at concentrations between 10–50 μmol/L. Variation of the [ATP] had no activating effect on the CT-L activity in 20S preparations (Fig. 2B,▪). When Mg2+ was chelated with EDTA, the activating effect of low [ATP] on 26S activity disappeared (Fig. 2B,○), thus indicating that ATP hydrolysis is required for activation. Consistent with this assumption, reduction of the Mg2+ concentration activated 26S activity and resulted in peak activities at 50 μmol/L Mg2+ in the presence of 2 mmol/L ATP, while reduction of Mg2+ down to the low μmol/L range did not affect 26S peptidase activity in the presence of 20 μmol/L ATP (Fig. 2E). Variation of the Mg2+ concentration had no effect on 20S peptidase activity (Fig. 2E,▪).
Quantification of constant amounts of 20S and 26S at various [ATP] showed that reduction of ATP from 5mmol/L to 2mmol/L resulted in a linear decrease of the 26S content by 60–75% in the presence and absence of 5mmol/L Mg2+. Further reduction of [ATP] did not result in a further decrease of the 26S content, suggesting that approximately 30% of 26S are stable at low [ATP], independent of the availability of ATP for hydrolysis (Fig. 2F). Variation of [ATP] did not affect 20S content (Fig. 2F).
To further confirm that a proportion of 26S is stable at low [ATP], 26S was gel-filtered in the presence of 50 μmol/L ATP,5 mmol/L Mg2+ (Fig. 2G). The elution profile of the 20S content showed an asymmetrical peak that corresponded to the native molecular mass of the 20S (700kDa; fraction #23) and a wide left sided shoulder at an elution position corresponding to the native molecular mass of the 26S (>1.5MDa; fraction #19). The latter contained approximately 30% of the total amounts of 20S. The elution profile of the proteasome peptidase activity showed the opposite distribution with maximal enzyme activities in fraction #19 (>1.5MDa) and a right sided shoulder at an elution position of 700kDa. Peptidase activity that eluted in fraction #19 was 3-fold higher than the activity eluted in fraction #23.
Discussion
The present study uncovers a direct relationship between the proteasome, tissue [ATP] and myocardial injury during CI. Furthermore, our observations imply that a subset of the 26S acts as a cell-destructive protease that is activated when the cellular energy supply declines.
The finding that ATP negatively regulates proteasome activities is consistent with previous measurements of proteasome inhibitor sensitive peptidase activities in crude heart extracts[15].
Activity measurements at the actual tissue [ATP] under normo- and hypothermic conditions after CI suggested significant activation of the cardiac proteasome as the tissue ATP content declines in the ischemic heart. As demonstrated by enzyme activity measurements at the physiological [ATP] in the present study, and by previous studies that assessed proteasome activity in ischemic hearts without adjusting [ATP] in the enzyme assays to the actual tissue [ATP][4–6], this activation remains concealed as long as the regulatory effect of ATP is not being considered.
Proteasome inhibition during CI with the specific inhibitor epoxomicin[13] resulted in a significant reduction of organ edema and preserved the ultrastructural integrity of the cardiomyocyte. This provided initial evidence that proteasome activation is a pathophysiologically relevant mechanism of ischemic myocardial injury.
Proteasome inhibitors have previously been shown to reduce cardiac ischemia reperfusion injury[16, 17]. However, information on the cardiac proteasome was not provided and beneficial effects of proteasome inhibitors were attributed to their immune modulatory and anti-inflammatory actions. The present study now provides initial evidence that proteasome inhibition has direct cardioprotective effects during CI.
The phenomenon that proteasome activation during CI was 4–5-fold higher when enzyme assays were performed at 4°C as compared to 37°C could have multiple explanations. Besides the possibility that quantification of the low fluorescence signals in peptidase assays with extracts from non-ischemic hearts at 4°C possibly underestimates the activity, the peptide substrate N-Suc-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin is not exclusively used by the proteasome as a substrate[18]. Thus, competing peptidases in the cardiac extracts might be more sensitive to hypothermia than the proteasome. Furthermore, degradation of natural proteins by the proteasome requires a complex series of protein-protein interactions[19]. Because hypothermia is likely to decelerate these interactions, which are not required for processing of the artificial peptidase test substrates, it may reduce the competing effect of natural protein substrates on proteasome peptidase activities in the cardiac extracts and accentuate the activating effect of low [ATP].
Although ATP dependent and proteasome inhibitor sensitive peptidase activities in tissue extracts have been attributed to the 26S[6, 15], the observation that proteasome activities are negatively regulated by ATP is difficult to interpret as the 26S disassembles when the ATP concentration is reduced[8, 10, 11]. To resolve this paradox, we employed purified 20S and 26S for a direct assessment of the influence of ATP on proteasome activity and stability. Consistent with previous findings on CT-L peptidase activity of purified 26S[20], activity measurements confirmed maximal activation of all three major 26S peptidase activities by ATP in the low μmol/L range, and also a dose-dependent down regulation of its peptidase activity when ATP was increased to its physiological concentration in the heart[14], as we detected in cardiac extracts.
Variation of the Mg2+ concentration in peptidase activity assays, quantification of the 20S/26S content at various [ATP] and gel-filtration experiments suggested that ATP hydrolysis is required for 26S activation and that approximately 30% of 26S are stable at very low levels of ATP. The latter is consistent with our previous findings from solid phase affinity immobilization experiments[11].
Proteasome activation by low [ATP] was detectable in extracts from hearts under physiological baseline conditions, during prolonged CI and in highly purified 26S derived from human blood cells. Thus, this regulation is likely to represent a general physiological mechanism that controls the activity of a major proteolytic system. Therefore, these data support the notion that the 26S is under direct control of the cellular energy status, that a subset of 26S is a cell destructive protease that is activated as ATP declines and that a sufficient energy supply prevents the tissue from autodestruction.
Polyubiquitylated proteins are the preferred 26S substrates[21]. The finding that high molecular mass ubiquitin-protein conjugates were decreased after CI could point towards their increased degradation when the 26S is activated as ATP declines. However, the effects of epoxomicin on the disappearance of ubiquitin-protein conjugates during CI were small and variable, which questions a causal relationship. Previous studies inferred that the ubiquitin-protein conjugate pool size results from direct control of ligation or disassembly rates, whereas potential modulation of the degradation rates would have little effects[22, 23]. Therefore, it is conceivable that the decrease in cardiac ubiquitin-protein conjugates after CI is rather the effect of altered function of ubiquitin-protein ligase systems and deubiquitylating enzymes. On the other hand, degradation of ubiquitin-protein conjugates might have occurred before epoxomicin was able to permeate into the cell and accumulate in sufficient concentrations.
The beneficial actions of epoxomicin in the present study support the concept of proteasome inhibitors as potential cardiac therapeutics, and suggest that this strategy could be used to improve organ preservation in cardiac transplantation. Nevertheless, it should be noted that the proteasome inhibitor MG132 decreased recovery of post-ischemic function in a perfused rat heart preparation[6]. Besides the possibility that these effects might be attributable to the limited specificity of MG132[24], its adverse effects on contractility in post-ischemic hearts could also be related to differential sensitivities of cardiac 20S subpopulations to the proteasome inhibitors MG132 and bortezomib[25]. Along with our finding of a subpopulation of 26S that is stable and activated at low [ATP], these data collectively point towards specific functions of proteasome subpopulations and imply that the side effect profile of proteasome inhibitors can be reduced if subpopulation specific inhibitors become available.
Supplementary Material
Representative photomicrographs showing H&E (A., 400×), Masson’s trichrome (B., 400×) staining and transmission electron microscopy (C. 6000×) in a non-ischemic heart after cardioplegic arrest (ctrl.), in the heart after cardioplegic arrest and 24h of CI in UW (CI24h) and after CI in UW supplemented with 50 μmol/L of epoxomicin (CI24h+Epox). cpe: cytoplasmatic and perinuclear edema. Scale bars: A. and B. − 20 μm; C. − 1 μm.
A. Ubiquitin pools in extracts from hearts after cardioplegic arrest and 0 or 24h of CI in UW ± epoxomicin. Top: The membrane was incubated with anti-ubiquitin; left image: exposure 50s, right image: exposure 500s; the membrane was cut at the 15kDa marker prior to exposure. Bottom: The membrane was incubated with anti-GAPDH (loading control). Lanes 1/5: ubiquitin, 10ng. Lanes 2/6: non-ischemic heart. Lanes 3/7: heart after 24h CI in UW. Lanes 4/8: heart after 24h CI in UW with 50 μmol/L epoxomicin. Lanes 2–4 and 6–8: 50 μg protein. B. Quantification of the chemiluminescence signals. Data are relative pixel densities when compared to non-ischemic hearts; n=4. The storage conditions are indicated in the graph. Light grey bars: free ubiquitin. Dark grey bars: ubiquitin-protein conjugates. *: p<0.05 vs. non-ischemic hearts.
Acknowledgments
Supported by grants AHA#0755604B, DFGMA2474/2-2 (both to MM) and NIHT32 GM008750 (to RLG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hicks M, Hing A, Gao L, Ryan J, Macdonald PS. Organ preservation. Methods Mol Biol. 2006;333:331–374. doi: 10.1385/1-59745-049-9:331. [DOI] [PubMed] [Google Scholar]
- 2.Willis MS, Patterson C. Into the heart: the emerging role of the ubiquitin-proteasome system. J Mol Cell Cardiol. 2006;41:567–579. doi: 10.1016/j.yjmcc.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 3.Yu X, Patterson E, Kem DC. Targeting proteasomes for cardioprotection. Curr Opin Pharmacol. 2009;9:167–172. doi: 10.1016/j.coph.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Majetschak M, Patel MB, Sorell LT, Liotta C, Li S, Pham SM. Cardiac proteasome dysfunction during cold ischemic storage and reperfusion in a murine heart transplantation model. Biochem Biophys Res Commun. 2008;365:882–888. doi: 10.1016/j.bbrc.2007.11.092. [DOI] [PubMed] [Google Scholar]
- 5.Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, Szweda LI. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001;276:30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 6.Powell SR, Wang P, Katzeff H, Shringarpure R, Teoh C, Khaliulin I, Das DK, Davies KJ, Schwalb H. Oxidized and ubiquitinated proteins may predict recovery of postischemic cardiac function: essential role of the proteasome. Antioxid Redox Signal. 2005;7:538–546. doi: 10.1089/ars.2005.7.538. [DOI] [PubMed] [Google Scholar]
- 7.Das S, Powell SR, Wang P, Divald A, Nesaretnam K, Tosaki A, Cordis GA, Maulik N, Das DK. Cardioprotection with palm tocotrienol: antioxidant activity of tocotrienol is linked with its ability to stabilize proteasomes. Am J Physiol Heart Circ Physiol. 2005;289:H361–367. doi: 10.1152/ajpheart.01285.2004. [DOI] [PubMed] [Google Scholar]
- 8.Eytan E, Armon T, Heller H, Beck S, Hershko A. Ubiquitin C-terminal hydrolase activity associated with the 26 S protease complex. J Biol Chem. 1993;268:4668–4674. [PubMed] [Google Scholar]
- 9.Babbitt SE, Kiss A, Deffenbaugh AE, Chang YH, Bailly E, Erdjument-Bromage H, Tempst P, Buranda T, Sklar LA, Baumler J, Gogol E, Skowyra D. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell. 2005;121:553–565. doi: 10.1016/j.cell.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 10.Liu CW, Li X, Thompson D, Wooding K, Chang TL, Tang Z, Yu H, Thomas PJ, DeMartino GN. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majetschak M, Sorell LT. Immunological methods to quantify and characterize proteasome complexes: development and application. J Immunol Methods. 2008;334:91–103. doi: 10.1016/j.jim.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Covarrubias L, Manning EW, 3rd, Sorell LT, Pham SM, Majetschak M. Ubiquitin enhances the Th2 cytokine response and attenuates ischemia-reperfusion injury in the lung. Crit Care Med. 2008;36:979–982. doi: 10.1097/CCM.0B013E318164E417. [DOI] [PubMed] [Google Scholar]
- 13.Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci U S A. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beer M, Seyfarth T, Sandstede J, Landschutz W, Lipke C, Kostler H, von Kienlin M, Harre K, Hahn D, Neubauer S. Absolute concentrations of high-energy phosphate metabolites in normal, hypertrophied, and failing human myocardium measured noninvasively with (31)P-SLOOP magnetic resonance spectroscopy. J Am Coll Cardiol. 2002;40:1267–1274. doi: 10.1016/s0735-1097(02)02160-5. [DOI] [PubMed] [Google Scholar]
- 15.Powell SR, Davies KJ, Divald A. Optimal determination of heart tissue 26S-proteasome activity requires maximal stimulating ATP concentrations. J Mol Cell Cardiol. 2007;42:265–269. doi: 10.1016/j.yjmcc.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell B, Adams J, Shin YK, Lefer AM. Cardioprotective effects of a novel proteasome inhibitor following ischemia and reperfusion in the isolated perfused rat heart. J Mol Cell Cardiol. 1999;31:467–476. doi: 10.1006/jmcc.1998.0880. [DOI] [PubMed] [Google Scholar]
- 17.Pye J, Ardeshirpour F, McCain A, Bellinger DA, Merricks E, Adams J, Elliott PJ, Pien C, Fischer TH, Baldwin AS, Jr, Nichols TC. Proteasome inhibition ablates activation of NF-kappa B in myocardial reperfusion and reduces reperfusion injury. Am J Physiol Heart Circ Physiol. 2003;284:H919–926. doi: 10.1152/ajpheart.00851.2002. [DOI] [PubMed] [Google Scholar]
- 18.Giguere CJ, Schnellmann RG. Limitations of SLLVY-AMC in calpain and proteasome measurements. Biochem Biophys Res Commun. 2008;371:578–581. doi: 10.1016/j.bbrc.2008.04.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahlmann B, Kuehn L, Reinauer H. Studies on the activation by ATP of the 26 S proteasome complex from rat skeletal muscle. Biochem J. 1995;309(Pt 1):195–202. doi: 10.1042/bj3090195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Depraetere V. Getting activated with poly-ubiquitination. Nat Cell Biol. 2001;3:E181. doi: 10.1038/35087116. [DOI] [PubMed] [Google Scholar]
- 22.Haas AL, Bright PM. The dynamics of ubiquitin pools within cultured human lung fibroblasts. J Biol Chem. 1987;262:345–351. [PubMed] [Google Scholar]
- 23.Patel MB, Majetschak M. Distribution and interrelationship of ubiquitin proteasome pathway component activities and ubiquitin pools in various porcine tissues. Physiol Res. 2007;56:341–350. doi: 10.33549/physiolres.931005. [DOI] [PubMed] [Google Scholar]
- 24.Kisselev AF, Goldberg AL. Proteasome inhibitors: from research tools to drug candidates. Chem Biol. 2001;8:739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- 25.Kloss A, Meiners S, Ludwig A, Dahlmann B. Multiple cardiac proteasome subtypes differ in their susceptibility to proteasome inhibitors. Cardiovasc Res. 2009 doi: 10.1093/cvr/cvp217. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative photomicrographs showing H&E (A., 400×), Masson’s trichrome (B., 400×) staining and transmission electron microscopy (C. 6000×) in a non-ischemic heart after cardioplegic arrest (ctrl.), in the heart after cardioplegic arrest and 24h of CI in UW (CI24h) and after CI in UW supplemented with 50 μmol/L of epoxomicin (CI24h+Epox). cpe: cytoplasmatic and perinuclear edema. Scale bars: A. and B. − 20 μm; C. − 1 μm.
A. Ubiquitin pools in extracts from hearts after cardioplegic arrest and 0 or 24h of CI in UW ± epoxomicin. Top: The membrane was incubated with anti-ubiquitin; left image: exposure 50s, right image: exposure 500s; the membrane was cut at the 15kDa marker prior to exposure. Bottom: The membrane was incubated with anti-GAPDH (loading control). Lanes 1/5: ubiquitin, 10ng. Lanes 2/6: non-ischemic heart. Lanes 3/7: heart after 24h CI in UW. Lanes 4/8: heart after 24h CI in UW with 50 μmol/L epoxomicin. Lanes 2–4 and 6–8: 50 μg protein. B. Quantification of the chemiluminescence signals. Data are relative pixel densities when compared to non-ischemic hearts; n=4. The storage conditions are indicated in the graph. Light grey bars: free ubiquitin. Dark grey bars: ubiquitin-protein conjugates. *: p<0.05 vs. non-ischemic hearts.