Abstract
The MADS-domain transcription factor SHORT VEGETATIVE PHASE plays a key role as a repressor of the transition to flowering and as a regulator of early floral development in Arabidopsis thaliana (Arabidopsis). However, no flowering-time repressors have been functionally identified in the model legume Medicago truncatula (Medicago). In this study, phylogenetic analysis of two closely-related MtSVP-like sequences, MtSVP1 and MtSVP2, showed that their predicted proteins clustered together within the eudicot SVP clade. To determine if the MtSVP-like genes have a role in flowering, they were functionally characterized in Medicago and Arabidopsis. Transcripts of both MtSVP genes were abundant and broadly expressed in vegetative tissues but were detected at much lower levels in flowers in Medicago. Over-expression of the MtSVP genes in Arabidopsis resulted in delayed flowering and flowers with many abnormal phenotypes such as leafy sepals, changes to floral organ number and longer pedicels than the wild type. By contrast, in transgenic Medicago, over-expression of MtSVP1 resulted in alterations to flower development, but did not alter flowering time, suggesting that MtSVP1 may not function to repress the transition to flowering in Medicago.
Key words: Arabidopsis, flowering, legume, MADS, Medicago, SVP.
Introduction
Plants precisely regulate the timing of the transition from vegetative to reproductive growth to optimize sexual reproduction and productivity (Putterill et al., 2004). The genetic pathways that regulate flowering time have been investigated in many plants, but are best understood in the monocot cereals such as rice, wheat, and barley and in the eudicot model plant, Arabidopsis thaliana (Arabidopsis) (Trevaskis et al., 2006; Kim et al., 2009; Srikanth and Schmid, 2011; Andres and Coupland, 2012). In Arabidopsis, six major pathways influence flowering time in response to external (photoperiod, vernalization, and thermosensory pathways) and endogenous signals (autonomous, age, and gibberellin) with carbohydrate status also playing a critical role in controlling the transition to flowering (Wahl et al., 2013). Flowering signals from these multiple pathways are integrated by floral integrator genes including FLOWERING LOCUS T (FT), TWIN SISTER OF FT (TSF), and SUPPRESSOR OF OVER-EXPRESSION OF CONSTANS1 (SOC1) that go on to promote the transition to flowering.
In Arabidopsis, the gene encoding the MADS-domain transcription factor SHORT VEGETATIVE PHASE (AtSVP) is expressed broadly during vegetative development including in leaves and shoot apices and functions as a flowering time repressor (Hartmann et al., 2000; Lee et al., 2007b ). Consistent with a role in flowering time regulation, Arabidopsis svp mutants are early flowering (Hartmann et al., 2000) while overexpression of SVP (35S:AtSVP) leads to a late-flowering phenotype (Masiero et al., 2004; Li et al., 2008). AtSVP functions, in part, by complexing with a second important flowering time repressor, the MADS-domain protein FLOWERING LOCUS C (FLC) and delays flowering by repressing FT, TSF, and SOC1 in the leaves and SOC1 at the apical meristem (Searle et al., 2006; Lee et al., 2007a ; Fujiwara et al., 2008; Li et al., 2008; Jang et al., 2009). The expression of AtSVP is down-regulated by the thermosensory, autonomous, and gibberellin flowering-time pathways and after the transition to flowering there is loss of AtSVP expression in the inflorescence meristem (Hartmann et al., 2000; Lee et al., 2007b ; Liu et al., 2007; Li et al., 2008).
However, AtSVP expression is detectable in floral primordia at the very earliest stages of their development (Hartmann et al., 2000; Liu et al., 2007). AtSVP together with the closely-related MADS-domain protein AGAMOUS-LIKE 24 (AGL24) and a third MADS factor SOC1 play redundant roles in promoting floral primordium proliferation during stage 1 and stage 2 of flower development by repressing the class E MADS domain floral homeotic gene, SEPALLATA 3 (SEP3) (Gregis et al., 2009; Liu et al., 2009). AtSVP and AGL24 also form heterodimers with the MADS domain protein APETALA1 (AP1) to repress class B and C floral organ identity genes via recruitment of the SEUSS-LEUNIG co-repressor complex (Gregis et al., 2009; Posé et al., 2012). After stage 2, AtSVP expression is repressed by SEP3, which then functions with LFY to activate the class B and class C genes (Kaufmann et al., 2009; Liu et al., 2009). Although floral development proceeds normally in svp and agl24 mutants, svp agl24 double mutants show floral abnormalities that increase in severity with increases in temperature (Gregis et al., 2006). Overexpression of AtSVP causes aberrant floral morphology in transgenic Arabidopsis such as the formation of secondary flowers, sepaloid petals, and flowers with shoot-like structures (Masiero et al., 2004; Liu et al., 2007).
While the role of SVP genes in flowering time and floral primordia development appear to be conserved in some plants, other SVP genes can have rather diverse functions. The SVP-like genes from Brassica campestris (BcSVP) (Lee et al., 2007a ), Eucalyptus grandis (EgrSVP) (Brill and Watson, 2004), and Poncirus trifoliate (PtSVP) (Li et al., 2010) are implicated both in floral transition and meristem identity based on their expression pattern and the overexpression phenotype in Arabidopsis. In Antirrhinum majus (snapdragon), the SVP-like gene AmINCOMPOSITA (INCO) has developed a more prominent role in floral meristem identity (Masiero et al., 2004). inco mutants have no flowering-time phenotype, but display abnormal floral morphology that includes the lack of repression of prophyll development. In monocots, the three SVP-like genes in Hordeum vulgare (barley): BARLEY MADS1 (HvBM1), VEGETATIVE TO REPRODUCTIVE TRANSITION GENE 2 (HvVRT2), and HvBM10 (HvVRT2-like) are expressed in vegetative tissues. RNA interference (RNAi) constructs targeting either BM1 or BM10 did not result in any phenotypic abnormalities or change in heading time, but ectopic expression caused floral reversion in transgenic barley plants (Trevaskis et al., 2007b ). In Oryza sativa (rice), there are three SVP-like genes: OsMADS22, OsMADS47, and OsMADS55. Knock-down and over-expression lines showed that these genes appear to function in brassinosteroid signalling and shoot development (Duan et al., 2006; Lee et al., 2008). Heterologous expression of OsMADS22 and OsMADS47 in Arabidopsis caused floral defects (Fornara et al., 2008) while that of OsMADS55 led to delayed flowering (Lee et al., 2012). In perennials, there is a role for SVP-like proteins in dormancy. Deletion of the six SVP-like genes [DORMANCY-ASSOCIATED MADS-BOX 1–6 (DAM1–6) in Prunus persica (peach) resulted in failure to enter dormancy under cold or short-day induction (Bielenberg et al., 2008; Jimenez et al., 2009). In Actinidia chinensis (kiwifruit), a perennial vine, all of the four SVP-like genes (AcSVP1–4) were implicated in bud dormancy and their ectopic expression in Arabidopsis caused floral abnormalities. However, only AcSVP1 and AcSVP3 were able to complement the Arabidopsis svp mutant (Wu et al., 2012).
SVP-like sequences were identified in another large and agronomically important plant family, the Fabaceae, but none have been functionally characterized (Hecht et al., 2005; Jung et al., 2012). Thus, the aim of this study was to investigate the role of SVP-like genes in regulating flowering time in the model legume, Medicago truncatula (Medicago). Medicago offers several advantages including a diploid and relatively modest genome of ~525Mb that has been sequenced, large collections of accessions with variation in flowering time, and extensive mutant populations for forward and reverse genetic screens (Julier et al., 2007; Rose, 2008; Tadege et al., 2008; He et al., 2009; Branca et al., 2011; Young et al., 2011). Similar to the Arabidopsis winter annual accessions and winter varieties of wheat and barley, flowering in Medicago is promoted by a long day (LD)-photoperiod and vernalization and Medicago FTa1, an orthologue of FT, plays an important role in promoting flowering under these inductive conditions (Laurie et al., 2011; Yeoh et al., 2011). However, Medicago lacks flowering-time repressors that belong to the FLC-MADS AFFECTING FLOWERING (MAF) clade in the Brassicaceae family or genes that are similar to the VERNALIZATION2 (VRN2) flowering time repressor from cereals that are directly or indirectly repressed by vernalization and other pathways such as the Arabidopsis autonomous pathway (Hecht et al., 2005; Trevaskis et al., 2007a ; Putterill et al., 2013; Ruelens et al., 2013).
Here, two Medicago SVP-like genes were identified and characterized by analysing their gene-expression patterns and by manipulating their expression in transgenic Arabidopsis and Medicago plants.
Materials and methods
Bioinformatics
The two Medicago (Medicago truncatula) SVP-like genes were identified by BLAST searches against the Medicago EST and BAC sequences. Alignment was performed using ClustalW in the Geneious software package [version 6.0.5 available from www.geneious.com (last accessed 1 November 2013) (Biomatters, Ltd.)]. A phylogenetic tree was generated using the Neighbor–Joining method via bootstrap resampling in the Geneious program. The accession numbers for the sequences used are as follows: Medicago MtSVP1 (Medtr5g032520), MtSVP2 (Medtr5g032150), Medtr5g066180; Arabidopsis AtSVP (At2G22540), AtAP1 (At1g69120), AtSOC1 (At2g45660), AtAGL14 (At4g11880), AtAGL19 (At4g22950), AtAGL24 (At4g24540); rice OsMADS22 (AB107957), OsMADS47 (AY345221), OsMADS55 (AY345223); barley HvBM10 (EF043040), HvVRT2 (DQ201168), HvBM1 (AJ249142); Lolium perenne (ryegrass) LpMADS10 (AAZ17549); Euphorbia esula (leafy spurge) EeDAM2 (ABY60423); peach PpDAM1 (DQ863253), PpDAM2 (DQ863255), PpDAM3 (DQ863256), PpDAM4 (DQ863250), PpDAM5 (DQ863251), PpDAM6 (DQ863252); Brassica campestris (Chinese cabbage) BcSVP (DQ922945); snapdragon AmINCOMPOSITA (CAG27846); Eucalyptus grandis EgrSVP (AY263809); Populus trichocarpa (poplar) PtSVP (XM_002310274); Lotus japonicus LjSVP (TC40194); Pisum sativum (garden pea) PsSVP (AY830919); Glycine max (soybean) GmSVP1 (Glyma01g02880), GmSVP2 (Glyma02g04710), GmSVP3 (Glyma06g10020), GmSVP4 (Glyma07g30040), GmSVP5 (Glyma08g07260), GmSVP6 (Glyma13g33040), GmSVP7 (Glyma15g06300), GmSVP8 (Glyma15g06310); kiwifruit AcSVP1 (JF838216), AcSVP2 (JF838217), AcSVP3 (JF838218), AcSVP4 (JF838219); and Phaseolus vulgaris (common bean) PvSVP (TC35086). These sequences were obtained from previous studies (Bielenberg et al., 2008; Fornara et al., 2008; Jung et al., 2012; Wu et al., 2012) and NCBI.
Plant material and growth conditions
Jester and R108_C3 (R108) are the two Medicago genotypes used in this study. They belong to the two subspecies of Medicago truncatula Gaertn (barrel medic), ssp. truncatula and ssp. tricycla, respectively. The reverse genetic lines for MtSVP2 were obtained from the Samuel Roberts Noble Foundation (Ardmore, Oklahoma). Scarification, germination, seed vernalization prior to growth in LD conditions (16/8h light/dark), and cultivation of Medicago plants were done as described previously (Laurie et al., 2011; Yeoh et al., 2013). For vernalization of seedlings, seeds germinated for 3 d at 21 °C were grown on soil in LD until a monofoliate and the first trifoliate leaf emerged (2-leaf stage of development, ~11 d in LD). Plants at this stage were vernalized at 4 °C for 2 weeks in controlled growth cabinets under short-day (SD) photoperiod (8/16h light/dark) with 40% humidity and then returned in LD. Flowering time of Medicago plants was measured in terms of the number of days from germination to flowering and/or the total number of nodes accumulated on the main axis at flowering.
The Arabidopsis wild type (WT) Columbia (Col) and svp-41 mutant in the Col background were grown in LD. The svp-41 mutant harbours a deletion of bases 193 and 194 in the AtSVP coding sequence (Hartmann et al., 2000). The resulting frameshift creates a premature stop codon at the 69th amino acid (aa), generating a truncated 69 aa protein compared with the wild-type AtSVP protein of 240 aa in length. This mutation causes the svp-41 mutant to flower earlier than WT Col plants.
RNA extraction and quantitative reverse transcriptase-PCR (qRT-PCR) analysis
RNA extraction, cDNA synthesis, and qRT-PCR were performed as previously described (Laurie et al., 2011; Yeoh et al., 2013). The identity of the PCR amplicons was checked by DNA sequencing. For gene expression analyses, leaf or shoot apical buds were harvested from three-leaf (fully-expanded) stage plants, unless otherwise stated. In the developmental time-course, flower buds were small unopened flowers, while flowers were open flowers. The data were normalized to the housekeeping gene, PROTODERMAL FACTOR 2 (PDF2) or TUBULIN. The primers used are shown in Supplementary Table S1 at JXB online.
Generation of transgenic Arabidopsis plants
In order to generate MtSVP overexpression (35S:MtSVP) constructs, the cDNAs of MtSVP1 and MtSVP2 were amplified using the Phusion Hot Start High-Fidelity DNA Polymerase (New England Biolabs) with gene-specific primers (Fig. 1A; see Supplementary Table S1 at JXB online) and then cloned in pCR8/GW/TOPO TA (TOPO) entry vector (Invitrogen, Corporation, USA). The entry clones were linearized with 5U XhoI and then recombined into the plant transformation vector, pB2GW7 (Karimi et al., 2002) to create expression clones using the GATEWAY TECHNOLOGY kit (Invitrogen, Corporation, USA) according to the manufacturer’s instructions. Entry and expression clones were subjected to restriction enzyme digestion and sequencing to check for the correct orientation and the absence of mutation in the constructs. Clones were transformed by heat shock into One Shot TOP10 Chemically Competent E. coli (Invitrogen, Corporation, USA) according to the manufacturer’s instructions. Agrobacterium tumefaciens strain GV3101 with expression clones was applied to Arabidopsis WT Col and svp-41 mutant flower buds as previously described by Martinez-Trujillo et al., (2004). Seeds were directly sown onto soil and transformants were selected using the Basta herbicide (10mg l–1).
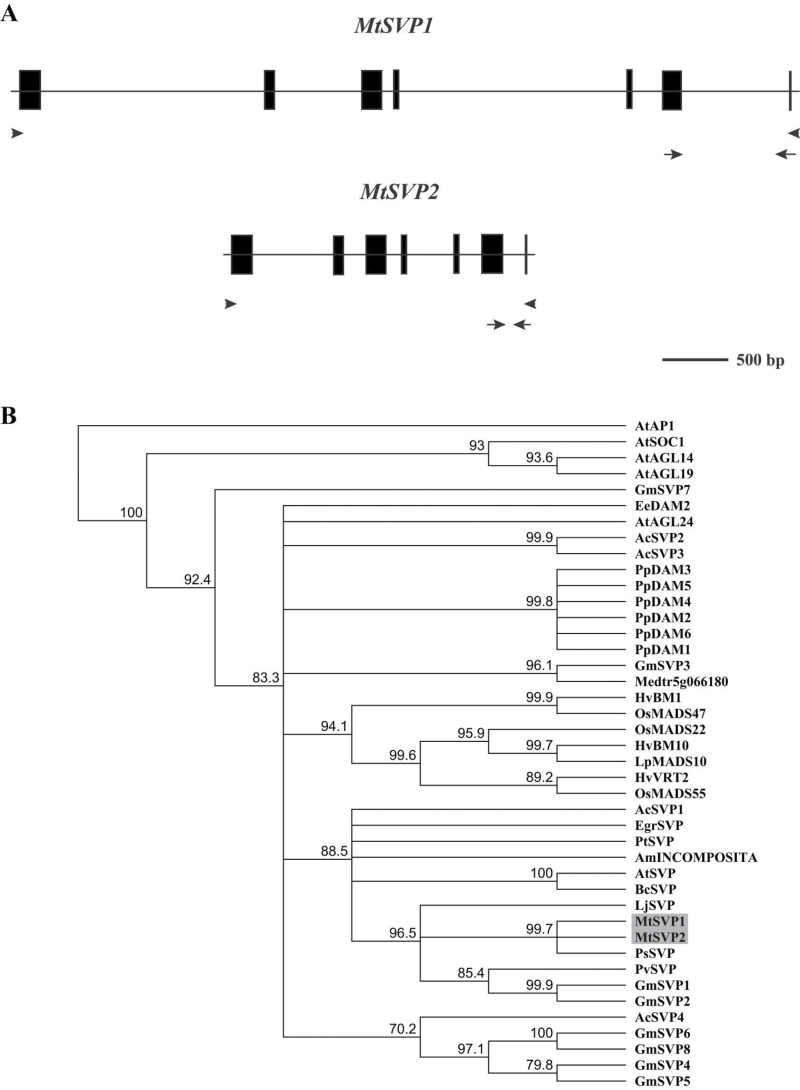
Fig. 1.
Genomic structures of MtSVP1 and MtSVP2 and phylogenetic analysis of SVP-like proteins from different species. (A) The exons are indicated by black boxes and introns by a thin line. The arrows indicate the positions of primers used for quantitative real-time PCR while the arrowheads represent the primers used for cloning the full-length MtSVP1 and MtSVP2 coding sequences from Medicago cDNA. (B) A consensus phylogenetic tree based on the amino acid alignment of MtSVP1 and MtSVP2 predicted proteins (shaded box) and SVP-like proteins from other species. The tree was generated using the Neighbor–Joining (NJ) method via bootstrap resampling with support threshold of 55% and rooted on AtAP1. The numbers indicate the bootstrap values based on 1000 replicates. Ac, Actinidia chinensis; Am, Antirrhinum majus; At, Arabidopsis thaliana; Bc, Brassica campestris; Ee, Euphorbia esula; Egr, Eucalyptus grandis; Gm, Glycine max; Hv, Hordeum vulgare; Lj, Lotus japonicas; Lp, Lolium perenne; Mt, Medicago truncatula; Os, Oryza sativa; Pp, Prunus persica; Ps, Pisum sativum; Pt, Poncirus trifoliate; and Pv, Phaseolus vulgaris. (This figure is available in colour at JXB online.)
Transformation and regeneration of Medicago R108
Transformation of Medicago R108 and regeneration via somatic embryogenesis was conducted as previously described by Laurie et al. (2011) with minor modifications. Two rounds of transformation were conducted with 35S:MtSVP1 or vector only (pB2GW7) as a control. The control group went through somatic embryogenesis on selective or non-selective medium as the negative control or the regeneration control, respectively. The transformant plants were selected with 1–2mg l–1 of dl-phosphinothricin (Sigma). Regenerated T0 transformed plantlets were transferred to soil and sprayed again with 120mg l–1 Basta (Kiwicare) to confirm their transgenic nature. PCR-genotyping also confirmed the presence of the transgenes.
Reverse genetic screening for Tnt1 insertions and linkage analysis
Mutant plants with Tnt1 insertions at the MtSVP genes were screened using a reverse genetic approach (Tadege et al., 2008). Tnt1 insertions were identified in the Noble Foundation, USA using Tnt1- and gene-specific primers. No lines were found to have Tnt1 insertion at the MtSVP1 locus while two different lines have Tnt1 sequences inserted within the MtSVP2 gene, one within intron 4 in line NF11029 and one within intron 6 in line NF13617. The lines were grown under LD and vernalized LD (VLD) conditions. Their gDNA was extracted and the nature and direction of Tnt1 insertion in the lines were confirmed using PCR. The expression levels of MtSVP2 were measured in the transgenic homozygous plants from the two lines using qRT-PCR. All the primers used are shown in Supplementary Table S1 at JXB online.
Results
Identification of two closely related SVP-like genes in Medicago
The top-scoring hits from interrogation of Medicago genomic and EST sequences using BLAST analyses with the AtSVP protein were two closely-related SVP-like genes named MtSVP1 and MtSVP2. MtSVP1 (Medtr5g032520) (Hecht et al., 2005) and MtSVP2 (Medtr5g032150) encode MADS-domain proteins with 89% aa identity to each other and 70% and 66% aa identity respectively, with the AtSVP protein. MtSVP1 and MtSVP2 were less similar to AGL24, which is the most closely related MADS domain protein to SVP (Parenicova et al., 2003), with 56% and 55% aa identity, respectively. Alignment with SVP/AGL24-like proteins indicates that the MADS-box, intervening (I-box) region, and keratin-like (K-box) motif are well conserved in MtSVP1 and MtSVP2 (see Supplementary Fig. S1 at JXB online) consistent with the features shared by the MADS-domain family of transcription factors in plants (Parenicova et al., 2003). MtSVP1 and MtSVP2 have a comparable genomic structure with 7 exons, but MtSVP1 is a bigger gene due to having larger introns (Fig. 1A).
In a Neighbor–Joining phylogenetic tree, MtSVP1 and MtSVP2 grouped with eudicot SVP/AGL24 proteins in a clade that is distinct from other Arabidopsis MADS-box proteins such as SOC1, AGL14 and AGL19 (Fig. 1B). The two MtSVP proteins and other legume SVP proteins formed a sister clade to AtSVP with MtSVP1 and MtSVP2 being most closely-related to pea PsSVP. The monocot SVP-like proteins from rice (OsMADS47, OsMADS55, and OsMADS22), barley (HvBM1, HvBM10, and HvVRT2) and ryegrass (LpMADS10) formed an independent sub-clade, distinct from the eudicot group.
A third candidate MtSVP/AGL24 gene, Medtr5g066180 (Hecht et al., 2005) encoded a protein that was less similar to AtSVP (48% aa identity) than were MtSVP1 and MtSVP2. The divergence of Medtr5g066180 compared with MtSVP1 and MtSVP2 was also observed in the phylogenetic tree as it fell outside the clade containing AtSVP, MtSVP1, and MtSVP2 (Fig. 1B). It was slightly more similar to AGL24 (54% aa identity) but did not fall into an AGL24 clade. Thus, further work focused on investigating the expression and function of MtSVP1 and MtSVP2.
Gene expression patterns of MtSVP1 and MtSVP2
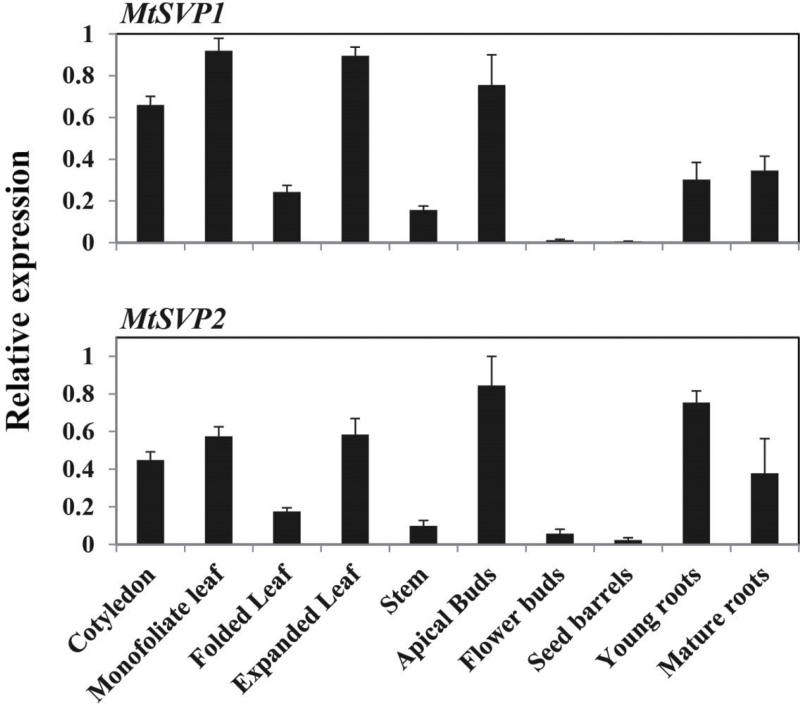
To gain insight into the possible roles of MtSVP1 and MtSVP2 in plant development, their gene expression profiles were analysed using qRT-PCR. First, transcript abundance in different tissues of vernalized long-day grown (VLD) Medicago wild type (WT) plants was compared (Fig. 2). Both MtSVP1 and MtSVP2 were expressed during vegetative development in all the tissues tested; most highly in cotyledons, monofoliate leaves, expanded trifoliate leaves, and apical buds. Transcripts of both genes were also present in folded leaves, stems, and young and mature roots and were detected at much lower levels in young flower buds and developing seed barrels.
Fig. 2.
Expression of MtSVP1 and MtSVP2 in different tissues of Medicago wild-type Jester plants. Plants were grown in LD after 2 weeks of vernalization at 4 °C in the dark. The young roots and cotyledons were from 5-d-old seedlings; monofoliate leaf, folded and expanded trifoliate leaves, stem, apical buds, and mature roots from 15-d-old plants; flower buds (young) from 23-d-old flowering plants; and seed barrels from 35-d-old plants. Gene expression was determined using qRT-PCR and the data are shown as the mean ±standard error (SE) of three biological replicates, which were normalized to Medicago PROTODERMAL FACTOR2 (PDF2). The data are presented relative to the highest value. Tissues were harvested at 2h after dawn [Zeitgeber time (ZT) 2]. (This figure is available in colour at JXB online.)
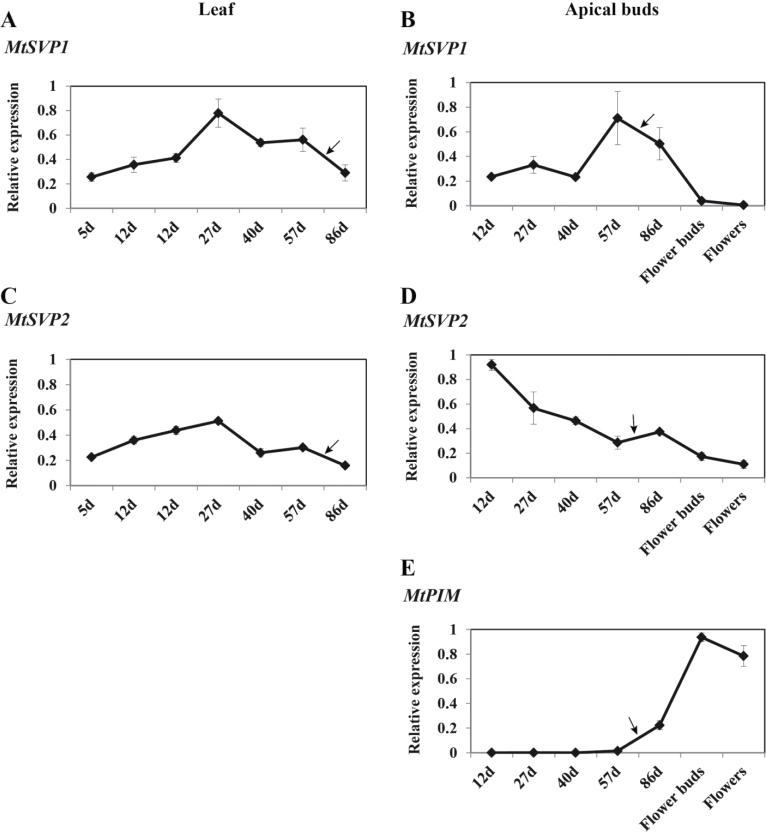
The expression of MtSVP1 and MtSVP2 in leaves and shoot apices was examined in more detail over a developmental time-course in Medicago WT R108 plants in LD conditions (Fig. 3). The expression of the AP1-like gene, PROLIFERATING INFLORESCENCE MERISTEM (PIM1) was used as a marker of the transition to flowering (Fig 3E) together with the appearance of the first visible floral buds. Consistent with the results shown in Fig. 2, both genes were expressed throughout development in leaves and shoot apices, with expression declining to its lowest level in mature flowers. In leaves, MtSVP1 gene expression increased 3–4-fold after 27 d of growth and then gradually declined back to the levels observed in young vegetative plants, while MtSVP2 gene expression was weakly elevated by about 2-fold at 27 d before decreasing back to the levels seen in young plants (Fig. 3A, C).
Fig. 3.
Developmental regulation of MtSVP1 and MtSVP2 in leaves and shoot apices of Medicago wild-type R108 plants. Relative expression levels of MtSVP1 and MtSVP2 in fully-expanded trifoliate leaves (A, C) and uppermost apical buds (B, D) harvested from plants of increasing developmental ages grown in LD. The arrows indicate the time of flowering. The floral meristem identity gene, PROLIFERATING INFLORESCENCE MERISTEM (MtPIM) was used as a molecular marker of the floral transition (E). Tissues were harvested at ZT2. Gene expression was determined using qRT-PCR and the data are shown as the mean ±standard error (SE) of three biological replicates, which were normalized to PDF2. The data are presented relative to the highest value in both tissues for each gene. (This figure is available in colour at JXB online.)
More marked differences in gene expression were observed between the two genes in their developmental pattern of expression in apical buds. MtSVP1 transcript was abundant in apical buds from the early stages of vegetative development, up-regulated further by around 3-fold just prior to the floral transition, and then gradually decreased after the onset of flowering with a strong decline in open flowers (Fig. 3B). MtSVP2 was highly expressed in apical buds in young vegetative plants and transcript levels declined throughout development with the lowest level detected in mature flowers (Fig. 3D). While at lower levels relative to other developmental stages, transcripts of both MtSVP genes were readily detectable in flower buds and open flowers. The effect of extended cold temperatures experienced by seedlings during vernalization treatment on the expression levels of the MtSVP genes was also investigated. However, no direct effects of cold temperatures on the transcript abundance of either of the MtSVP genes in leaves and apical buds were seen (see Supplementary Fig. S2 at JXB online).
Overexpression of the two Medicago SVP-like genes in Arabidopsis delays flowering and causes floral defects
To assess the potential roles of the two MtSVP genes in flowering-time control and the regulation of flower development, they were overexpressed in the heterologous plant Arabidopsis. cDNA clones of the MtSVP genes were generated from total RNA of Jester wild-type plants, fused to the cauliflower mosaic virus (CaMV) 35S promoter (35S:MtSVP1 and 35S:MtSVP2) and introduced into Arabidopsis WT Col plants. Nine T1 35S:MtSVP1 and 35S:MtSVP2 plants with single locus insertions were identified (see Supplementary Table S2A at JXB online).
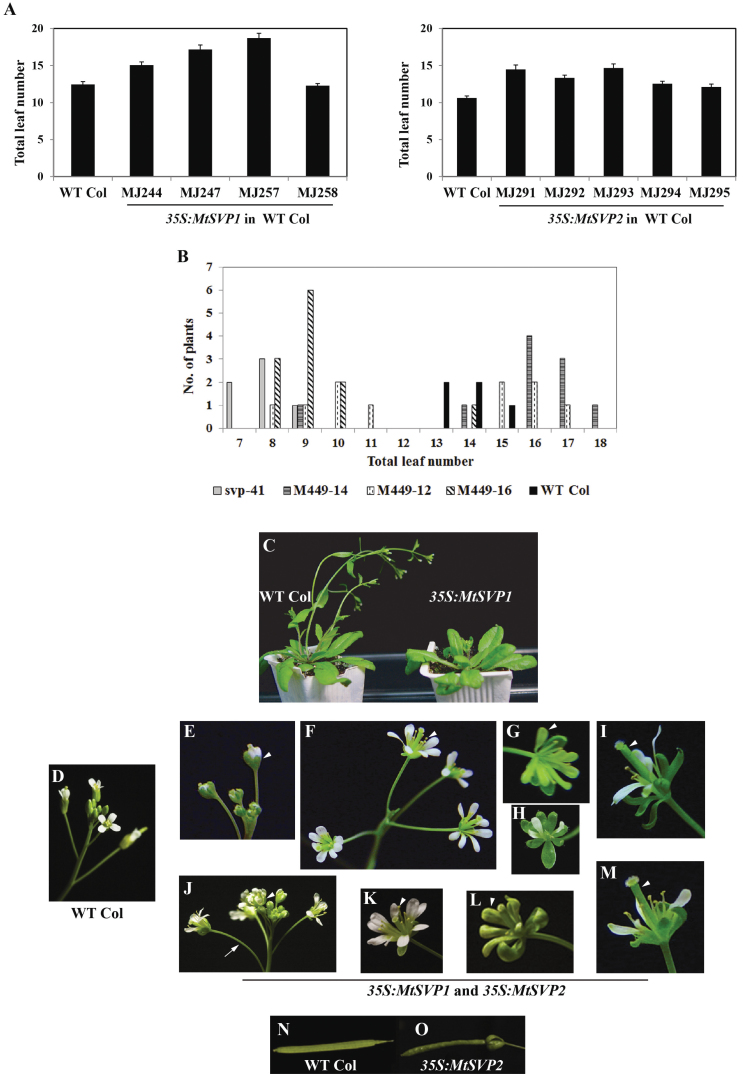
The T2 progeny of eight of these transgenic lines had delayed flowering compared with WT Col plants (Fig. 4A, C) as well as floral defects (Fig. 4D–O), with the severity of these correlating with the delay in flowering time (see Supplementary Table S2 at JXB online). At the onset of flower formation, the transgenic plants were easily distinguished from WT Col (Fig. 4D) by the appearance of enlarged sepals surrounding a more splayed arrangement of floral organs (Fig. 4E, J). At later stages, other defects became more apparent and the most extreme ones were characterized by irregular symmetry of floral organ arrangement, alterations in petal and anther number, elongated carpels, deformed siliques, sepalloid (green) petals and long pedicels (Fig. 4F–I, K–M). In some cases, the sepals remained larger and much greener than the wild type and did not abscise even when siliques had developed (Fig. 4H, L, O).
Fig. 4.
Flowering time and floral phenotypes of transgenic Arabidopsis plants overexpressing MtSVP1 and MtSVP2. (A) Graphs showing the effect of ectopic expression of MtSVP1 (left) and MtSVP2 (right) on the flowering time of Arabidopsis WT Col. The flowering time of transgenic T2 plants from four independent 35S:MtSVP1 Col lines (n=17–19) and five independent 35S:MtSVP2 Col lines (n=15–18) is shown. Col WT plants were grown as a control (n=9–24). Plants were grown in LD until they flowered. Flowering time was measured as the total number of rosette and cauline leaves at flowering and is shown as the mean ±SE. The flowering time of the 35S:MtSVP lines was significantly different from WT (P <0.01; t-test) except for MJ258. (B) Overexpression of MtSVP2 complements the Arabidopsis svp-41 mutation. Histograms showing the flowering time measured as the total number of rosette and cauline leaves at flowering of svp-41 mutants (n=6), WT Col (n=5), and three segregating T2 populations from the overexpression of MtSVP2 in the Arabidopsis svp-41 background (n=10–12). (C) Photograph showing the delayed flowering phenotype of transgenic Arabidopsis plants overexpressing MtSVP1 (right) compared with WT Col of the same age. (D–O) Photographs of inflorescence, flowers, and siliques of WT Col (D, N) and transgenic plants from 35S:MtSVP1 (E–I) and 35S:MtSVP2 (J–M, O) Col lines. Plants overexpressing the MtSVP genes displayed variable floral abnormalities that include failure of sepals to enclose the floral bud at the early stages of development (arrowheads in E and J); irregular number of petals (F, K), sepals (H), and anthers (arrowheads in F and K); splayed arrangement of floral organs (I, M); enlarged carpels (arrowheads in I and M); sepaloid (green) petals (arrowheads in G and L); longer pedicel than WT (arrow in J); and lack of sepal abscission in wrinkled, narrow siliques (O).
MtSVP2 was also overexpressed in the Arabidopsis early flowering mutant, svp-41. Two out of three independent transgenic lines showed rescue of the flowering-time phenotype in the T2 generation (Fig. 4B). As observed for 35S:MtSVP plants in the Col background, some MtSVP2 svp-41 T2 plants flowered later than WT Col plants (Fig. 4B) and had floral abnormalities (data not shown).
Overexpression of MtSVP1 in Medicago results in longer pedicel length and other changes in flower development
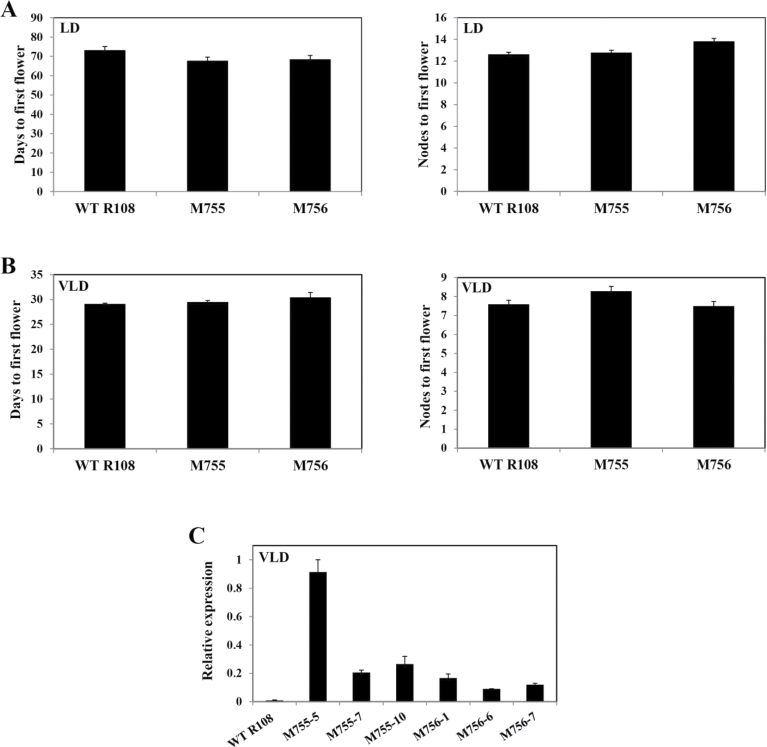
Next, the effect of ectopic MtSVP1 expression on Medicago flowering time and floral development was investigated. WT R108 leaflets were transformed by co-cultivation with Agrobacterium tumefaciens containing the 35S:MtSVP1 construct and plants were regenerated from three different leaflets via somatic embryogenesis (Cosson et al., 2006). The average flowering time of the T1 progeny from two independent T0 regenerated 35S:MtSVP1 lines was similar to the WT R108 plants grown in LD and VLD conditions (Fig. 5A, B) The T1 35S:MtSVP1 plants flowered at the same time as WT R108 despite elevated expression of MtSVP1 in leaves (Fig. 5C).
Fig. 5.
Overexpression of MtSVP1 in Medicago has no effect on flowering time. (A) Flowering time in days to first flower and nodes to first flower of WT R108 (n=8) and transgenic T1 Medicago plants from two independent 35S:MtSVP1 R108 lines (n=11–14) grown in LD. (B) Flowering time in days to first flower and nodes to first flower of WT R108 (n=8) and transgenic 35S:MtSVP1 T1 plants from two independent lines (n=7–10) grown in LD conditions after 14 d of vernalization (VLD). The flowering time in (A) and (B) is shown as the mean ±SE. (C) MtSVP1 transcript accumulation in fully expanded trifoliate leaves of WT R108 and six 35S:MtSVP1 T1 plants from two independent T0 transformants grown in VLD. Gene expression was determined using qRT-PCR and the data are shown as the mean ±SE of two biological replicates, which were normalized to TUBULIN. The data are presented relative to the highest value. The leaf tissues were harvested at ZT2. (This figure is available in colour at JXB online.)
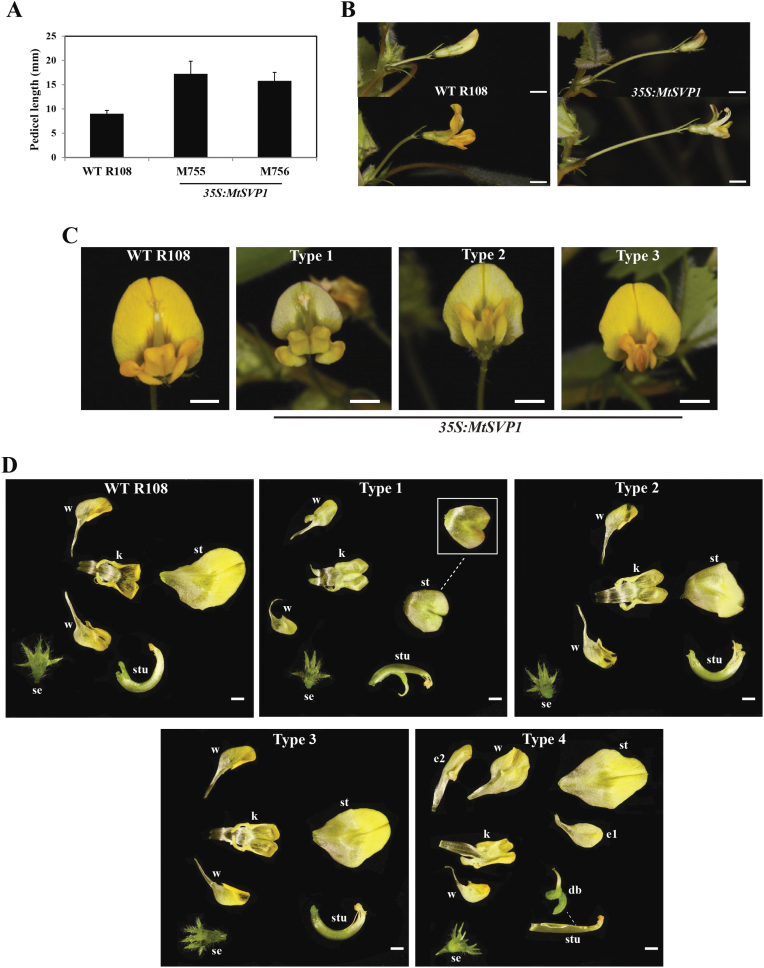
Some changes to floral development were observed in the transgenic lines. Flowers of T1 plants from both 35S:MtSVP1 lines had significantly longer pedicels than WT plants (Fig. 6A, B). In addition, some of the flowers from the MtSVP1 plants were smaller than WT flowers with altered petal shape and colour (Fig. 6C, D) and, in a single case, a flower with extra petals and sepals was observed amongst the 35S:MtSVP1 transformants (Fig. 6D).
Fig. 6.
Overexpression of MtSVP1 in Medicago results in longer pedicels and smaller and abnormal flowers. (A) Average pedicel length of WT R108 and 35S:MtSVP1 T1 transformants from two lines, M755 and M756. The data are shown as the mean ±SE of 51 flowers from WT R108 (n=5 plants), 54 flowers from M755 (n=5 plants), and 52 flowers from M756 (n=7 plants). The plants were grown in LD after 14 d of cold treatment. The pedicel length was measured from the base of a pedicel to the base of the attached open flower. (B) Photographs showing flowers of WT R108 and transgenic 35S:MtSVP1 plants at the same magnification. (C, D) Flower phenotypes of WT R108 and transgenic Medicago 35S:MtSVP1 T1 plants. Five types of flowers were observed in 35S:MtSVP1 transformants compared with WT R108. Flowers were either similar to WT R108 or had abnormalities (Type 1–Type 4). Type 1 flowers were the smallest flowers observed and were paler in colour compared with WT R108. There was also a purple patch at the back side of standard petal, which appeared to be in a more open form. Type 2 flowers were paler in colour, slightly bigger than Type 1 flowers but still smaller compared with WT R108. Type 3 flowers were the same colour as WT R108, slightly bigger than Type 1 flowers, but still smaller than WT R108. The photos are shown in the same scale. (D) Photographs of floral organs from WT R108 and the different types of flowers from 35S:MtSVP1 transformants as explained above. The inset in Type 1 is the back side of the standard petal showing the purple patch. Another type of flower with extra petals (e1 and e2) and sepal (se) was also seen amongst the 35S:MtSVP1 transformants (Type 4). St, standard petal; w, wing petal; k, keel petal; stu, staminal tube surrounding the carpel; db, developing seed barrel. The photos are shown in the same scale. Bars=2mm in (B), 1.29mm in (C), and 1mm in (D).
Analysis of plants carrying Tnt1 retroelement insertions in MtSVP2
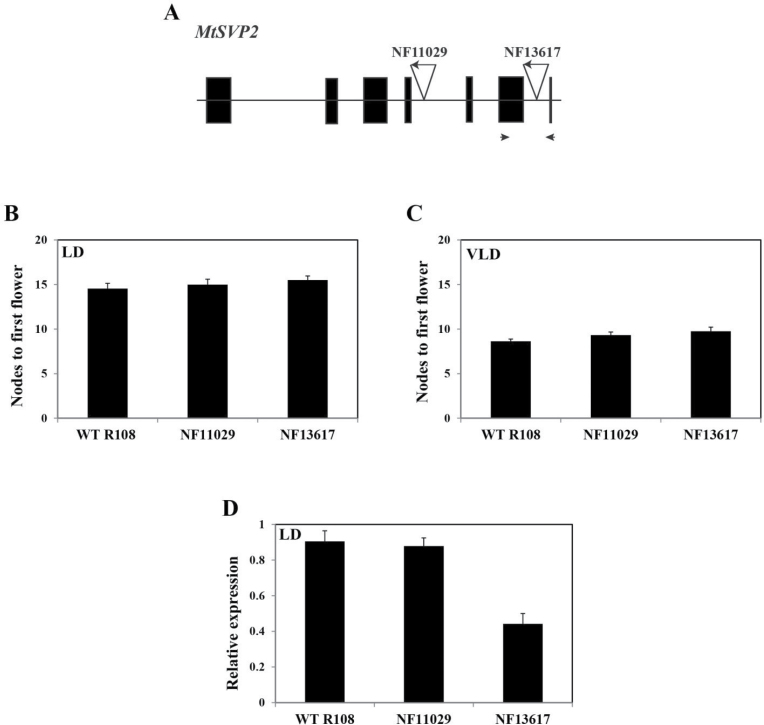
As part of the effort to elucidate the role of the MtSVP genes in Medicago flowering, a reverse genetic approach was used on a population of R108 Tnt1 retrotransposon-tagged mutants (Tadege et al., 2008) with the aim of finding plants carrying insertion mutations in the two MtSVP genes. No lines were found with a Tnt1 insertion at the MtSVP1 locus. Two lines were identified with Tnt1 insertions within the MtSVP2 gene. Line NF11029 had an insertion in intron 4 and line NF13617 had one within intron 6 (Fig. 7A). MtSVP2 expression was unaffected in NF11029, but partially knocked down in NF13617 (Fig. 7D). The flowering time (Fig. 7B, C) and flower development (data not shown) were unaffected in both Tnt1 insertion lines.
Fig. 7.
Flowering time and gene expression of Medicago R108 plants with Tnt1 insertions at MtSVP2. (A) Schematic diagram of the MtSVP2 gene with the positions of Tnt1 insertions in the two lines, NF11029 (Tnt1 inserted within intron 4) and NF13617 (intron 6). Exons are shown as black boxes and introns as thin lines. The arrowheads indicate the positions of primers used for qRT-PCR. (B) Flowering time of WT R108 (n=11) and transgenic plants with homozygous Tnt1 insertion at MtSVP2 in lines NF11029 (n=8) and NF13617 (n=14) under LD conditions. (C) Flowering time of WT R108 (n=11) and transgenic homozygous plants from lines NF11029 (n=3) and NF13617 (n=4) under vernalized LD (VLD) conditions. The flowering time in (B) and (C) was measured in nodes to first flower and is shown as the mean ±SE. (D) Relative expression levels of MtSVP2 in fully expanded trifoliate leaves of WT R108 and homozygous plants from lines NF11029 and NF13617 grown under LD conditions (46-d-old). Gene expression was determined using qRT-PCR and the data are shown as the mean ±SE of three biological replicates, which were normalized to PDF2. The data are presented relative to the highest value. Tissues were harvested at ZT2. (This figure is available in colour at JXB online.)
Discussion
In this study, two Medicago SVP-like genes, MtSVP1 and MtSVP2, were identified whose predicted proteins clustered within the eudicot SVP clade separately from AGL24 and other MADS-domain transcription factors. They are more closely related to pea PsSVP than to other legume SVP-like sequences consistent with the high degree of synteny between pea and Medicago (Choi et al., 2004). The presence of two SVP-like genes in Medicago, as opposed to one gene in Arabidopsis, is likely to have resulted from the proposed whole genome duplication event in the papilionoid subfamily of the Fabaceae that occurred ~58 million years ago, after the divergence of the fabids and malvids (Bell et al., 2010; Young et al., 2011).
In Medicago, MtSVP1 and MtSVP2 are both highly expressed in vegetative tissues and are detected, but at much lower levels in young unopened flower buds and open flowers. This pattern has similarity to AtSVP and is consistent with a possible role of the MtSVP genes as flowering-time repressors during the vegetative phase. In addition, both MtSVP genes delayed flowering when ectopically expressed in Arabidopsis and MtSVP2 rescued the early flowering phenotype of svp-41 mutants, indicating that the two Medicago SVP genes have a similar function to SVP as repressors of flowering in the Arabidopsis heterologous system.
The 35S:MtSVP transgenic Arabidopsis plants also had aberrant flowers with all the four whorls affected in number and/or morphology and pedicels that were longer than in the wild type. Some flowers displayed vegetative features including green petals and leafy sepals that did not abscise. These floral abnormalities resembled, but were not identical to the phenotypes seen when AtSVP or other SVP-like genes from other plant species were overexpressed in Arabidopsis. The increase in petal and anther number was similar to 35S:BcSVP transgenic plants (Lee et al., 2007a ) while the leafy sepals and/or green petals were also seen in the overexpression phenotype of AtSVP (Masiero et al., 2004), LpMADS10 (Petersen et al., 2006), HvBM1, HvBM10 (Trevaskis et al., 2007b ), OsMADS22, OsMADS47 (Fornara et al., 2008), and AcSVP3 (Wu et al., 2012), among others. Thus, despite having similar effects on flowering time in transgenic Arabidopsis, these differences in floral phenotypes indicate that the Arabidopsis and Medicago SVP proteins do not have identical biochemical functions in Arabidopsis. The enhanced vegetative features of flowers in MtSVP-overexpressing Arabidopsis lines may be the result of disrupting floral meristem identity and ensuing development due to ectopic expression of MtSVP in the inflorescence meristem and/or prolonged expression of the MtSVP genes in flower buds.
The similar sequences, Medicago expression patterns, and effects on flowering time and floral development in Arabidopsis suggest that the two MtSVP genes may have overlapping functions in Medicago. Interestingly, in contrast to transgenic Arabidopsis, overexpression of MtSVP1 did not alter flowering time in Medicago. Thus, it is possible that the absence of key SVP protein interaction partners in Medicago such as FLC means that the Medicago SVP genes do not have a major role in regulating flowering time in WT Medicago. In addition, expression of the two MtSVP genes is not regulated by extended exposure to cold temperatures as is FLC, indicating that the MtSVP genes themselves are not directly targeted by the vernalization pathway in Medicago. On the other hand, some floral phenotypes were observed in the 35S:MtSVP1 transgenic Medicago plants. Floral defects included flowers with longer pedicels, as seen in transgenic Arabidopsis and, in some cases, novel phenotypes such as smaller flowers than WT with aberrant petal sizes, shapes, and colours. These results hint at a role for MtSVP1 in Medicago flower development; a possibility given that the MtSVP genes are expressed in flowers at different stages of development. Alternatively, ectopic expression of MtSVP1 may have interfered with the proper make-up of floral meristem and floral organ homeotic patterning complexes involving MADS-domain transcription factors (De Folter et al., 2005; Lee et al., 2007a ; Fornara et al., 2008).
As the two MtSVP genes are likely to be partially redundant, functional studies of the MtSVP genes could be developed further by analysing transgenic Medicago plants with knockdown of expression of both MtSVP genes. Another exciting approach would be to analyse MtSVP regulatory targets at a genome-wide level using techniques such as ChiP-sequencing (ChIP-seq), which have recently revealed that AtSVP directly regulates the expression of a large number of Arabidopsis genes implicated in many different plant processes (Tao et al., 2012; Gregis et al., 2013).
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Multiple sequence alignment of SVP-like proteins from different plant species.
Supplementary Fig. S2. Effect of vernalization on the transcript levels of MtSVP1 and MtSVP2.
Supplementary Table S1. List of oligonucleotide primers (5′–3′) used in this study.
Supplementary Table S2. Flowering time of T2 transgenic plants from the overexpression of MtSVP1 and MtSVP2 in WT Col under LD conditions and classification of floral phenotypes.
Acknowledgements
We thank Erika Varkonyi-Gasic for her suggestions on the manuscript and Rongmei Wu for providing the svp-41 mutant seeds. The research was funded by the New Zealand Foundation for Research Science and Technology (www.msi.govt.nz/, last accessed 1 November 2013) contract numbers C10X0816 MeriNET and C10X0704, the New Zealand Marsden Fund (www.royalsociety.org.nz/programmes/funds/marsden/, last accessed 1 November 2013) contract 10-UOA-200, and the University of Otago Doctoral Scholarship to MJ.
References
- Andres F, Coupland G. 2012. The genetic basis of flowering responses to seasonal cues. Nature Reviews Genetics 13, 627–639 [DOI] [PubMed] [Google Scholar]
- Bell CD, Soltis DE, Soltis PS. 2010. The age and diversification of the angiosperms re-revisited. American Journal of Botany 97, 1296–1303 [DOI] [PubMed] [Google Scholar]
- Bielenberg D, Wang Y, Li Z, Zhebentyayeva T, Fan S, Reighard G, Scorza R, Abbott A. 2008. Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genetics and Genomes 4, 495–507 [Google Scholar]
- Branca A, Paape TD, Zhou P, et al. 2011. Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula . Proceedings of the National Academy of Sciences, USA 108, E864–E870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill EM, Watson JM. 2004. Ectopic expression of a Eucalyptus grandis SVP orthologue alters the flowering time of Arabidopsis thaliana . Functional Plant Biology 31, 217–224 [DOI] [PubMed] [Google Scholar]
- Choi H-K, Mun J-H, Kim D-J, et al. 2004. Estimating genome conservation between crop and model legume species. Proceedings of the National Academy of Sciences, USA 101, 15289–15294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson V, Durand P, d’Erfurth I, Kondorosi A, Ratet P. 2006. Medicago truncatula transformation using leaf explants. Methods in Molecular Biology 343, 115–128 [DOI] [PubMed] [Google Scholar]
- De Folter S, Immink RGH, Kieffer M, et al. 2005. Comprehensive interaction map of the Arabidopsis MADS box transcription factors. The Plant Cell Online 17, 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Li L, Hu P, Xu S-P, Xu Z-H, Xue H-W. 2006. A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. The Plant Journal 47, 519–531 [DOI] [PubMed] [Google Scholar]
- Fornara F, Gregis V, Pelucchi N, Colombo L, Kater M. 2008. The rice StMADS11-like genes OsMADS22 and OsMADS47 cause floral reversions in Arabidopsis without complementing the svp and agl24 mutants. Journal of Experimental Botany 59, 2181–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Oda A, Yoshida R, et al. 2008. Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis . The Plant Cell 20, 2960–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Andres F, Sessa A, et al. 2013. Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis . Genome Biology 14, R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM. 2006. AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis . The Plant Cell Online 18, 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Dorca-Fornell C, Kater MM. 2009. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. The Plant Journal 60, 626–637 [DOI] [PubMed] [Google Scholar]
- Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, Huijser P. 2000. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis . The Plant Journal 21, 351–360 [DOI] [PubMed] [Google Scholar]
- He J, Benedito V, Wang M, Murray J, Zhao P, Tang Y, Udvardi M. 2009. The Medicago truncatula gene expression atlas web server. BMC Bioinformatics 10, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Foucher F, Ferrandiz C, et al. 2005. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiology 137, 1420–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Torti S, Coupland G. 2009. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis . The Plant Journal 60, 614–625 [DOI] [PubMed] [Google Scholar]
- Jimenez S, Lawton-Rauh A, Reighard G, Abbott A, Bielenberg D. 2009. Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach. BMC Plant Biology 9, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julier B, Huguet T, Chardon F, Ayadi R, Pierre J-B, Prosperi J-M, Barre P, Huyghe C. 2007. Identification of quantitative trait loci influencing aerial morphogenesis in the model legume Medicago truncatula . Theoretical and Applied Genetics 114, 1391–1406 [DOI] [PubMed] [Google Scholar]
- Jung CH, Wong CE, Singh MB, Bhalla PL. 2012. Comparative genomic analysis of soybean flowering genes. PLoS ONE 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY(TM) vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC. 2009. Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biology 7, e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-H, Doyle MR, Sung S, Amasino RM. 2009. Vernalization: winter and the timing of flowering in plants. Annual Review of Cell and Developmental Biology 25, 277–299 [DOI] [PubMed] [Google Scholar]
- Laurie RE, Diwadkar P, Jaudal M, et al. 2011. The Medicago truncatula Flowering Locus T homologue, MtFTa1, is a key regulator of flowering time. Plant Physiology 156, 2207–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Park SH, Ahn JH. 2012. Functional conservation and diversification between rice OsMADS22/OsMADS55 and Arabidopsis SVP proteins. Plant Science 185–186, 97–104 [DOI] [PubMed] [Google Scholar]
- Lee JH, Park SH, Lee JS, Ahn JH. 2007a. A conserved role of SHORT VEGETATIVE PHASE (SVP) in controlling flowering time of Brassica plants. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression 1769, 455–461 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH. 2007b. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis . Genes and Development 21, 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Choi SC, An G. 2008. Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. The Plant Journal 54, 93–105 [DOI] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H. 2008. A repressor complex governs the integration of flowering signals in Arabidopsis . Developmental Cell 15, 110–120 [DOI] [PubMed] [Google Scholar]
- Li Z-M, Zhang J-Z, Mei L, Deng X-X, Hu C-G, Yao J-L. 2010. PtSVP, an SVP homolog from trifoliate orange (Poncirus trifoliata L. Raf.), shows seasonal periodicity of meristem determination and affects flower development in transgenic Arabidopsis and tobacco plants. Plant Molecular Biology 74, 129–142 [DOI] [PubMed] [Google Scholar]
- Liu C, Xi W, Shen L, Tan C, Yu H. 2009. Regulation of floral patterning by flowering time genes. Developmental Cell 16, 711–722 [DOI] [PubMed] [Google Scholar]
- Liu C, Zhou J, Bracha-Drori K, Yalovsky S, Ito T, Yu H. 2007. Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134, 1901–1910 [DOI] [PubMed] [Google Scholar]
- Martinez-Trujillo M, Limones-Briones V, Cabrera-Ponce JL, Herrera-Estrella L. 2004. Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Molecular Biology Reporter 22, 63–70 [Google Scholar]
- Masiero S, Li M-A, Will I, Hartmann U, Saedler H, Huijser P, Schwarz-Sommer Z, Sommer H. 2004. INCOMPOSITA: a MADS-box gene controlling prophyll development and floral meristem identity in Antirrhinum . Development 131, 5981–5990 [DOI] [PubMed] [Google Scholar]
- Parenicova L, de, Folter S, Kieffer M, et al. 2003. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. The Plant Cell 15, 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K, Kolmos E, Folling M, Salchert K, Storgaard M, Jensen CS, Didion T, Nielsen KK. 2006. Two MADS-box genes from perennial ryegrass are regulated by vernalization and involved in the floral transition. Physiologia Plantarum 126, 268–278 [Google Scholar]
- Posé D, Yant L, Schmid M. 2012. The end of innocence: flowering networks explode in complexity. Current Opinion in Plant Biology 15, 45–50 [DOI] [PubMed] [Google Scholar]
- Putterill J, Zhang L, Yeoh CC, Balcerowicz M, Jaudal M, Gasic EV. 2013. FT genes and regulation of flowering in the legume Medicago truncatula . Functional Plant Biology , http://dx.doi.org/10.1071/FP13087 [DOI] [PubMed] [Google Scholar]
- Putterill J, Laurie R, Macknight R. 2004. It’s time to flower: the genetic control of flowering time. BioEssays 26, 363–373 [DOI] [PubMed] [Google Scholar]
- Rose RJ. 2008. Medicago truncatula as a model for understanding plant interactions with other organisms, plant development and stress biology: past, present and future. Functional Plant Biology 35, 253–264 [DOI] [PubMed] [Google Scholar]
- Ruelens P, de Maagd RA, Proost S, Theißen G, Geuten K, Kaufmann K. 2013. FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nature Communications 4 [DOI] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G. 2006. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis . Genes and Development 20, 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A, Schmid M. 2011. Regulation of flowering time: all roads lead to Rome. Cellular and Molecular Life Sciences 68, 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, et al. 2008. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula . The Plant Journal 54, 335–347 [DOI] [PubMed] [Google Scholar]
- Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H. 2012. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis . The Plant Journal 70, 549–561 [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Dennis ES, Peacock WJ. 2007a. The molecular basis of vernalization-induced flowering in cereals. Trends in Plant Science 12, 352–357 [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES. 2006. HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiology 140, 1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Tadege M, Hemming MN, Peacock WJ, Dennis ES, Sheldon C. 2007b. Short Vegetative Phase-like MADS-box genes inhibit floral meristem identity in barley. Plant Physiology 143, 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M. 2013. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana . Science 339, 704–707 [DOI] [PubMed] [Google Scholar]
- Wu R-M, Walton EF, Richardson AC, Wood M, Hellens RP, Varkonyi-Gasic E. 2012. Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. Journal of Experimental Botany 63, 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh C, Balcerowicz M, Laurie R, Macknight R, Putterill J. 2011. Developing a method for customized induction of flowering. BMC Biotechnology 11, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh CC, Balcerowicz M, Zhang L, Jaudal M, Brocard L, Ratet P, Putterill J. 2013. Fine mapping links the FTa1 flowering time regulator to the dominant spring1 locus in Medicago . PLoS ONE 8, e53467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debelle F, Oldroyd GED, et al. 2011. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.