Abstract
Objective
The objective of this study was to identify genetic variants associated with angiotensin-converting enzyme (ACE) inhibitor-associated angioedema.
Participants and methods
We carried out a genome-wide association study in 175 individuals with ACE inhibitor-associated angioedema and 489 ACE inhibitor-exposed controls from Nashville (Tennessee) and Marshfield (Wisconsin). We tested for replication in 19 cases and 57 controls who participated in Ongoing Telmisartan Alone and in Combination with Ramipril Global Endpoint Trial (ONTARGET).
Results
There were no genome-wide significant associations of any single-nucleotide polymorphism (SNP) with angioedema. Sixteen SNPs in African Americans and 41 SNPs in European Americans were associated moderately with angioedema (P<10−4) and evaluated for association in ONTARGET. The T allele of rs500766 in PRKCQ was associated with a reduced risk, whereas the G allele of rs2724635 in ETV6 was associated with an increased risk of ACE inhibitor-associated angioedema in the Nashville/Marshfield sample and ONTARGET. In a candidate gene analysis, rs989692 in the gene encoding neprilysin (MME), an enzyme that degrades bradykinin and substance P, was significantly associated with angioedema in ONTARGET and Nashville/Marshfield African Americans.
Conclusion
Unlike other serious adverse drug effects, ACE inhibitor-associated angioedema is not associated with a variant with a large effect size. Variants in MME and genes involved in immune regulation may be associated with ACE inhibitor-associated angioedema.
Keywords: adverse drug event, angioedema, angiotensin-converting enzyme, neprilysin
Introduction
Angiotensin-converting enzyme (ACE) inhibitors reduce mortality in patients with congestive heart failure, risk factors for coronary artery disease, and renal disease [1,2]. In rare cases, ACE inhibitors cause angioedema – swelling of the lips, face, tongue, pharynx, and even bowel [3]. When severe, angioedema can lead to airway compromise and death. The mechanism by which ACE inhibitors cause angioedema remains uncertain, but is presumed to relate to impaired degradation of vasoactive peptides that are substrates for ACE, such as bradykinin and substance P. The incidence of ACE inhibitor-associated angioedema has been reported to be from 0.1 to 0.7% in large epidemiological studies to as high as 2.8–6% in randomized- controlled trials of ACE inhibitors [3–5]. The risk of angioedema is increased in African Americans, women, older patients, smokers, individuals with seasonal allergies, and patients taking immunosuppressants [6–9]. In contrast, the risk of ACE inhibitor-associated angioedema is decreased in diabetics and in individuals of Asian ancestry [7,8,10].
The observation that rates of ACE inhibitor-associated angioedema differ among racial groups suggests that genetic factors influence risk. However, the clinical presentation of angioedema often occurs long after the initiation of an ACE inhibitor [11], suggesting that gene– environment interactions may confound attempts to detect genetic predictors of angioedema. In recent years, genome-wide association studies (GWAS) have been used to detect genetic predictors of serious adverse drug events including statin-induced myopathy [12], abacavir-induced hypersensitivity reaction, [13] elevated alanine aminotransaminase during ximelagatran [14], and osteonecrosis of the jaw in bisphosphonate-exposed patients [15]. Although serious adverse drug events may be rare, GWAS have detected genetic associations in small samples because effect sizes have been large.
An alternative approach to identifying genetic predictors of ACE inhibitor-associated angioedema is to test for associations between genetic variants in candidate genes and the adverse event. Studies in rodent models suggest that two vasoactive peptide substrates of ACE contribute toward angioedema – bradykinin and substance P [16,17]. In addition, bradykinin contributes toward hereditary angioedema, resulting from the deficiency of C1 esterase inhibitor [18]. On the basis of this biology, candidate genes for ACE inhibitor-associated angioedema include those genes encoding enzymes involved in the degradation or actions of bradykinin or substance P when ACE is inhibited.
In the present study, we carried out a GWAS in 175 individuals with ACE inhibitor-associated angioedema and 489 ACE inhibitor-exposed controls without angioedema from Nashville (Tennessee) and Marshfield (Wisconsin), the largest reported sample of ACE inhibitor-associated angioedema and ACE inhibitor-exposed controls for which DNA is available. We carried out a replication study in patients who developed ACE inhibitor-associated angioedema and ACE inhibitor-exposed controls from among participants in the Ongoing Telmisartan Alone and in Combination with Ramipril Global Endpoint Trial (ONTARGET) study [19]. We also carried out a candidate gene study using the smaller ONTARGET case–control study as the discovery sample and the Nashville/Marshfield study as the replication sample.
Methods
Participants and samples in the GWAS
The initial GWAS was carried out using samples collected as part of a case–control study at Vanderbilt and as part of the Marshfield Clinic Personalized Medicine Research Project [20,21]. The Institutional Review Boards of Vanderbilt and Marshfield approved each study and informed consent was obtained from all participants. Cases were defined as having ACE inhibitor-associated angioedema if they had swelling of the lips, pharynx, or face while taking an ACE inhibitor, but had never had angioedema while not taking an ACE inhibitor. Controls were individuals who had been exposed to an ACE inhibitor for at least 6 months but had never developed angioedema.
At Vanderbilt, patients were identified at presentation with angioedema, by referral from their primary physician at a later date, or by search of the electronic medical record and subsequent confirmation with their primary physician. Hereditary angioedema was excluded by the measurement of C1 esterase activity if appropriate. Cases and controls were matched with respect to sex, race, and smoking status. All Vanderbilt patients were interviewed by a research nurse or physician and the medical history was confirmed using a detailed case report form. In Marshfield, cases were identified by electronic medical records and the medical history of each potential study patient was reviewed by a trained research coordinator, as described previously [20]. A case report form, similar to the one used in Nashville, was completed.
Testing for a consistent association in ONTARGET
Briefly, the randomized double-blind ONTARGET trial compared the effect of the ACE inhibitor ramipril, the angiotensin receptor blocker telmisartan, and the combination of the two drugs in patients with vascular disease or high-risk patients with diabetes [19]. Overall, 8576 participants were assigned to receive 10mg of ramipril per day, 8542 participants were assigned to receive 80mg of telmisartan per day, and 8502 participants were assigned to receive both drugs (combination therapy). All participants were exposed to an ACE inhibitor during the run-in period. A detailed description of the trial can be found elsewhere [19]. 7293 ONTARGET participants of 17 118 agreed to have their DNA collected for genetic analysis. During a median of 56 months of follow-up, angioedema was diagnosed in 19 participants for whom DNA was available. Each angioedema case was matched on the basis of sex, age (within 5 years), and self-reported ancestry to three ACE-inhibitor exposed controls free of angioedema.
Genotyping
For the GWAS, all cases and controls were genotyped using a 610Quadv1.B BeadChip (Illumina, San Diego, California, USA). We excluded participants with sex errors (four), duplicates (14), or those who were related (six). Quality control procedures were used to remove copy number variants and any technical failures. We excluded single-nucleotide polymorphisms (SNPs) with a call rate of less than 0.98 or a minor allele frequency of less than 0.0001, and samples with a call rate of less than 0.98. From 620 901 raw SNPs, we excluded 21 890 copy number variants, 5443 monomorphic SNPs, 6592 technical failures, and 7632 with a call rate less than 0.98 or a minor allele frequency less than 0.0001, such that 579 344 total SNPs were analyzed.
For the candidate gene study, cases and controls were genotyped for polymorphisms in genes encoding the bradykinin-degrading or substance P-degrading enzymes carboxypeptidase N (CPN), neprilysin (MME), amino-peptidase P (XPNPEP2) and dipeptidyl peptidase IV (DPP4), the bradykinin B2 receptor (BDKRB2), the bradykinin B1 receptor (BDKRB1), and the NK1 receptor (TACR1) [22–28]. Within these candidate genes, we chose a total of 33 SNPs that had been associated previously with a phenotype or function. We also looked for an association between ACE inhibitor-associated angioedema and variants at the chromosome 1q13 and chromosome 17q21 loci that have been associated with asthma, as well as SNPs that have been associated with circulating IgE concentrations [29,30]. Supplementary Table 1 (http://links.lww.com/FPC/A622) lists the candidate gene SNPs genotyped in the ONTARGET case–control study. Genotyping was performed using the VeraCode genotyping platform as implemented on a BeadXpress reader by Illumina. SNPs identified as significant in ONTARGET were then genotyped in the Nashville/ Marshfield sample using the mid-throughput sequenome genotyping platform and tested for replication.
Statistical analysis
Participant characteristics are summarized as means±SD of the mean or as frequencies. SNPs were evaluated for deviation from Hardy–Weinberg equilibrium using a χ2-test. For genetic association studies, allele frequency differences were assessed for cases and controls. Conditional logistic regression analyses were carried out to estimate the odds ratios (ORs) and their corresponding 95% confidence intervals (CIs), with cases and controls matched on age, sex, and ancestry. The regression models were analyzed under the assumption of different genetic models (dominant, additive, and recessive). Inflation factor was calculated for genomic control analysis using PLINK (v1.07). Principal components were estimated using Smartpca 8000, Eigensoft package; none of the principle components was significant or included in the model. Genotype association analyses were stratified by ancestry (European American and African American). In the GWAS, a P-value less than 5×10− 8 was considered a statistically significant genome-wide association. Any SNP with a P-value less than 10−4 was evaluated for association in the ONTARGET study. In the candidate gene study, in which the ONTARGET sample served as the discovery dataset, a P-value less than 0.05 was considered significant because of the small number of cases and we did not adjust P-values for multiple comparisons. Similarly, a P-value less than 0.05 was considered significant for SNPS in candidate genes first identified in ONTARGET that were also associated with angioedema in the Vanderbilt/Marshfield study. Liberal P-value thresholds were used as we relied on a consistent association to confirm results.
Results and discussion
Characteristics of cases and controls in the GWAS and the ONTARGET study
Table 1 provides the characteristics of the cases and controls in the initial GWAS. One hundred and forty cases and 327 controls were recruited at Vanderbilt and 35 cases and 162 controls were recruited at Marshfield (for a total of 175 cases and 489 controls). Within each site, cases and control were intentionally matched with respect to sex, race, and smoking status. Controls were significantly more likely to be taking an ACE inhibitor at the time of ascertainment (P<0.001), as many cases were ascertained after their ACE inhibitor was stopped because of adverse effects. As reported previously [7], cases were more likely to have a history of seasonal allergies (P<0.001). Table 2 provides the characteristics of the ONTARGET study participants.
Table 1.
Participant characteristics of angioedema cases and ACE inhibitor-exposed controls in the Nashville/Marshfield GWAS
| Cases (175) [n (%)] | Controls (489) [n (%)] | |
|---|---|---|
| Age (years) | 58.4±14.1 | 61.7±13.1 |
| Sex (male : female) | 79 (45.1) : 96 (54.9) | 229 (46.8) : 260 (53.2) |
| Race (unknown : African : European) | 0 : 66 (37.7) : 109 (62.3) | 2 (0.4) : 157 (32.1) : 330 (67.5) |
| Current smoker (unknown : no : yes) | 28 (16.0) : 110 (62.9) : 37 (21.1) | 110 (22.5) : 288 (59.0) : 91 (18.6) |
| Diabetic (missing : no : yes) | 1 (0.6) : 116 (66.3) : 58 (33.1) | 7 (1.4) : 300 (61.4) : 182 (37.2) |
| Seasonal allergy (missing : no : yes) | 47 (26.9) : 43 (24.6) : 85 (48.6) | 152 (31.1) : 183 (37.4) : 154 (31.5) |
| Current ACE inhibitor (missing : no : yes) | 1 (0.6) : 117 (66.9) : 57 (32.6) | 7 (1.4) : 143 (29.4) : 339 (69.3) |
At Vanderbilt, some cases were ascertained during their acute episode of angioedema and were thus taking an ACE inhibitor at the time of ascertainment. ACE, angiotensin-converting enzyme.
Table 2.
Participant characteristics of angioedema cases and ACE inhibitor-exposed controls in ONTARGETACE, angiotensin-converting enzyme; ONTARGET, Ongoing Telmisartan Alone and in Combination with Ramipril Global Endpoint Trial.
| Cases (19) [n (%)] | Controls (57) [n (%)] | |
|---|---|---|
| Sex (male : female) | 12 (63.2) : 7 (36.8) | 36 (63.2) : 21 (36.8) |
| Age (years) | 64.5±7.5 | 64.5±7.3 |
| Race (African : European : other) | 1 (5.3) : 16 (84.2) : 2 (10.5) | 3 (5.3) : 48 (84.2) : 6 (10.5) |
| Treatment (ramipril : telmisartan : combination) | 15 (78.9) : 0 : 4 (21.1) | 20 (35.1) : 18 (31.6) : 19 (33.3) |
Results of the GWAS
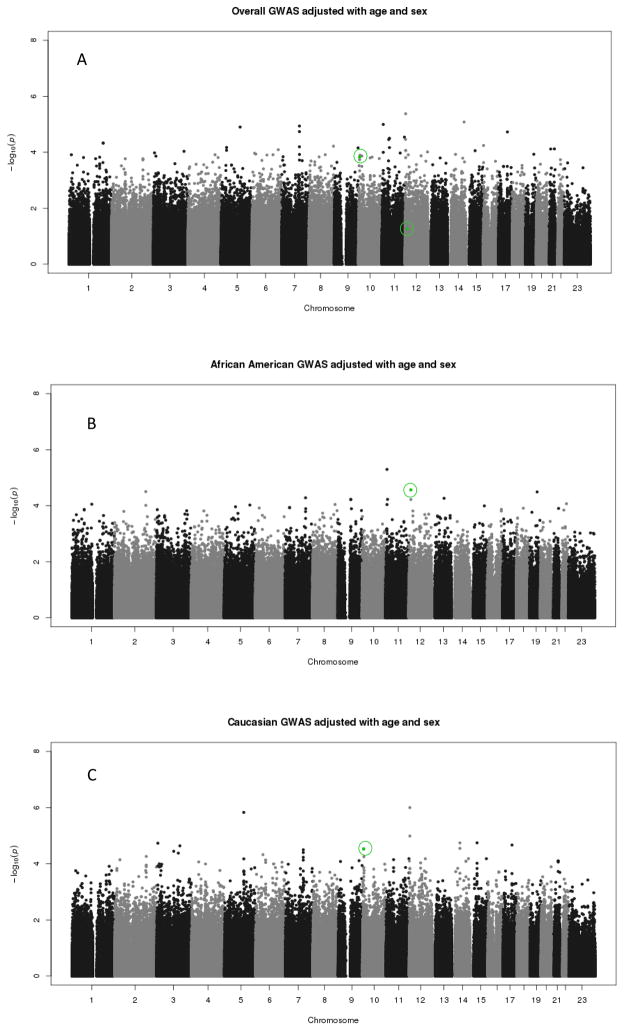
We carried out stratified GWAS in African Americans and European Americans, respectively (Fig. 1) (Q-Q plots appear in Fig. 1 of the supplement, http://links.lww.com/FPC/A622). There were no genome-wide significant associations (P<5 ×10−8) of any SNP with ACE inhibitor-associated angioedema in either African Americans or those of European Ancestry. For sixteen SNPs in African Americans (Supplementary Table 2, http://links.lww.com/FPC/A622) and 41 SNPs in those of European Ancestry (Supplementary Table 3, http://links.lww.com/FPC/A622), there was moderate evidence of an association with angioedema. The SNPs listed in Supplementary Table 2 (http://links.lww.com/FPC/A622) and Supplementary Table 3 (http://links.lww.com/FPC/A622) are the SNPs that were evaluated in the replication dataset.
Fig. 1.
(a) The results of tests for a trend in the association between angiotensin-converting enzyme (ACE) inhibitor-associated angioedema and each single-nucleotide polymorphism (SNP) measured in the GWAS in participants Nashville/Marshfield study (irrespective of ethnicity). The analysis was adjusted for age and sex. Rs2724635 and Rs500766, indicated in green, were associated with ACE inhibitor-associated angioedema in ONTARGET. (b) The results of tests for a trend in the association between ACE inhibitor-associated angioedema and each SNP measured in the GWAS in African Americans from the Nashville/Marshfield study. The analysis was adjusted for age and sex. Rs2724635, indicated in green, was associated with ACE inhibitor-associated angioedema in ONTARGET. (c) The results of tests for a trend in the association between ACE inhibitor-associated angioedema and each SNP measured in the GWAS in European Americans from the Nashville/Marshfield study. The analysis was adjusted for age and sex. Rs500766, indicated in green, was associated with ACE inhibitor-associated angioedema in ONTARGET.
We next sought to replicate these findings in cases and controls from the ONTARGET study. Of the SNPs that were associated modestly with ACE inhibitor-associated angioedema in the discovery sets, two were significantly associated with ACE inhibitor-associated angioedema (rs500766 and rs2724635, Table 3) at the level of P less than 0.05. Rs500766 is a polymorphism in the gene encoding for protein kinase C θ (PRKCQ). In both the Nashville/Marshfield sample (OR 0.42, 95% CI 0.28–0.63, P=2.97×10 − 5 in the additive model and OR 0.42, 95% CI 0.26–0.67, P=3.04×10 − 4 in the dominant model) and in ONTARGET (OR 0.28, 95% CI 0.09–0.89, P=0.03 in the dominant genetic model), the Tallele was significantly associated with a reduced risk of ACE inhibitor-associated angioedema. Rs2724635 is a polymorphism in ETS variant gene 6 (ETV6), also known as TEL (or translocation ets leukemia), and the G allele was associated with an increased risk of ACE inhibitor-associated angioedema in both African Americans in the Nashville/Marshfield sample (OR 2.78, 95% CI 1.67–4.00, P=2.73×10−5 in the additive model; OR 3.23, 95% CI 1.75–6.25, P=2.11×10−4 dominant model; OR 5.56, 95% CI 1.85–16.6, P=2.01×10−3 recessive mode) and in the ONTARGET sample (OR 3.27, 95% CI 1.03–10.35, P=0.044 recessive model). This polymorphism was not associated with angioedema in European Americans in the Nashville/Marshfield sample (Supplementary Fig. 2, http://links.lww.com/FPC/A622). Because of the small number of cases and controls in ONTARGET, this analysis was not stratified by race or ethnic group. If we limited analysis of ONTARGET to those of European ancestry, rs2724635 remained significant (OR 3.68, 95% CI 1.04–13.02, P=0.043 recessive model), whereas rs500766 was no longer significant (P=0.068 in the dominant model).
Table 3.
SNPs identified through Nashville/Marshfield sample GWAS that was replicated in ONTARGET
| Chromosome | SNP | Minor allelea | Cases | Controls | P-value Hardy–Weinberg equilibrium | Odds ratio additive genetic model (95% CI) | P-value | Odds ratio dominant genetic model (95% CI) | P-value | Odds ratio recessive genetic model (95% CI) | P-value | Gene |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | rs500766 | PRKCQ | ||||||||||
| Caucasians in the GWAS | T | 0.15 | 0.31 | 0.05 | 0.42 (0.28–0.63) | 2.97E – 05 | 0.42 (0.26–0.67) | 3.04E – 04 | 0.07 (0.01–0.49) | 7.97E – 03 | ||
| ONTARGET | T | 0.42 | 0.54 | 0.65 | 0.56 (0.25–1.25) | 0.16 | 0.28 (0.09–0.89) | 0.03 | 1.11 (0.31–4.01) | 0.87 | ||
| 12 | rs2724635 | ETV6 | ||||||||||
| African Americans in the GWAS | G | 0.72 | 0.50 | 0.75 | 2.78 (1.72–4.00) | 2.73E – 05 | 3.23 (1.75–6.25) | 2.11E – 04 | 5.56 1.85–16.67 | 2.01E – 03 | ||
| ONTARGET | G | 0.63 | 0.46 | 0.50 | 2.02 (0.91–4.48) | 0.09 | 1.70 (0.44–6.52) | 0.44 | 3.27 (1.03–10.35) | 0.04 | ||
CI, confidence interval; ETV6, ETS variant gene 6; GWAS, genome-wide association studies; ONTARGET, Ongoing Telmisartan Alone and in Combination with Ramipril Global Endpoint Trial.
Minor allele based on allele frequencies in the Nashville/Marshfield study.
Candidate gene analysis
In addition to carrying out GWAS, we carried out an analysis of the association between SNPs in candidate genes and ACE inhibitor-associated angioedema. In this case, the ONTARGET study was used as the discovery dataset and the Vanderbilt/Marshfield sample was used for replication. As noted in detail in the methods, candidate genes included those involved in the degradation or the action of the vasoactive peptides bradykinin and substance P, as well as a few genes implicated in atopic diseases. In the ONTARGET dataset, polymorphisms in the gene encoding neprilysin (MME, rs989692) and in CRB1 (rs2786098) were associated significantly with ACE inhibitor-associated angioedema (Table 4). The same SNP in MME (rs989692) significantly associated with ACE inhibitor-associated angioedema in individuals of African descent in the Vanderbilt/Marshfield sample (Table 5). In both cases, the G allele was associated with an increased risk of ACE inhibitor-associated angioedema.
Table 4.
SNPs identified using the candidate gene approach in ONTARGET
| Chromosome | SNP | Allele | Cases | Controls | P-value Hardy–Weinberg equilibrium | Odds ratio additive genetic model (95% CI) | P-value | Odds ratio dominant genetic model (95% CI) | P-value | Odds ratio recessive genetic model (95% CI) | P-value | Gene |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | rs989692 | T | 0.21 | 0.09 | 0.59 | 2.73 (0.92–8.12) | 0.07 | 4.27 (1.03–17.73) | 0.046 | 3.00 (0.19–47.96) | 0.44 | MME (neprilysin) |
| 1 | rs1924518 | A | 0.50 | 0.68 | 1.00 | 0.47 (0.22–1.03) | 0.06 | 0.47 (0.12–1.78) | 0.27 | 0.27 (0.07–1.02) | 0.05 | DENND1B |
| 1 | rs2786098 | T | 0.36 | 0.57 | 0.25 | 0.36 (0.14–0.94) | 0.036 | 0.36 (0.09–1.35) | 0.13 | 0.16 (0.02–1.21) | 0.08 | CRB1 |
CI, confidence interval; ONTARGET, Ongoing Telmisartan Alone and in Combination with Ramipril Global Endpoint Trial; SNP, single-nucleotide polymorphism.
Table 5.
Association of candidate gene SNPs identified from ONTARGET in the Nashville/Marshfield sample
| Chromosome | SNP | Allele | Cases | Controls | P-value Hardy–Weinberg equilibrium | Odds ratio additive genetic model (95% CI) | P-value | Odds ratio dominant genetic model (95% CI) | P-value | Odds ratio recessive genetic model (95% CI) | P-value | Gene |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| African Americans | ||||||||||||
| 3 | rs989692 | T | 0.56 | 0.54 | 0.30 | 0.66 (0.36–1.19) | 0.16 | 1.18 (0.48–2.86) | 0.72 | 3.23 (1.33–7.69) | 0.009 | MME (neprilysin) |
| 1 | rs1924518 | A | 0.03 | 0.03 | 1.00 | 1.00 (0.19–5.28) | 1.00 | 1.00 (0.19–5.28) | 1.00 | Null | NA | DENND1B |
| 1 | rs2786098 | T | 0.13 | 0.11 | 1.00 | 1.21 (0.51–2.89) | 0.67 | 1.04 (0.41–2.65) | 0.94 | 0.00 (0.00-∞) | 0.99 | CRB1 |
| European Americans | ||||||||||||
| 3 | rs989692 | T | 0.57 | 0.50 | 0.91 | 1.35 (0.93–1.96) | 0.11 | 1.35 (0.77–2.38) | 0.29 | 0.58 (0.30–1.13) | 0.11 | MME |
| 1 | rs1924518 | A | 0.20 | 0.18 | 0.59 | 1.09 (0.70–1.72) | 0.70 | 1.08 (0.63–1.85) | 0.78 | 0.76 (0.22–2.58) | 0.66 | DENND1B |
| 1 | rs2786098 | T | 0.21 | 0.21 | 0.63 | 0.95 (0.61–1.47) | 0.81 | 0.94 (0.55–1.61) | 0.83 | 1.11 (0.34–3.57) | 0.86 | CRB1 |
CI, confidence interval; ONTARGET, Ongoing Telmisartan Alone and in Combination with Ramipril Global Endpoint Trial; SNP, single-nucleotide polymorphism.
Discussion
In contrast to other studies of serious adverse drug effects of comparable size, we found no associations of genome-wide significance, indicating that no single gene had a large effect size and suggesting that ACE inhibitor-associated angioedema behaves more like a complex trait than other serious adverse drug effects. Using GWAS, we identified two SNPs (rs500766 and rs2724635) that were modestly associated with ACE inhibitor-associated angioedema in the Nashville/Marshfield study and associated with angioedema in cases and controls from the ONTARGET trial. These two SNPs occur in genes involved in the regulation of immune function. Using a candidate gene approach, we found that a polymorphism in intron one of the gene encoding for neprilysin was associated with an increased risk of angioedema in ACE inhibitor-exposed participants in the ONTARGET trial and in African American ACE inhibitor users in a Nashville/Marshfield case–control study.
Studies in rodents suggest that both bradykinin and substance P contribute toward angioedema and plasma extravasation during ACE inhibition [16,17]. Normally, bradykinin and substance P are degraded primarily by ACE. When ACE is inhibited, however, other endogenous enzymes contribute toward the degradation and inactivation of bradykinin and substance P, including neprilysin [22]. Large clinical studies support a role for neprilysin in the pathogenesis of ACE inhibitor-associated angioedema. In the Omapatrilat Cardiovascular Treatment versus Enalapril (OCTAVE) trial, patients treated with omapatrilat, a combined inhibitor of ACE and neprilysin, had a three-fold higher risk of angioedema compared with patients treated with the ACE inhibitor enalapril [31]. The present study suggests the possibility that genetic variation in neprilysin could also contribute toward the risk of ACE inhibitor-associated angioedema.
Bradykinin induces plasma extravasation through direct effects at its B2 receptor and through the indirect, B2 receptor-dependent release of substance P [23]. Substance P, in turn, causes plasma extravasation through the NK1 receptor [32]. Previous small studies have not supported an association between SNPs in the B2 receptor and ACE inhibitor-associated angioedema. We also did not find an association between genetic polymorphisms in the B2 receptor and angioedema in the present study. In addition, when ACE is inhibited, bradykinin can be inactivated primarily by other enzymes, including membranous aminopeptidase P (APP), an enzyme encoded by the X-linked gene XPNPEP2 [25,33]. An SNP, – 2399C>A (rs3788853, C-2399A) has been associated with APP activity and with ACE inhibitor-associated angioedema in men, but not in women, in the Nashville case–control study [21]. We did not find an association between ACE inhibitor-associated angioedema and XPNPEP2 by GWAS in men and women combined.
Our study also implicated inflammatory pathways in the pathogenesis ACE inhibitor-associated angioedema. We found consistent associations between polymorphisms in two genes involved in immune regulation and ACE inhibitor-associated angioedema. PKCθ is involved in the activation of T lymphocytes [34]. ETV6 (ETS variant gene 6, also known as TEL or translocation ets leukemia) is a transcriptional repressor that is disrupted by translocation in 26–47% of childhood pre-B-cell acute lymphocytic leukemias [35,36], but also in some T-cell leukemias. ETV6 also regulates interleukin 18 (IL-18), IL-10, and IL-4, [36,37] cytokines involved in the clonal expansion of TH1 (IL-18 and IL-10) and TH2 (IL-4) subsets of CD4+ helper T cells [38].
The potential association of genes involved in immune regulation with ACE inhibitor-associated angioedema is intriguing, given clinical risk factors for ACE inhibitor-associated angioedema. For example, a history of seasonal allergies is associated with an increased risk of ACE inhibitor-associated angioedema [7]. In addition, several studies suggest that transplant and immunosuppressant use are associated with an increased risk of ACE inhibitor-associated angioedema [9,39]. The mTOR inhibitors commonly used in transplant patients cause a significant decrease in TH1 relative to TH2 cells [40]. This may also explain the observation that DPP4 (CD26) antigen and activity are decreased during acute ACE inhibitor-associated angioedema [41], as TH1 cells express three to six times more CD26 than do TH2 cells [42]. Taken together, these data suggest the hypothesis that ACE inhibitor-associated angioedema is associated with environmental or genetic factors that reduce the ratio of TH1 to TH2 cells.
A major limitation of this study is a small sample size. This is not unusual in studies of rare adverse drug reactions. Nevertheless, the study population recruited for this GWAS is the largest sample available for ACE inhibitor-associated angioedema of which we are aware. By comparison, in the OCTAVE trial, there were 86 cases of ACE inhibitor-associated angioedema [31]. Because of the small sample size, however, genome-wide significance could not be achieved in the current study. The availability of the ONTARGET enabled us to use a more liberal P-value threshold in the GWAS discovery analysis and to rely on a consistent association of the signals. However, because no other sample of ACE inhibitor-associated angioedema cases and controls rivals the size of the Nashville/Marshfield cohort, the sample size in ONTARGET was inevitably smaller than that used in the discovery dataset, also a limitation.
A second important limitation is the lack of individuals of African descent in the replication group. The finding of a common association between rs2724635 in ETV6 in the GWAS in African Americans and in the Caucasian patients in ONTARGET could suggest a common mechanism independent of race. A concern, however, is the lack of a similar association between this polymorphism and ACE inhibitor-associated angioedema in the GWAS in Caucasians. The association of other SNPs in Chr12p13 in the GWAS in Caucasians highlights this region as an area for future study.
Conclusion
Angioedema is a potentially life-threatening side effect of ACE inhibitors that is considered to result from decreased degradation of vasoactive peptides such as bradykinin and substance P. Patient characteristics that have been associated with an increased risk of ACE inhibitor-associated angioedema include African ancestry, smoking, a history of seasonal allergies, and the concurrent use of neprilysin [31] or DPP4 inhibitors [43]. In contrast to other studies of serious adverse drug effects of comparable size, we found no associations of genome-wide significance, suggesting that ACE inhibitor-associated angioedema behaves more like a complex trait than do other serious adverse drug effects. Nevertheless, genetic variation in neprilysin and in genes involved in immune regulation may be associated with ACE inhibitor- associated angioedema.
Clinical trials indicate that pharmacological inhibition of neprilysin concurrently increases the risk of ACE inhibitor-associated angioedema [31]. The results of the GWAS study also suggest a potential role for immune regulation in the pathogenesis of ACE-associated angioedema. Although a biological explanation of these results is plausible on the basis of clinical observations of associations between ACE inhibitor-associated angioedema and a history of seasonal allergies or immunosuppressant use, these findings must be viewed as hypothesis generating.
Supplementary Material
Acknowledgments
The authors thank Elizabeth Stone, RN, for her work in ascertaining and interviewing patients in Nashville and Terrie Kitchner for her work in abstracting data from the electronic medical record in Marshfield (Wisconsin). They also thank Dr Matthew McQueen for his input into the manuscript.
This study was supported by NIH grants HL079184, RR024975, HL065962, and GM007569, Japanese Ministry of Education, Culture, Sports, Science and Technology, Boehringer Ingelheim Pharmaceuticals.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (www.pharmacogeneticsandgenomics.com).
Conflicts of interest
There are no conflicts of interest.
References
- 1.Brown NJ, Vaughan DE. Angiotensin-converting enzyme inhibitors. Circulation. 1998;97:1411–1420. doi: 10.1161/01.cir.97.14.1411. [DOI] [PubMed] [Google Scholar]
- 2.Baker WL, Coleman CI, Kluger J, Reinhart KM, Talati R, Quercia R, et al. Systematic review: comparative effectiveness of angiotensin-converting enzyme inhibitors or angiotensin II-receptor blockers for ischemic heart disease. Ann Intern Med. 2009;151:861–871. doi: 10.7326/0003-4819-151-12-200912150-00162. [DOI] [PubMed] [Google Scholar]
- 3.Byrd JB, Adam A, Brown NJ. Angiotensin-converting enzyme inhibitor-associated angioedema. Immunol Allergy Clin North Am. 2006;26:725–737. doi: 10.1016/j.iac.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Agodoa LY, Appel L, Bakris GL, Beck G, Bourgoignie J, Briggs JP, et al. Effect of ramipril vs amlodipine on renal outcomes in hypertensive nephrosclerosis: a randomized controlled trial. JAMA. 2001;285:2719–2728. doi: 10.1001/jama.285.21.2719. [DOI] [PubMed] [Google Scholar]
- 5.Slater EE, Merrill DD, Guess HA, Roylance PJ, Cooper WD, Inman WH, et al. Clinical profile of angioedema associated with angiotensin converting-enzyme inhibition. JAMA. 1988;260:967–970. [PubMed] [Google Scholar]
- 6.Brown NJ, Ray WA, Snowden M, Griffin MR. Black americans have an increased rate of angiotensin converting enzyme inhibitor-associated angioedema. Clin Pharmacol Ther. 1996;60:8–13. doi: 10.1016/S0009-9236(96)90161-7. [DOI] [PubMed] [Google Scholar]
- 7.Kostis JB, Kim HJ, Rusnak J, Casale T, Kaplan A, Corren J, et al. Incidence and characteristics of angioedema associated with enalapril. Arch Intern Med. 2005;165:1637–1642. doi: 10.1001/archinte.165.14.1637. [DOI] [PubMed] [Google Scholar]
- 8.Miller DR, Oliveria SA, Berlowitz DR, Fincke BG, Stang P, Lillienfeld DE. Angioedema incidence in US veterans initiating angiotensin-converting enzyme inhibitors. Hypertension. 2008;51:1624–1630. doi: 10.1161/HYPERTENSIONAHA.108.110270. [DOI] [PubMed] [Google Scholar]
- 9.Byrd JB, Woodard-Grice A, Stone E, Lucisano A, Schaefer H, Yu C, et al. Association of angiotensin-converting enzyme inhibitor-associated angioedema with transplant and immunosuppressant use. Allergy. 2010;65:1381–1387. doi: 10.1111/j.1398-9995.2010.02398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morimoto T, Gandhi TK, Fiskio JM, Seger AC, So JW, Cook EF, et al. An evaluation of risk factors for adverse drug events associated with angiotensin-converting enzyme inhibitors. J Eval Clin Pract. 2004;10:499–509. doi: 10.1111/j.1365-2753.2003.00484.x. [DOI] [PubMed] [Google Scholar]
- 11.Brown NJ, Snowden M, Griffin MR. Recurrent angiotensin-converting enzyme inhibitor-associated angioedema. JAMA. 1997;278:232–233. doi: 10.1001/jama.278.3.232. [DOI] [PubMed] [Google Scholar]
- 12.Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, et al. SLCO1B1 variants and statin-induced myopathy – a genomewide study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 13.Mallal S, Nolan D, Witt C, Masel G, Martin AM, Moore C, et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359:727–732. doi: 10.1016/s0140-6736(02)07873-x. [DOI] [PubMed] [Google Scholar]
- 14.Kindmark A, Jawaid A, Harbron CG, Barratt BJ, Bengtsson OF, Andersson TB, et al. Genome-wide pharmacogenetic investigation of a hepatic adverse event without clinical signs of immunopathology suggests an underlying immune pathogenesis. Pharmacogenomics J. 2008;8:186–195. doi: 10.1038/sj.tpj.6500458. [DOI] [PubMed] [Google Scholar]
- 15.Sarasquete ME, Garcia-Sanz R, Marin L, Alcoceba M, Chillon MC, Balanzategui A, et al. Bisphosphonate-related osteonecrosis of the jaw is associated with polymorphisms of the cytochrome P450 CYP2C8 in multiple myeloma: a genome-wide single nucleotide polymorphism analysis. Blood. 2008;112:2709–2712. doi: 10.1182/blood-2008-04-147884. [DOI] [PubMed] [Google Scholar]
- 16.Emanueli C, Grady EF, Madeddu P, Figini M, Bunnett NW, Parisi D, et al. Acute ACE inhibition causes plasma extravasation in mice that is mediated by bradykinin and substance P. Hypertension. 1998;31:1299–1304. doi: 10.1161/01.hyp.31.6.1299. [DOI] [PubMed] [Google Scholar]
- 17.Sulpizio AC, Pullen MA, Edwards RM, Brooks DP. The effect of acute angiotensin-converting enzyme and neutral endopeptidase 24. 11 inhibition on plasma extravasation in the rat. J Pharmacol Exp Ther. 2004;309:1141–1147. doi: 10.1124/jpet.103.064105. [DOI] [PubMed] [Google Scholar]
- 18.Zuraw BL. Clinical practice. Hereditary angioedema. N Engl J Med. 2008;359:1027–1036. doi: 10.1056/NEJMcp0803977. [DOI] [PubMed] [Google Scholar]
- 19.The ONTARGET Investigators. Telmisartan, ramipril, or both in patients at high risk for vascular events. N Engl J Med. 2008;358:1547–1559. doi: 10.1056/NEJMoa0801317. [DOI] [PubMed] [Google Scholar]
- 20.McCarty CA, Wilke RA, Giampietro PF, Wesbrook SD, Caldwell MD. Marshfield Clinic Personalized Medicine Research Project (PMRP): design, methods and recruitment for a large population-based biobank. Pers Med. 2005;2:49–79. doi: 10.1517/17410541.2.1.49. [DOI] [PubMed] [Google Scholar]
- 21.Woodard-Grice AV, Lucisano AC, Byrd JB, Stone ER, Simmons WH, Brown NJ. Sex-dependent and race-dependent association of XPNPEP2 C-2399A polymorphism with angiotensin-converting enzyme inhibitor-associated angioedema. Pharmacogenet Genomics. 2010;20:532–536. doi: 10.1097/FPC.0b013e32833d3acb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu B, Figini M, Emanueli C, Geppetti P, Grady EF, Gerard NP, et al. The control of microvascular permeability and blood pressure by neutral endopeptidase. Nat Med. 1997;3:904–907. doi: 10.1038/nm0897-904. [DOI] [PubMed] [Google Scholar]
- 23.Kopp UC, Farley DM, Smith LA. Bradykinin-mediated activation of renal sensory neurons due to prostaglandin-dependent release of substance P. Am J Physiol. 1997;272:R2009–R2016. doi: 10.1152/ajpregu.1997.272.6.R2009. [DOI] [PubMed] [Google Scholar]
- 24.Blais CJ, Rouleau JL, Brown NJ, Lepage Y, Spence D, Munoz C, et al. Serum metabolism of bradykinin and des-Arg9-bradykinin in patients with angiotensin-converting enzyme inhibitor-associated angioedema. Immunopharmacology. 1999;43:293–302. doi: 10.1016/s0162-3109(99)00133-2. [DOI] [PubMed] [Google Scholar]
- 25.Duan QL, Nikpoor B, Dube MP, Molinaro G, Meijer IA, Dion P, et al. A variant in XPNPEP2 is associated with angioedema induced by angiotensin I-converting enzyme inhibitors. Am J Hum Genet. 2005;77:617–626. doi: 10.1086/496899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouchard L, Faucher G, Tchernof A, Deshaies Y, Lebel S, Hould FS, et al. Comprehensive genetic analysis of the dipeptidyl peptidase-4 gene and cardiovascular disease risk factors in obese individuals. Acta Diabetol. 2009;46:13–21. doi: 10.1007/s00592-008-0049-4. [DOI] [PubMed] [Google Scholar]
- 27.Cui J, Melista E, Chazaro I, Zhang Y, Zhou X, Manolis AJ, et al. Sequence variation of bradykinin receptors B1 and B2 and association with hypertension. J Hypertens. 2005;23:55–62. doi: 10.1097/00004872-200501000-00013. [DOI] [PubMed] [Google Scholar]
- 28.Seneviratne C, Ait-Daoud N, Ma JZ, Chen G, Johnson BA, Li MD. Susceptibility locus in neurokinin-1 receptor gene associated with alcohol dependence. Neuropsychopharmacology. 2009;34:2442–2449. doi: 10.1038/npp.2009.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4:e1000166. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 31.Kostis JB, Packer M, Black HR, Schmieder R, Henry D, Levy E. Omapatrilat and enalapril in patients with hypertension: the Omapatrilat Cardiovascular Treatment vs. Enalapril (OCTAVE) trial. Am J Hypertens. 2004;17:103–111. doi: 10.1016/j.amjhyper.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Baluk P, Thurston G, Murphy TJ, Bunnett NW, McDonald DM. Neurogenic plasma leakage in mouse airways. Br J Pharmacol. 1999;126:522–528. doi: 10.1038/sj.bjp.0702323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blais CJ, Marc-Aurele J, Simmons WH, Loute G, Thibault P, Skidgel RA, et al. Des-Arg9-bradykinin metabolism in patients who presented hypersensitivity reactions during hemodialysis: role of serum ACE and aminopeptidase P. Peptides. 1999;20:421–430. doi: 10.1016/s0196-9781(99)00020-0. [DOI] [PubMed] [Google Scholar]
- 34.Salek-Ardakani S, So T, Halteman BS, Altman A, Croft M. Differential regulation of Th2 and Th1 lung inflammatory responses by protein kinase C theta. J Immunol. 2004;173:6440–6447. doi: 10.4049/jimmunol.173.10.6440. [DOI] [PubMed] [Google Scholar]
- 35.Bohlander SK. ETV6: a versatile player in leukemogenesis. Semin Cancer Biol. 2005;15:162–174. doi: 10.1016/j.semcancer.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Boily G, Larose J, Langlois S, Sinnett D. Identification of transcripts modulated by ETV6 expression. Br J Haematol. 2007;136:48–62. doi: 10.1111/j.1365-2141.2006.06377.x. [DOI] [PubMed] [Google Scholar]
- 37.Sakurai T, Yamada T, Kihara-Negishi F, Teramoto S, Sato Y, Izawa T, et al. Effects of overexpression of the Ets family transcription factor TEL on cell growth and differentiation of K562 cells. Int J Oncol. 2003;22:1327–1333. [PubMed] [Google Scholar]
- 38.Bocsi J, Richter M, Hambsch J, Barten MJ, Dahnert I, Schneider P, et al. Transient Th1/Th2 disbalance indicates postoperative effusions and edema after cardiopulmonary bypass in children. Cytometry A. 2006;69:165–168. doi: 10.1002/cyto.a.20213. [DOI] [PubMed] [Google Scholar]
- 39.Duerr M, Glander P, Diekmann F, Dragun D, Neumayer HH, Budde K. Increased incidence of angioedema with ACE inhibitors in combination with mtor inhibitors in kidney transplant recipients. Clin J Am Soc Nephrol. 2010;5:703–708. doi: 10.2215/CJN.07371009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Byrd JB, Touzin K, Sile S, Gainer JV, Yu C, Nadeau J, et al. Dipeptidyl peptidase IV in angiotensin-converting enzyme inhibitor associated angioedema. Hypertension. 2008;51:141–147. doi: 10.1161/HYPERTENSIONAHA.107.096552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willheim M, Ebner C, Baier K, Kern W, Schrattbauer K, Thien R, et al. Cell surface characterization of T lymphocytes and allergen-specific T cell clones: correlation of CD26 expression with T(H1) subsets. J Allergy Clin Immunol. 1997;100:348–355. doi: 10.1016/s0091-6749(97)70248-3. [DOI] [PubMed] [Google Scholar]
- 43.Brown NJ, Byiers S, Carr D, Maldonado M, Warner BA. Dipeptidyl peptidase-IV inhibitor use associated with increased risk of ACE inhibitor-associated angioedema. Hypertension. 2009;54:516–523. doi: 10.1161/HYPERTENSIONAHA.109.134197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.