Abstract
Symbiosis between legume plants and soil rhizobia culminates in the formation of a novel root organ, the ‘nodule’, containing bacteria differentiated as facultative nitrogen-fixing organelles. MtNF-YA1 is a Medicago truncatula CCAAT box-binding transcription factor (TF), formerly called HAP2-1, highly expressed in mature nodules and required for nodule meristem function and persistence. Here a role for MtNF-YA1 during early nodule development is demonstrated. Detailed expression analysis based on RNA sequencing, quantitiative real-time PCR (qRT-PCR), as well as promoter–β-glucuronidase (GUS) fusions reveal that MtNF-YA1 is first induced at the onset of symbiotic development during preparation for, and initiation and progression of, symbiotic infection. Moreover, using a new knock-out mutant, Mtnf-ya1-1, it is shown that MtNF-YA1 controls infection thread (IT) progression from initial root infection through colonization of nodule tissues. Extensive confocal and electronic microscopic observations suggest that the bulbous and erratic IT growth phenotypes observed in Mtnf-ya1-1 could be a consequence of the fact that walls of ITs in this mutant are thinner and less coherent than in the wild type. It is proposed that MtNF-YA1 controls rhizobial infection progression by regulating the formation and the wall of ITs.
Key words: CCAAT box-binding factor, infection, infection thread, Medicago truncatula, NF-YA, nodule development, rhizobium, symbiosis.
Introduction
Plants within the legume family possess the remarkable pro- perty of endosymbiotic interaction with a group of bacteria collectively referred to as ‘rhizobia’. Symbiotic development culminates in the formation of a new root organ called the nodule, which is colonized by rhizobia that fix atmospheric dinitrogen to ammonia, which is then assimilated by the host plant. A molecular dialogue between the two partners initiates nodule development. Beginning prior to infection, a phase often referred to as the pre-infection phase, plant-derived flavonoids stimulate rhizobia to secrete lipochito-oligosaccharide signal molecules called Nod factors (NFs), which are essential for the initiation of symbiotic infection and nodule organogenesis. NFs reorient root hair growth to induce a curl or ‘shepherd’s crook’. Entrapped within the curl, or sometimes between two adjacent root hairs, a microcolony of rhizobia is formed (Gage, 2004) from which rhizobia enter the root hair following local degradation of the root hair cell wall (Ridge and Rolfe, 1985; van Spronsen et al., 1994), a process that involves both rhizobial cellulase and legume pectate lyase genes (Robledo et al., 2008; Xie et al., 2012). Bacterial entry is accompanied by remodelling of the plasma membrane and primary root hair cell wall (Brewin, 2004; Robledo et al., 2008), leading to the formation of a tube-like apoplastic compartment called the infection thread (IT). Bacteria migrate through the IT’s extracellular matrix towards the root cortex and developing nodule organ by a combination of cell divisions and sliding movements (Fournier et al., 2008). The IT extracellular matrix contains plant glycoproteins, as is typical of extracellular matrices, while the IT wall is similar in composition to that of other plant cell walls, containing cellulose, xyloglucans, and methyl-esterified and non-esterified pectins (Rae et al., 1992). Coincident with infection, cell divisions are activated in root cortical cells that subtend the growing IT. Early cell divisions signify the nascent nodule primordium that subsequently develops into a fully active meristematic tissue. In legumes such as Medicago truncatula, cell divisions are initially observed in pericycle cells (Timmers et al., 1999) but then predominantly in inner cortical cells, and the resulting indeterminate meristem drives nodule growth. In their mature state, indeterminate nodules are composed of characteristic zones of development, with each zone composed of specialized tissues and cell types (Vasse et al., 1990). Zone 1 is the apical zone, characterized by meristematic activity and the absence of rhizobia. Zone 2 is the pre-fixation zone containing numerous ITs that continually re-infect meristem-derived cells and from which bacteria are released into small cytoplasmic vesicles known as symbiosomes. Interzone 2–3 is a narrow, amyloplast-rich cell layer preceding full nitrogen fixation and within which key plant and bacterial symbiotic genes are activated (Soupene et al., 1995). Zone 3 is the fixation zone or nodule ‘central tissue’, where host cells and rhizobia complete differentiation processes that were initiated in the proximal part of zone 2, including endoreduplication and the acquisition of morphological features characteristic of the nitrogen-fixing organelle, the ‘bacteroid’.
Forward and reverse genetic approaches have identified genes and contribute to knowledge of the signalling pathway(s) required for successful infection and nodule organogenesis (Murray, 2011; Oldroyd, 2013). The perception of NF by LysM receptor kinases, including Nod factor perception (MtNFP) (Ben Amor et al., 2003; Arrighi et al., 2006), and the subsequent generation and deciphering of nuclear calcium spikes, lead to a developmental cascade that also involves new host gene transcription. Signalling and symbiotic development downstream of NF receptors requires the anion channel Doesn’t make infection1 (DMI1) (Ane et al., 2004), the receptor kinase DMI2 (Catoira et al., 2000; Endre et al., 2002), and the calcium calmodulin kinase DMI3 (Mitra et al., 2004a). A second LysM receptor kinase, LYK3, is essential for proper root hair curling and IT progression (Catoira et al., 2001; Smit et al., 2007). Several genes encoding transcription factors (TFs) are regulators of rhizobial infection and progression, including Nodulation signaling pathway1 (NSP1) and NSP2, two GRAS TFs (Oldroyd and Long, 2003; Smit et al., 2005), ERF required for Nodulation 1 (ERN1) and ERN2, and nodule inception (NIN). Mutants in these genes are typically blocked at the microcolony stage and have absent or significantly reduced numbers of epidermal ITs. The nuclear protein IPD3 (called Cyclops in Lotus japonicus), a phosphorylation substrate of DMI3, is also essential for rhizobial colonization (Yano et al., 2008; Horvath et al., 2011) as is the case for the nuclear coiled-coil protein RPG (Arrighi et al., 2008) and the Ubox/WD40 protein LUMPY INFECTIONS (LIN) (Kiss et al., 2009).
Here the involvement of another type of TF during rhizobial infection, the CCAAT box-binding factor MtNF-YA1, is shown. MtNF-YA1 belongs to a family of TFs called nuclear factor Y (NF-Y), also called CCAAT box-binding factors (CBFs) or haem adhesion proteins (HAPs). NF-Y homologues occur in all eukaryotes. NF-Y binds CCAAT boxes in promoters as a heterotrimeric complex, composed of a specific CCAAT box-binding factor known as NF-YA and two histone-like proteins NF-YB and NF-YC. While in animals each subunit is encoded by a single gene, in plants structural diversification has led to the appearance of gene families of ~10 members, each with specialized functions in diverse developmental or stress-responsive processes (for reviews, see Petroni et al., 2012; Laloum et al., 2013). In previous reports, MtNF-YA1 (formerly called MtHAP2-1; see Laloum et al., 2013) was identified as a rhizobial-induced, symbiosis-specific NF-YA subunit. MtNF-YA1 is expressed in zones 1 and 2 of mature root nodules (El Yahyaoui et al., 2004; Combier et al., 2006; Moreau et al., 2011), regulated by microRNA169 and the small peptide uORF1p, and required for nodule meristem persistence and function (Combier et al., 2006, 2008). Here it is demonstrated that the expression of this TF is induced within hours after rhizobial inoculation, significantly before the initiation of nodule organogenesis. Moreover, it is demonstrated that MtNF-YA1 expression is correlated with the initiation and progression of the symbiotic infection in roots and in nodule tissues, and that a new null mutant allele, Mtnf-ya1-1, is strongly affected in IT progression.
Materials and methods
Plant growth and bacterial strains
After scarification and surface sterilization as described in Barker et al. (2007), M. truncatula cultivar A17 Jemalong and nf-ya1-1 mutant seedlings were germinated and were grown in different conditions depending on the experiment performed. (1) Aeroponic caissons were used when large quantities of infections or nodules or precise synchrony of initial infection steps were required (such as in Figs 2, 4, 5, and Supplementary Figs S1, S2 available at JXB online) as described in Barker et al. (2007). Plants were inoculated 5 d post-germination with Sinorhizobium meliloti GMI6526 as described in Cerri et al. (2012). (2) Sepiolite medium-containing pots were used when fully grown, nitrogen-fixing nodules were required (such as in Figs 3, 6, and Supplementary Figs S3–S6). Seedlings were grown in sepiolite (Brenntag SA, St Sulpice, France) supplemented with nitrogen-free caisson medium (Journet et al., 2001). Composite plants with roots expressing the promoter–β-glucuronidase (GUS) or complementation constructs (see below) were obtained using A17 Jemalong seedlings, as described in Boisson-Dernier et al. (2001) and inoculated with S. meliloti GMI6526 as described in Cerri et al. (2012).
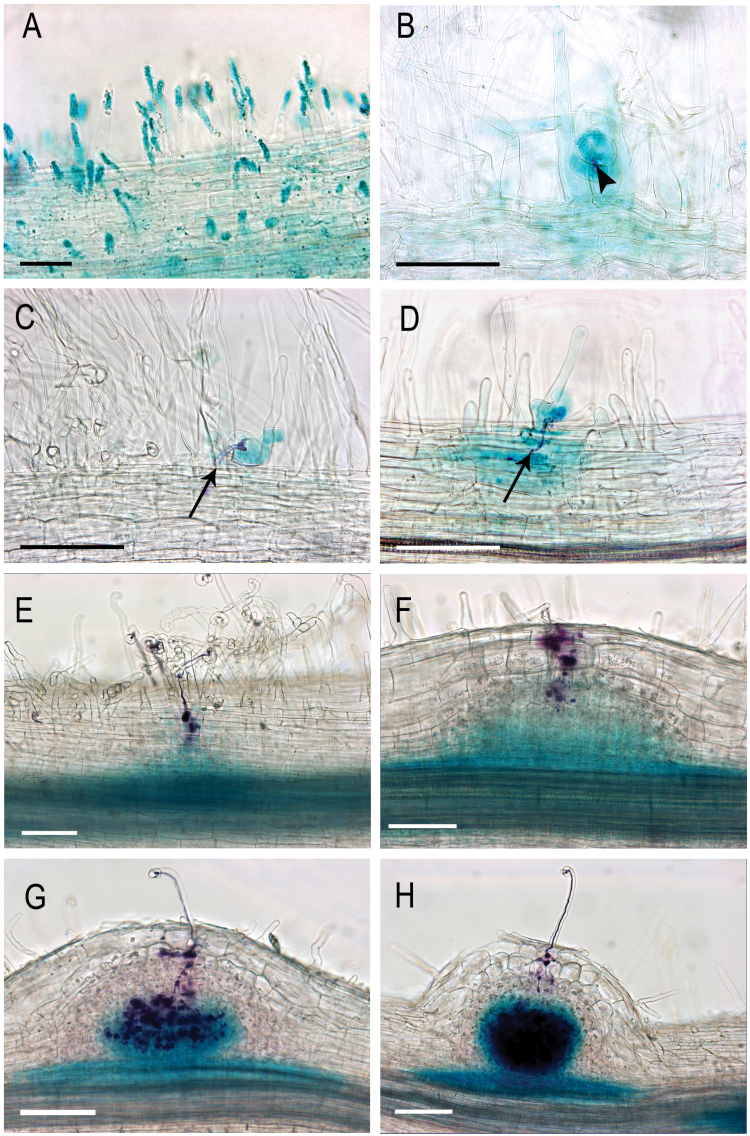
Fig. 2.
Expression analysis of MtNF-YA1 during early stages of the symbiotic interaction between Medicago truncatula and Sinorhizobium meliloti using a promoter–GUS reporter gene. Double staining using Magenta-Gal and X-Gluc allowing the visualization of the infecting S. meliloti in purple and MtNF-YA1 expression in blue. (A) Root hairs in the pre-infection zone, 24hpi. (B and C) Microcolony (black arrowhead, B) in the centre of a curled root hair 48hpi expressing MtNF-YA1. (C) Infected root hair crossed by an infection thread (IT) (black arrow, C) and expressing MtNF-YA1, 48h post-inoculation. (D and E) ITs (black arrows) that are reaching the cortex are shown; 72hpi, MtNF-YA1 expression is mainly found associated with infected root hairs or cortical cells or cells in contact with the infected cells. (E) Note also the expression of MtNF-YA1 ahead of the IT in dividing cells of the inner cortex. (F) At 72hpi, MtNF-YA1 expression is shown in a nodule primordium facing an infection site, and around ITs reaching the inner cortical layers. (G and H) At 96hpi, MtNF-YA1 is expressed strongly in the central part of developing nodules in cells in contact with ITs or released bacteria. Bars represent 100 μm.
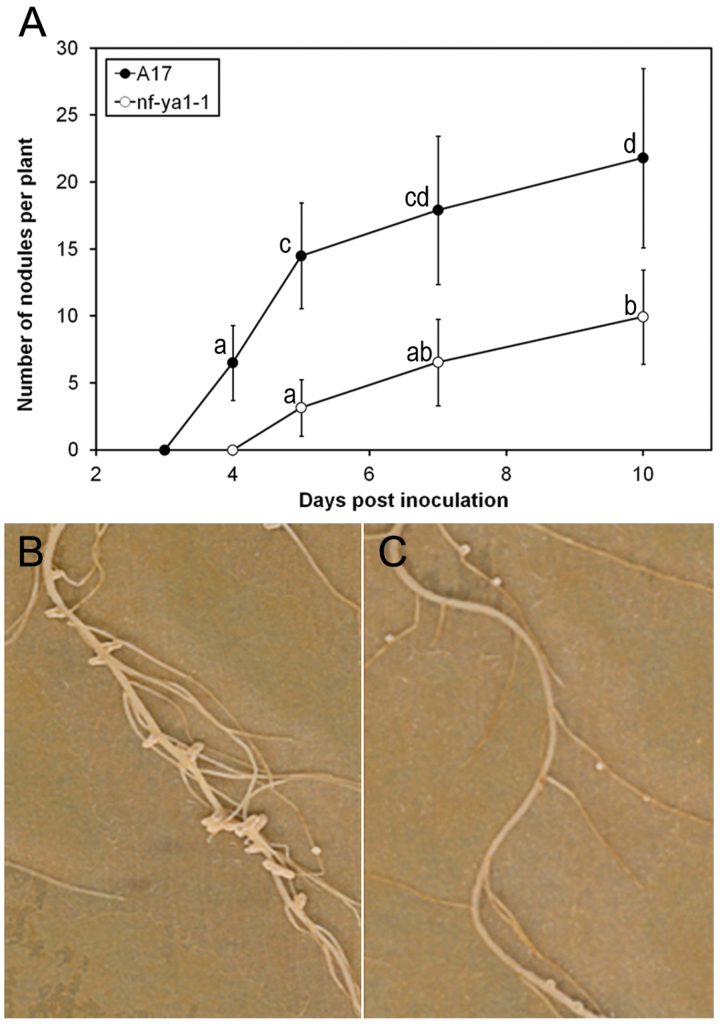
Fig. 4.
Nodule development in A17 WT and in the Mtnf-ya1-1 mutant. (A) WT and Mtnf-ya1-1 plants were grown aeroponically, and inoculated with Sinorhizobium meliloti. The roots were then harvested and the number of nodules per plants was counted 4, 5, 7, and 10 days post-inoculation. Each point is the average of 13 plants. An ANOVA modelling followed by a multiple comparison using a P-value of 0.05 was applied to these data. Letters from a to d illustrate the different groups of statistically different data points. (B and C) Picture of nodulated root systems, 21 dpi in WT (B) and Mtnf-ya1-1 (C) roots. Note the reduced number and size of mutant nodules as well as their different position, more on side roots and less on main roots, in the Mtnf-ya1-1 mutant compared with the WT.
Fig. 5.
Infection phenotype of the Mtnf-ya1-1 mutant during early stages of the symbiotic interaction between Medicago truncatula and Sinorhizobium meliloti. The presence of bacteria in blue is visualized by a β-Gal assay. Light microscopy pictures of WT (A, C, E) and Mtnf-ya1-1 mutant (B, D, F) roots inoculated by S. meliloti. (A and B) Epidermal infection threads (ITs) observed 48hpi; note the swollen and branched aspect of mutant ITs. (C and D) ITs observed 72hpi; note the difference in progression of ITs between the WT and mutant. (E and F) ITs observed 96hpi. Note again the swollen and bulbous aspect of mutant ITs also in the cortex and the delay in IT progression and nodule organogenesis. (G–J) Brightfield (G, I) and confocal images (H, J) of WT (G, H) and Mtnf-ya1-1 mutant (I, J), root hairs, 48hpi by S. meliloti and illustrating the abnormal swollen and bulbous phenotype of ITs in mutant root hairs. Fluorescent bacteria can be observed in green, while the red colour is the autofluorescence of the wall, in confocal images H and J. White bars represent 100 μm for A–F and 10 μm for G–J. (K) Quantification of infection points in WT (A17) and Mtnf-ya1-1 mutant roots. Plants were grown aeroponically, inoculated with S. meliloti, and harvested 5 dpi; bacterial infections were revealed by β-Gal assays, counted under the light microscope, and this number was reported relative to the length of the root. While most infections had progressed to the cortex in WT roots at this stage, most infections were present in the epidermis in Mtnf-ya1-1 mutant roots. Note the 6.3-fold higher number of infections in the roots of mutants compared with the WT. Data presented are the average obtained for 20 root systems. A Wilcoxon rank sum test showed that the two data sets are statistically different with a highly significant P-value of 1.45e-11.
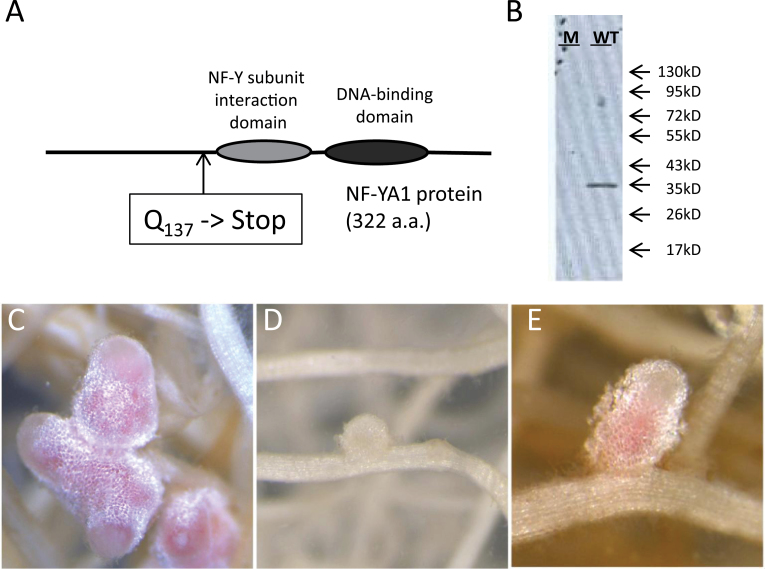
Fig. 3.
Mtnf-ya1-1 mutant description and complementation. (A) Schematic view of the non-synonymous mutation in Mtnf-ya1-1 leading to a premature stop codon at amino acid position 137. (B) Western blot analysis of wild-type (WT) and Mtnf-ya1-1 mutant (M) nodule proteins using an anti-NF-YA1 antibody and showing the absence of MtNF-YA1 protein (36kDa) as well as the absence of significant amounts of a shorter, truncated version of the protein in Mtnf-ya1-1. (C–E) Complementation experiment. A17 WT plants (C) and mutant Mtnf-ya1-1 plants (D, E) were transformed with empty vector control (C, D) or with MtNF-YA1 expressed under the control of its own promoter (E) using Agrobacterium rhizogenes transformation. Composite plants were subsequently inoculated with Sinorhizobium meliloti and roots were harvested for examination at 21 dpi.
Fig. 6.
Analysis of infection thread (IT) ultrastructure using transmission electron microscopy. (A and B) Cross-section through an IT of the infection zone of a nodule 21 dpi in WT A17 (A) and in the Mtnf-ya1-1 mutant (B). Note the clear difference in IT wall thickness which is less in the mutant compared with the WT (white arrowheads). (C and D) Section through an IT of the fixation zone of a nodule 21 dpi in WT A17 (C) and in the Mtnf-ya1-1 mutant (D). Note that the IT wall is not only thinner in the mutant but also appears less coherent and more fragile, with fibres that come off the wall into the cytoplasm of the infected host cell (black arrow). (E) Quantification of IT wall thickness in the infection zone of WT A17 nodules (dark grey) and Mtnf-ya1-1 mutant nodules (light grey). The data shown are the average of 24 measurements. A Wilcoxon rank sum test showed that the two data sets are statistically different with a highly significant P-value of 8.86e-08.
MtNF-YA1 expression using RNA-seq and qRT-PCR
For the RNA-seq experiment, total RNA was isolated from root tissues using the GeneAll HybridR+ kit (GeneAll, Seoul, Korea) and cDNA libraries were synthesized using the TruSeq™ RNA sample preparation kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. The library clusters were sequenced on the Illumina HiSeq2000 sequencer using the TruSeq™ SBS kit v3-HS to generate 100bp single-end sequences.
Quantitative real-time PCR (qRT-PCR) analyses were done following the MIQE Guidelines (Bustin et al., 2009) (http://www.rdml.org/miqe.php) as previously described (Riely et al., 2013). Primers used to quantify MtNF-YA1 expression were: TTGATAAAGCGTAACAAGCCA (fwd) and TCCTCTTGGTCT ACGCATTG (rev). Two reference genes were used: encoding a 26S proteasome regulatory subunit S5A (Medicago Gene Index: TC108192) and an ubiquitin carrier protein (Medicago Gene Index: TC176441). Similar results were obtained in both cases so data presented here represent relative expression values calculated using TC108192, using primers TGGCAGGAAAGGGTGTTC (fwd) and GCCACCTGAATACCAGCAG (rev).
DNA constructs
Promoter–GUS construct
The promoter of MtNF-YA1 (a 2.2kb fragment upstream of the first alternative ATG) was amplified from M. truncatula A17 Jemalong genomic DNA by PCR using Phusion DNA polymerase (Thermo Fisher Scientific, USA) and recombined into the Gateway® vector pDONRP4-P1R according to the manufacturer’s instructions (Invitrogen). Entry clones for the GUS open reading frame (ORF) and the 3′ untranslated region (UTR) of MtNF-YA1 were obtained in the Gateway vectors pDONR207 and pDONRP2R-P3, respectively. Subsequently entry clones were recombined in the binary vector pK7m34GW (Karimi et al., 2002).
Complementation construct
The ORF of MtNF-YA 1 was amplified from M. truncatula nodule cDNA by PCR using Phusion DNA polymerase and recombined in the pDONR207 vector. Entry clones were recombined with the same promoter and 3′UTR construct used for the promoter–GUS construct into pK7m34GW.
Primer sequences used were: attB4 pMtNF-YA1, GGGGAC AACTTTGTA TAGAAAAGTTGGTGCCAAATT CAGAGATAC TACTTCC; attB1r pMtNF-YA1, GGGGACTGCTTTTTTG TAC AAACTTGATTCAAGTA CTATGTTCTTCTCTATTC; attB1 GUS, GGGGACAAGTTTGT ACAAAAAAGCAGGCTTCATG TTAC GTCCTGTAGAAACCCCAAC; attB2 GUS, GGGGACC ACTTTGTA CAAGAAAGCTGGGTCTTATTGTTT GCCTCCC TGCTGCGGT; attB2r NF-YA1_3′UTR, GGGGACAGCTT TCT TGTACAAAGTGGGGGTTTCGATTC AGAAAGGAAACAAG TG; and attB3 NF-YA1_3′UTR, GGGGACAACTTTGT ATAA TAAAGTTGGTTACAGAATCCCAA GCCAACATGGTGTTG.
GUS and β-galactosidase assays
Histochemical staining for GUS activity and bacterial β-galactosidase (β-Gal) activity were performed as described in Cerri et al. (2012).
mRNA in situ hybridization
Medicago truncatula mature nodules [35 days post-inoculation (dpi)] were fixed, embedded, and sectioned for in situ hybridization as described in Valoczi et al. (2006). Hybridization was performed at 50 °C using sense (control) and antisense MtNF-YA1-specific RNA probes synthesized as follows: a 302bp fragment from the 3′UTR of the gene was amplified from nodule cDNA using the following primers: NF-YA1T7senseFor, TGTAATACGACT CACTATAGGGCGTAATATTTA GTAGTATTGTCATT GTCTT TCC; NF-YA1senseRev, AGAGTCTGA AAATAAGAGGTT CT TATAC; NF-YA1antisenseFor, GTAATATTTA GTAGTATTGTC ATT GTCTTTCC; and NF-YA1T7antisenseRev, TGTAATACGA CTCACTATAGGGCAGAGTCTGAAAAT AAGAGGTTCTTAT AC. Digoxigenin (DIG)-labelled sense and antisense RNA probes were then obtained by in vitro transcription with T7 polymerase using the DIG-RNA labeling kit (Roche Diagnostics). After overnight hybridization, slides were washed in 2× SSC/50% formamide at 50 °C and then treated with RNase A to eliminate non-specific background. Finally the presence of RNA was assessed using alkaline phosphatase–anti-DIG antibodies.
Acetylene reduction assay
Nitrogen fixation was assayed using nodules from plants grown on sepiolite substrate and harvested 35 dpi with S. meliloti strain 2011 (Sm2011). The fresh weight of nodules was measured and their nitrogenase activity was subsequently determined by an acetylene reduction assay (Hardy et al., 1968).
Mtnf-ya1-1 mutant description and complementation
The Mtnf-ya1-1 mutant was identified by a TILLING (Targeting Local Induced Lesions In Genomes) approach within a A17 Jemalong EMS (ethyl methanesulphonate)-mutagenized population. Two sequential backcrosses with A17 were then performed and homozygous lines selected. Root transformation was performed on the Mtnf-ya1-1 mutant using an empty vector control or the complementation constructs, and kanamycin-selected composite plants (2 weeks) were transferred to the greenhouse (16h light/8h dark, 22 °C, relative humidity 60–70%) on sepiolite substrate imbibed with nitrogen-free caisson medium for 7 d. Transgenic roots were inoculated with 20ml of an Sm2011 suspension (OD600 nm=0.05) per plant. Nodule development was observed at 21 dpi. Three biological experiments were performed with a minimum of 30 independent plants.
Anti-MtNF-YA1 antibody production and western blot analysis
Anti-NF-YA1 polyclonal antibodies were raised in rabbit by Eurogentec (http://www.eurogentec.be) against two peptides at the N-terminus of MtNF-YA1; namely, 1MAMQPVYLKEHEGNV15 and 64APSKNLVRGVEQLFD78. To analyse protein expression, wild-type (WT) and MtMtnf-ya1-1 mutant plants were grown aeroponically, inoculated with S. meliloti, and nodules were then harvested and ground in liquid nitrogen. An aliquot of 50–100mg of powder was then resuspended in 100 μl of extraction buffer [50mM TRIS-HCl at pH 7.4, 150mM NaCl, 10% glycerol (v/v), 1mM dithiothreitol (DTT), 1mM phenylmethylsulphonyl fluoride (PMSF), and 1% plant protease inhibitor cocktail (Sigma)] and centrifuged at 10 000 g for 10min at 4 °C. The protein concentration in the supernatant was determined with the Bradford protein assay kit (Bio-Rad), using bovine serum albumin (BSA) as a standard. A 50 μg aliquot of total protein was separated on NuPage 10% BIS-TRIS gels (Invitrogen) according to the manufacturer’s instructions and transferred onto Protran BA85 nitrocellulose membranes (Schleicher & Schuell) by wet electroblotting (Mini-Protean II system; Bio-Rad). For detection of MtNF-YA1, blots were incubated with anti-NF-YA1 polyclonal antibodies (1:10 000 dilution). After incubation with the secondary antibody (anti-rabbit IgG–peroxidase, 1:15 000; Sigma), bands were visualized using the ECL Plus kit (Amersham Pharmacia Biotech) under standard conditions.
Microscopic observations
Root or nodule tissues were observed with a light microscope (Axiophot, Carl Zeiss, Oberkochen, Germany) and photographed using an M-1300-HS CCD camera (Princeton Instruments, Evry, France). Green fluorescent protein (GFP)-expressing rhizobia in ITs were imaged in roots of Mtnf-ya1-1 or sunn-2 supernodulant plants as previously described (Cerri et al., 2012). The images were processed using the Leica confocal and Fiji softwares (Schindelin et al., 2012), and maximal projections of selected planes of a z-stack are shown.
Electron microscopy
Nodules were harvested 4 weeks following inoculation, fixed with a solution of 2.5% glutaraldehyde in 0.1M potassium phosphate (pH 7.2) under vacuum, and post-fixed with a 2% OsO4 aqueous solution. The histological organization of nodules was observed on semi-thin sections (1 μm) of Epon-embedded nodules, stained by 0.2% toluidine blue. Observations were performed using bright field microscopy with AxioPlan Imaging (Zeiss, Jena, Germany). Ultrastructure was studied on ultrathin sections (70nm, Ultracut, Reichert, Germany) of Epon-embedded nodules stained with uranyl acetate and observed with a Hitachi HT7700 electron microscope (Hitachi, Tokyo, Japan). Measurements of IT wall thickness were performed using the ImageJ software (Perkin Elmer). Four to five measurements were made on the entire circumference of each IT measured; oblique sections in which the thickness of walls was clearly an artefact were avoided.
Statistical analysis
Analysis of variance (ANOVA) modellization followed by a multiple comparison test (Yandell, 1997) was applied on quantitative expression and nodule number data using a P-value threshold of 0.05.
A Wilcoxon rank sum test (Hollander and Wolfe, 1973) was used to compare the number of ITs and cell wall width between the A17 WT and the MtNF-YA1-1 mutant.
Results
MtNF-YA1 expression is associated with the symbiotic infection process
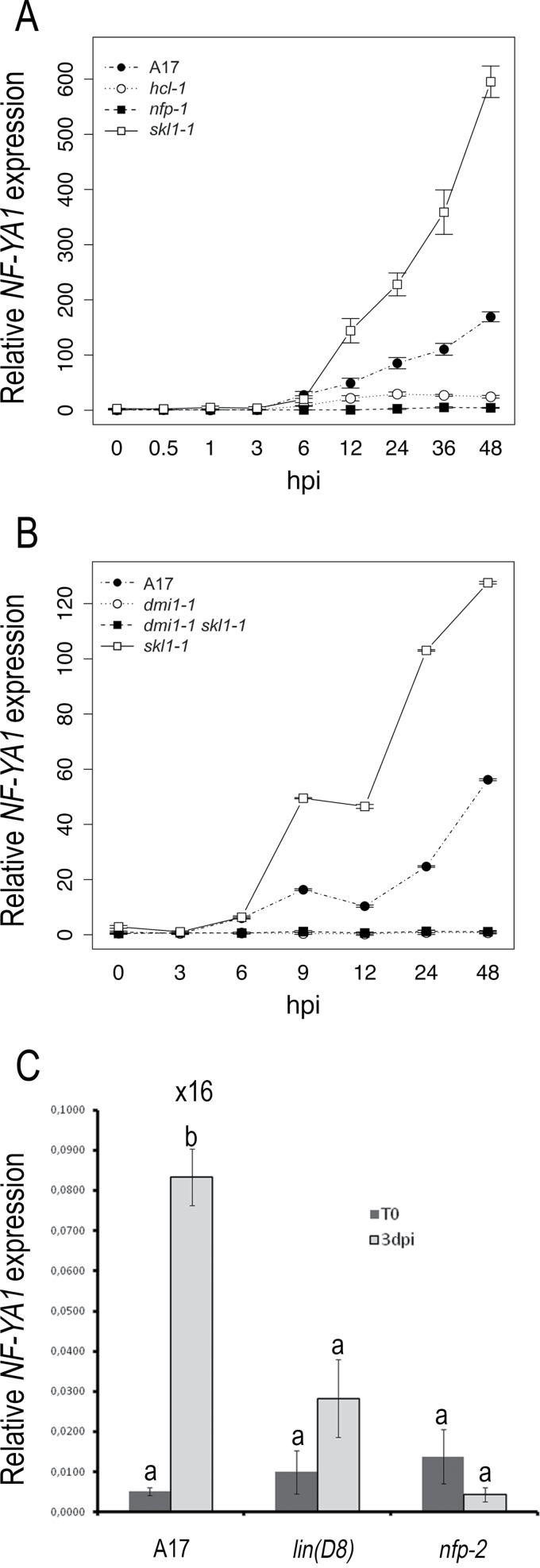
In previous reports, it was shown that MtNF-YA1 (previously called HAP2-1) is highly and specifically expressed in mature nodules, specifically in non-infected meristematic cells and in cells of the infection zone (El Yahyaoui et al., 2004; Combier et al., 2006). To determine if MtNF-YA1 is expressed during earlier phases of the symbiotic interaction, especially during the initial stages of the root infection process, a detailed expression time course was conducted using RNA-seq. As shown in Fig. 1A, MtNF-YA1 was strongly up-regulated in whole roots as early as 6h after inoculation, preceding infection by S. meliloti which is first evident at 48h. MtNF-YA1 expression levels rose continuously from 6h to 48h post-inoculation, throughout the phases of pre-infection, infection, and early nodule morphogenesis. Ethylene-insensitive ein2 (skl1-1) mutants of M. truncatula show a dramatic increase in the number of sustained infections (Penmetsa et al., 2008) and a corresponding substantial up-regulation of MtNF-YA1 was observed in skl plants relative to WT M. truncatula A17 (Penmetsa and Cook, 1997). In contrast, levels of expression remained unchanged in the NF-insensitive nfp-1 (C31) mutant (Ben Amor et al., 2003), and induction was significantly impaired in the lyk3 mutant allele hcl-1 (B56) that is blocked for bacterial entry and impaired for NF-induced gene expression (Limpens et al., 2003; Smit et al., 2007). To confirm this early induction and to assess the relationship between NF and ethylene perception pathways in MtNF-YA1 expression, qRT-PCR was used to quantify expression in four M. truncatula genotypes: A17, skl1-1, the nod-minus mutant dmi1-1 (C71), and a double mutant dmi1-1 skl1-1. As shown in Fig. 1B, qRT-PCR revealed the expected strong and early up-regulation of MtNF-YA1 in WT A17 and its superinduction in skl. MtNF-YA1 induction was not detected in either dmi1-1 or the double mutant dmi1-1 skl. Taken together, these results demonstrate that MtNF-YA1 expression is up-regulated early (6h) upon rhizobial inoculation, that this up-regulation is NF dependent, that superinduction in skl is dependent on the NF pathway, and that there are temporal and quantitative correlations between rhizobial infection and the level of MtNF-YA1 expression.
Fig. 1.
Expression analysis of MtNF-YA1 during early stages of the symbiotic interaction between Medicago truncatula and Sinorhizobium meliloti, using qRT-PCR and RNA-seq. (A) Relative expression of MtNF-YA1 in M. truncatula A17, lyk3 (hcl-1), nfp (nfp-1), and skl roots at various times after inoculation [hours post-inoculation (hpi)] as estimated by RNA-seq. Values represent the average of four biological replicates; bars represent standard errors. The graph was generated using the sciplot package in R (cran.r-project.org/package=sciplot). (B) Relative expression of MtNF-YA1 in M. truncatula A17, dmi1 (dmi1-1), skl, and dmi1 skl roots at various times after inoculation (hpi) as estimated by qRT-PCR. Values represent the average of three technical and three biological replicates; bars represent standard errors. The graph was generated using the sciplot package in R. (C) qRT-PCR analysis of MtNF-YA1 expression in entire roots, before inoculation (T0, dark grey boxes) and 3 days post-inoculation (3 dpi, light grey boxes), in three different genetic backgrounds, WT A17, the lumpy infection mutant (lin, allele D8), and the nod-factor perception mutant (nfp, allele nfp-2). Data shown are the average of three biological repeats. Bars represent standard errors. An ANOVA modelling followed by a multiple comparison test showed that using a P-value of 0.05, only the 3 dpi value (b) for WT A17 was statistically different from all the other values (a).
A related experiment, focusing on later time points, was conducted using qRT-PCR to evaluate expression in non-inoculated roots and at 3 dpi, a stage at which, in the hydroponic culture system used here, numerous ITs are found in the epidermis and outer cortical layers and cortical cell divisions are evident in the inner cortex. As shown in Fig. 1C, strong induction of MtNF-YA1 expression (16-fold) was observed in WT roots at 3 dpi. Parallel analyses were conducted with two M. truncatula mutant lines, namely the NF- insensitive LysM RLK mutant nfp (nfp-2) and the nodulation-defective mutant lin, which encodes a U-box/WD40 protein. lin plants exhibited a strong reduction in MtNF-YA1 induction relative to the WT, which correlates with the 4-fold reduction in the number of infections in lin, all of which arrest in the root epidermis, and nodule primordia that initiate but then fail to develop a persistent meristem (Kuppusamy et al., 2004; Kiss et al., 2009). In contrast, as determined above, nfp mutants failed to induce MtNF-YA1 expression upon S. meliloti inoculation.
To describe MtNF-YA1 expression in greater detail, particularly during early infection stages, a reporter construct was produced bearing 2.2kb of the MtNF-YA1 promoter sequence fused to the GUS gene (pMtNF-YA1-GUS). This promoter was functionally validated by complementation of the Mtnf-ya1-1 mutant, as described below (Fig. 3E). The 3′UTR of MtNF-YA1 has been shown previously (Combier et al., 2006) to contain a recognition site for MIR169, which is important for its proper temporal and spatial regulation, and thus this 3′UTR sequence was also included in the reporter construct. pMtNF-YA1-GUS was transformed into M. truncatula roots using Agrobacterium rhizogenes-mediated root transformation. MtNF-YA1 expression was weak (barely detectable) in non-inoculated roots, with the notable exceptions of the main root tips and early developing lateral roots (Supplementary Fig. S1A–D at JXB online).
As already shown in Fig. 1, pMtNF-YA1-GUS expression was induced well in advance of S. meliloti infection, which in the system used here was not evident until 48h post-inoculation. Root hairs of the pre-infection zone of transgenic roots strongly and specifically expressed pMtNF-YA1-GUS by 24h after inoculation (Fig. 2A). While this up-regulation was observed in many root hairs of the pre-infection zone, pMtNF-YA1-GUS expression was predominantly expressed in curled and infected root hairs during subsequent stages. MtNF-YA1 expression was observed in individual root hairs as soon as they curled, forming the typical ‘shepherd’s crook’ and entrapping a microcolony of S. meliloti at 24–48h post-inoculation (Fig. 2B). This tight association between MtNF-YA1 expression and the infection process was maintained throughout IT growth, first through the epidermis (Fig. 2C), subsequently through the different cortical layers (Fig. 2D, E), and finally during the initial stages of nodule development (Fig. 2F–H). Within young nodules, expression was primarily in the region of IT growth (Fig. 2G) and in cells after release into symbiosomes (Fig. 2H), while in fully differentiated nodules expression was confined to the apical region (Supplementary Fig. S2G, H at JXB online). MtNF-YA1 expression was also observed in dividing cells of nodule primordia at a distance from infecting bacteria (Fig. 2E, F). The up-regulation of MtNF-YA1 expression during rhizobial infection, however, appeared relatively transient and linked to initial phases of infection, as can be observed in Fig. 2E–H, in which the epidermal and cortical cells crossed by ITs no longer express MtNF-YA1. During subsequent steps of nodule development, MtNF-YA1 was expressed strongly and uniformly in the central part of growing nodules (Fig. 2G, H; Supplementary Fig. S4), before gradually becoming more expressed in the apical zone as the different nodule tissues differentiated (Supplementary Fig. 2G, H). Expression at the base of young developing nodules was, in addition, consistently observed across experiments but faded in mature nodules. This further illustrates the expression of MtNF-YA1 in internal tissues activated by rhizobia. In mature nodules, pMtNF-YA1-GUS expression was restricted to the distal end. Expression followed a gradient of decreasing intensity from the meristematic zone down to the proximal part of the infection zone (Supplementary Figs S2, S3). Non-radioactive mRNA in situ hybridization analyses of mature nodules confirmed, with higher cellular resolution, the localization of MtNF-YA1 mRNA within the meristem and infection zones (i.e. zones 1 and 2; Supplementary Fig. S3), as observed previously in Combier et al. (2006).
Identification and characterization of the Mtnf-ya1-1 null mutant
To assess the function of MtNF-YA1, an EMS mutant population of M. truncatula was screened using the TILLING approach (Henikoff et al., 2004). Among the alleles obtained, a non-synonymous mutation converting the glutamine in position 137 into a premature stop codon was identified and the corresponding mutant was named Mtnf-ya1-1. The stop codon in Mtnf-ya1-1 occurs upstream of the two main conserved regions found in NF-YA proteins across kingdoms, and which have been shown to be essential for the interaction of NF-YA with the NF-YB and NF-YC components and for binding to CCAAT boxes within promoters, respectively (Fig. 3A) (Mantovani, 1999). Western blot experiments in nodule tissues using anti-peptide antibodies raised against the N-terminal part of the protein showed that neither the 322 amino acid (36.4kDa) MtNF-YA1 full-length protein nor the 136 amino acid (15.2kDa) putative truncated protein that should in theory be produced in the mutant was detectable in the Mtnf-ya1-1 mutant. The absence of the truncated protein is possibly a consequence of nonsense-mediated mRNA decay (NMD) (Kervestin and Jacobson, 2012) (Fig. 3B). The Mtnf-ya1-1 mutant thus appears to be a null mutant allele of MtNF-YA1 and, as shown below, this mutant is strongly affected in nodule development, comparable with previous results obtained with RNA interference (RNAi) transcript suppression (Combier et al., 2006). In order to confirm that the symbiotic phenotypes observed in the Mtnf-ya1-1 mutant were truly due to the absence of MtNF-YA1, Mtnf-ya1-1 plants were complemented with ectopic MtNF-YA1, introduced by means of A. rhizogenes, and expressed under the same native promoter region used for the promoter–GUS experiment above. Complemented plants developed fully grown pink nodules (Fig. 3E) equivalent to those observed in A17 roots (Fig. 3C) and unlike the small and white nodules characteristic of the Mtnf-ya1-1 mutant (Fig. 3D).
As shown in Fig. 4, Mtnf-ya1-1 mutants have delayed and reduced nodule development. Under the growth conditions used here, nodules emerge between 3 and 4 dpi in WT A17, but were not evident until 5 dpi in Mtnf-ya1-1 plants (Fig. 4A). In addition to a delay in the initiation of nodule development, the number of nodules formed in Mtnf-ya1-1 mutants was significantly reduced compared with WT plants (Fig. 4). This was most obvious at 5 dpi when on average 4.6-fold fewer nodules had formed in the Mtnf-ya1-1 mutant compared with A17; this difference was 2.4-fold, yet significant, at 10 dpi (Fig. 4A). No effect on root length was observed in the Mtnf-ya1-1 mutants (Supplementary Fig. S4 at JXB online), but the distribution of nodules on the root system was different between A17 and the mutant, with most nodules present on the main root in A17 and preferentially on lateral roots in Mtnf-ya1-1 mutants (Fig. 4B, C). Moreover, Mtnf-ya1-1 nodule morphology was strongly affected (Supplementary Fig. S5A) (Vasse et al., 1990) and nitrogen fixation was strongly altered in these mutant nodules; indeed Mtnf-ya1-1 nodules were not fully fix– but fixed nitrogen at rates significantly below those of the WT when compared on a fresh weight basis (Supplementary Fig. S5B). In Mtnf-ya1-1, light and electron microscopy revealed reduced symbiosome formation, though in the few cases when bacterial release occurred differentiated bacteroids could be observed (Supplementary Fig. S6).
MtNF-YA1 controls rhizobial infection
The expression of MtNF-YA1 during early steps of the symbiotic interaction with S. meliloti, together with the observed delay in nodule development in the Mtnf-ya1-1 mutant, led to examination of whether this mutant was also affected in symbiotic infection. Using an S. meliloti strain constitutively expressing the lacZ reporter gene, the infection process was compared in WT A17 and Mtnf-ya1-1 mutant roots. Progression of ITs was clearly delayed and abnormal in the Mtnf-ya1-1 mutant, during all symbiotic steps investigated. Indeed, compared with WT ITs that mainly progress through root hairs by forming a thin and smooth tubular structure (Fig. 5A), ITs in the mutant line appear thicker, bulbous, and branched, with frequent signs of arrested growth at the epidermal layer and often multiple ITs emerging from the bulbs (Fig. 5B). In most cases, these ITs did not progress towards the cortical layers of the roots but stayed arrested in the epidermal layer, either at the microcolony stage or mainly inside root hairs (Fig. 5C, D). In contrast, infection points (infected root hairs and more rarely microcolonies) appeared much more numerous in roots of the Mtnf-ya1-1 mutant than in the WT (Fig. 5A–D). As shown in Fig. 5K for 5 dpi roots, a significant 6.3-fold increase in the number of infection points per cm of root was observed in Mtnf-ya1-1 compared with A17, confirming the observation of more frequent arrested infection events in the mutant. In addition, by 3 dpi, most ITs had reached the cortex in WT A17, and cortical cells situated under the cortical threads had started dividing (Fig. 5C). Comparable analyses in Mtnf-ya1-1 failed to detect signs of cortical division under epidermal ITs. Occasionally, ITs reached the outer cortex in Mtnf-ya1-1, but they possessed the same abnormal swelling phenotype observed in less progressed infections, and nodule initiation and development were significantly retarded compared with A17 (Fig. 5E, F).
Confocal microscopy was used to examine IT morphology further in the Mtnf-ya1-1 mutant by means of GFP-tagged S. meliloti as described in Fournier et al. (2008). As shown in Fig. 5G–J, ITs in the WT were thin and straight, with a single column of infecting bacteria at early stages, whereas Mtnf-ya1-1 mutant ITs appeared as thickened sac-like swellings, densely filled with bacteria but without polarity, connected by thin regions of apparent transient polar growth. The ultrastructure of WT and mutant ITs was then compared using transmission electron microscopy. Transverse sections through an IT within infection zone 2 of a 10-day-old nodule revealed a significantly thicker IT cell wall in WT compared with mutant plants (Fig. 6A, B; Supplementary Fig. S6 at JXB online). IT cell wall thickness was measured in 24 sections of both the WT and mutant (Fig. 6E), revealing a statistically significant 2-fold decrease in wall thickness of mutant ITs. Moreover, the ultrastructure of the mutant IT wall appeared less coherent and more fragile, with fibres that protrude from the wall into the cytoplasm of the infected host cell (Fig. 6C, D).
Discussion
MtNF-YA1 was first described as a symbiosis-up-regulated TF of the CCAAT box-binding family expressed in the meristematic zone of mature nodules and thought to play a role in nodule meristem functioning (Combier et al., 2006). In this study, using detailed expression analysis and functional data coming from a new null mutant allele, Mtnf-ya1-1, evidence is presented for an additional role for MtNF-YA1 during infection of M. truncatula by S. meliloti.
The present RNA-seq, qRT-PCR, and promoter–GUS expression data document that MtNF-YA1 expression is up-regulated by S. meliloti as early as 6h post-inoculation, but most substantially between 12h and 48h and 3 dpi. Thus MtNF-YA1 is among the earliest known rhizobium-induced symbiotic TFs, consistent with a role during early steps of the symbiotic interaction. During these initial phases, MtNF-YA1 is up-regulated specifically in root hairs of the susceptible zone of roots, which are the first root cells known to respond to rhizobial signals and rhizobia. The fact that this up-regulation is not observed in the nfp and dmi1 mutants demonstrates that induction of MtNF-YA1 expression depends on NF perception and signal transduction machinery. Interestingly, MtNF-YA1 is also induced, albeit at low levels, in the lyk3 mutant hcl-1, a genotype that lacks infection but exhibits strong NF-dependent root hair deformation responses (Ben Amor et al., 2003). Taken together, these expression data support the notion that MtNF-YA1 may function prior to infection in WT plants, and indeed the delayed infection and nodule development phenotypes of the Mtnf-ya1-1 mutant are consistent with this idea. Thus it is speculated that MtNF-YA1 is important during pre-infection phases, potentially in preparation for and/or initiation of infection.
Beyond this potential early role during infection initiation, MtNF-YA1 transcript increases in abundance throughout the early interaction, despite the fact that the domain of MtNF-YA1 expression becomes increasingly focused and ultimately concentrated in root hairs containing infection and then in nodules. The kinetics of this expression and the tight correlation between rhizobial infection and MtNF-YA1 expression in infected cells suggest a role for this TF during rhizobial infection, potentially as an extension of the pre-infection role postulated above. Thus throughout early symbiotic development, from the microcolony stage, through root hair penetration and cortical cell invasion, to bacterial release in the nodule infection zone, MtNF-YA1 is continually expressed in conjunction with bacterial infection. The strongly reduced up-regulation of MtNF-YA1 in lin, a mutant that has fewer infections and is blocked in IT progression but not in rhizobial entry nor the formation of nodule primordia, further suggests a role for MtNF-YA1 during rhizobial infection. This correlation between levels of MtNF-YA1 expression and the extent of rhizobial infection is extended by the superinduction of MtNF-YA1 in the hyperinfected mutant skl. The relatively transient expression pattern upon inoculation of MtNF-YA1 provides additional evidence for a role for this TF in initial phases of infection. However, the strongest evidence for an infection-related function of MtNF-YA1 comes from the marked infection phenotype of the Mtnf-ya1-1 null mutant allele. In particular, a 6-fold increase was observed in the average number of ITs in root hairs of Mtnf-ya1-1 compared with the WT, but IT morphology was abnormal and IT growth was typically arrested. Such an increase in unsuccessful epidermal infections in mutants controlling rhizobial infection has been observed before, for example in Vapyrin mutants (Murray et al., 2011). Indeed, most of the ITs observed in the Mtnf-ya1-1 mutant both in the epidermis and in cortical layers, when ITs manage to reach there, were swollen and showed signs of frequent arrest and/or erratic orientation. These observations implicate MtNF-YA1 in control of IT progression, while other aspects of infection such as bacterial release from ITs into symbiosomes appear rather normal (though reduced in frequency). Instead, ultrastructural analyses revealed that the cell wall of ITs is altered in the Mtnf-ya1-1 mutant, with thinner and friable IT walls in both the epidermis and cortex. Modified IT cell wall integrity could explain the bulbous and erratic IT growth phenotypes observed in Mtnf-ya1-1, with the IT being too fragile to contain growing bacteria properly.
Little is known about the composition of IT cell walls. Immunocytochemistry suggests that the IT walls in pea (Pisum sativum) closely resemble primary cell walls of the surrounding cells (Rae et al., 1992), at least in mature ITs, but no study describing the precise carbohydrate or protein composition of this compartment, especially during its formation, has been performed. Nevertheless, plant cell wall remodelling is integral to early events of the legume–rhizobium symbiosis (Brewin, 2004). Indeed, many nodule up-regulated genes are specialized members of gene families encoding cell wall structural proteins, especially proline-rich, arabinogalactan proteins, while others encode enzymes implicated in cell wall synthesis or modification (Brewin, 2004; El Yahyaoui et al., 2004). Despite this wealth of expression data, genetic evidence demonstrating a function for such genes is lacking and there is relatively little known about genes that control IT initiation and progression. Recently Xie et al. (2012) documented a role for L. japonicus. nodule pectate lyase (NPL) in the initiation of rhizobial infection, but it is uncertain whether this enzyme participates directly in IT formation. Given the present hypothesis that MtNF-YA1 controls rhizobial infection via effects on IT structure, it is of particular interest to explore the expression of cell wall remodelling enzymes in the Mtnf-ya1 mutant background.
Nodule organogenesis and rhizobial infection can be separated genetically, as shown by the L. japonicus hyperinfected mutant (hit1) in which large numbers of ITs can be observed in the cortex in the absence of cortical cell division (Murray et al., 2007), and also by the gain-of-function mutations in CCamK (DMI3) (Gleason et al., 2006; Tirichine et al., 2006) and in the cytokinin receptor gene LHK1 (Murray et al., 2007; Tirichine et al., 2007), in which spontaneous nodulation can be triggered in the absence of infecting rhizobia. Conversely, several reports show the interdependence of the two genetic programmes. In pea the characterization of the sym33 mutant indicates that successful infection is required for full elaboration of the indeterminate nodule meristem (Voroshilova et al., 2009). In L. japonicus, reciprocal crosses between the two gain-of-function alleles snf1 and snf2, leading to spontaneous nodulation, and several loss-of-function mutants blocked in the infection process reveal the existence of cross-talk between pathways for organogenesis and infection and suggest that activated CCamK (Ca2+/calmodulin-dependent kinase) and Cyclops mediate cross-pathway signalling between the two pathways (Hayashi et al., 2010; Madsen et al., 2010). More recently, in M. truncatula the analysis of the lin-4 mutant allele, which is blocked in IT initiation at the microcolony stage and forms abnormal nodules with centralized vasculature, suggests that IT initiation is required for normal nodule development (Guan et al., 2013). Here it is shown that in addition to being strongly expressed in the apical tissues of mature nodules and regulating nodule meristem function and thus nodule organogenesis (Combier et al., 2006, 2008), MtNF-YA1 is also expressed during, and controls, rhizobial infection in Medicago, both during initial steps of root infection and later during the infection of nodule tissues.
Pinpointing the primary effect of a mutation in the MtNF-YA1 gene is difficult, especially given the potential of TFs to regulate multiple genes and processes. Thus a perturbed infection process could impede cortical cell divisions and nodule meristem formation, or vice versa, or MtNF-YA1 could directly impact both phenomena as its expression pattern suggests. Indeed, such a dual role has been previously shown for other symbiotic genes. Weak alleles or RNAi lines only partially reducing gene expression have shown, for example, that NFP, LYK3, and DMI2 also play a role during rhizobial infection beyond their role in initiating symbiosis, though both early and late phenotypes of these genes could derive from their impacts on NF perception and signal transduction (Limpens et al., 2005; Arrighi et al., 2007; Smit et al., 2007). This is also the case for other symbiotic TFs such as MtNIN, which was shown to be required for autoactive CCamK-induced nodule organogenesis (Marsh et al., 2007). However, nin mutants are blocked in infection initiation (Schauser et al., 1999) and NIN has also been shown to control the symbiotic expression of NPL that is required for rhizobial infection in L. japonicus. Furthermore, the GRAS-type TFs NSP1 and NSP2 also both control infection and nodule organogenesis (Mitra et al., 2004b; Kalo et al., 2005; Smit et al., 2005). NSP1 and NSP2 have been shown to dimerize and to bind a promoter region called the infection box in the MtEnod11 promoter, but also to activate the expression of ERN1, another TF involved in both rhizobial infection and nodule organogenesis (Andriankaja et al., 2007; Middleton et al., 2007; Cerri et al., 2012). Many of these TFs probably interact and influence each other’s expression in complex regulatory circuits. While it has recently been shown in L. japonicus that NIN directly controls the expression of NF-YA1 (Soyano et al., 2013), the relationship between MtNF-YA1 and NSP1 and NSP2 remains to be established, and unravelling these pathways should lead to a better understanding of the cross-talk between nodule development and infection.
Compared with the drastic infection phenotypes of ern1, nsp1/2, and nin mutants, the infection phenotype of the Mtnf-ya1-1 mutants is less pronounced, as ITs occasionally reach the cortex in this mutant. However, a recent phylogenetic analysis of plant NF-YA proteins revealed that NF-YA1 belongs to a legume-specific subgroup of NF-YA proteins predominantly expressed in roots and nodules and containing a closely related paralogue in Medicago called MtNF-YA2 (77% overall amino acid identity and 100% in the conserved DNA- and subunit-binding domains) (Laloum et al., 2013). The existence of a close paralogue of NF-YA1 in Medicago raises the possibility of partial functional redundancy between MtNF-YA1 and MtNF-YA2 during nodulation. If this is indeed the case, a double Mtnf-ya1/Mtnf-ya2 mutant could present a much stronger infection phenotype than the single Mtnf-ya1-1 mutant.
NF-YA1 belongs to a family of DNA-binding proteins that interact in a heterotrimeric TF complex with two histone-like subunits NF-YB and NF-YC. Together these proteins bind CCAAT boxes, as so-called CCAAT box-binding factors (CBFs) (Mantovani, 1999; Petroni et al., 2012; Laloum et al., 2013). Interestingly, in common bean, an NF-YC gene playing a role in both the infection and the organogenetic pathway, with very similar phenotypes to MtNF-YA1 upon silencing, has recently been described (Zanetti et al., 2010). Identifying NF-YC and NF-YB partners of NF-YA1 involved in infection and nodule organogenesis in Medicago, as well as additional proteins interacting with NF-YA1-containing heterotrimeric CBF complexes, would certainly provide new insights into the signalling and developmental pathways controlled by MtNF-YA1. The identification of the Mtnf-ya1-1 null mutant allele will also be instrumental in investigating the molecular targets of MtNF-YA1-containing CBF complexes during rhizobial symbiosis.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Expression patterns of MtNF-YA1 in roots using a pMtNF-YA1-GUS reporter construct.
Fig. S2. Expression pattern of MtNF-YA1 during nodule development using a pMtNF-YA1-GUS reporter construct.
Fig. S3. Non-radioactive in situ MtNF-YA1 mRNA hybridization in 21 dpi nodules: the presence of DIG-labelled RNA probes was assessed using alkaline phosphatase–anti-DIG antibodies.
Fig. S4. Main root-length measurements.
Fig. S5. Morphology and N2 fixation capacity of WT and mutant nf-ya1-1 nodules 53 days post-inoculation.
Fig. S6. Morphology of bacteroids in infected cells observed by transmission electron microscopy.
Fig. S7. Transmission electron microscopy analysis of infection threads (ITs).
Acknowledgements
This work was funded by the ANR-09-BLAN-0033-01 HAPIHUB project, including a post-doctoral grant to PL. This work is part of the ‘Laboratoire d’Excellence’ (LABEX) entitled TULIP (ANR-10-LABX-41). TEM was performed in the Electron Microscopy Applied to Biology Center at the Université Paul Sabatier, Toulouse. Fluorescence microscopy analyses benefited from the FR40 (now called FR AIB) microscopy/cell imaging platform equipment. The Mtnf-ya1-1 mutant was identified by a TILLING approach within a A17 Jemalong EMS-mutagenized population, in the frame of the FP6-GLIP (Grain Legume Integrated Project) project. EL is a recipient of the Marie Curie International Outgoing Fellowships for Career Development within the 7th European Union Framework Program (FP7-PEOPLE-2009). This work was carried out with the support of the ‘Cooperative Research Program for Agricultural Science & Technology Development (Project no. PJ906910)’, Rural Development Administration, Republic of Korea, to JHM and DRC. We express our gratitude to Mael Baudin for sharing his immense skills in the use of Photoshop.
References
- Andriankaja A, Boisson-Demier A, Frances L, Sauviac L, Jauneau A, Barker D, de Carvalho-Niebel F. 2007. AP2-ERF transcription factors mediate nod factor-dependent MtENOD11 activation in root hairs via a novel cis-regulatory motif. The Plant Cell 19, 2866–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ane JM, Kiss GB, Riely BK, et al. 2004. Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303, 1364–1367 [DOI] [PubMed] [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, et al. 2006. The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiology 142, 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, et al. 2007. The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes (erratum). Plant Physiology 143, 1078–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Godfroy O, de Billy F, Saurat O, Jauneau A, Gough C. 2008. The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proceedings of the National Academy of Sciences, USA 105, 9817–9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DG, Pfaff T, Moreau D, et al. 2007. Growing M. truncatula: choice of substrates and growth conditions. The Medicago truncatula handbook (http://www.noble.org/medicagohandbook). [Google Scholar]
- Ben Amor B, Shaw S, Oldroyd G, Maillet F, Penmetsa R, Cook D, Long S, Denarie J, Gough C. 2003. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. The Plant Journal 34, 495–506 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Becard G, Rosenberg C, Barker DG. 2001. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Molecular Plant-Microbe Interactions 14, 695–700 [DOI] [PubMed] [Google Scholar]
- Brewin NJ. 2004. Plant cell wall remodelling in the rhizobium–legume symbiosis. Critical Reviews in Plant Sciences 23, 293–316 [Google Scholar]
- Bustin SA, Vandesompele J, Pfaffl MW. 2009. Standardization of qPCR and RT-qPCR. Genetic Engineering and Biotechnology News 29, 40–43 [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Denarie J. 2000. Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. The Plant Cell 12, 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Timmers ACJ, Maillet F, Galera C, Penmetsa RV, Cook D, Denarie J, Gough C. 2001. The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development 128, 1507–1518 [DOI] [PubMed] [Google Scholar]
- Cerri MR, Frances L, Laloum T, Auriac MC, Niebel A, Oldroyd GED, Barker DG, Fournier J, de Carvalho-Niebel F. 2012. Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiology 160, 2155–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier JP, de Billy F, Gamas P, Niebel A, Rivas S. 2008. Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development. Genes and Development 22, 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier J-P, Frugier F, de Billy F, et al. 2006. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula . Genes and Development 20, 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yahyaoui F, Kuster H, Ben Amor B, et al. 2004. Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiology 136, 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB. 2002. A receptor kinase gene regulating symbiotic nodule development. Nature 417, 962–966 [DOI] [PubMed] [Google Scholar]
- Fournier J, Timmers ACJ, Sieberer BJ, Jauneau A, Chabaud M, Barker DG. 2008. Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiology 148, 1985–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ. 2004. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiology and Molecular Biology Reviews 68, 280–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason C, Chaudhuri S, Yang T, Munoz A, Poovaiah B, Oldroyd G. 2006. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441, 1149–1152 [DOI] [PubMed] [Google Scholar]
- Guan D, Stacey N, Liu CW, et al. 2013. Rhizobial infection is associated with the development of peripheral vasculature in nodules of Medicago truncatula . Plant Physiology 162, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RWF, Holsten RD, Jackson EK, Burns RC. 1968. Acetylene–ethylene assay for N2 fixation—laboratory and field evaluation. Plant Physiology 43, 1185–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Banba M, Shimoda Y, Kouchi H, Hayashi M, Imaizumi-Anraku H. 2010. A dominant function of CCaMK in intracellular accommodation of bacterial and fungal endosymbionts. The Plant Journal 63, 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Till BJ, Comai L. 2004. TILLING. Traditional mutagenesis meets functional genomics. Plant Physiology 135, 630–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander M, Wolfe DA. 1973. Nonparametric statistical methods. New York: John Wiley & Sons, 68–75 [Google Scholar]
- Horvath B, Yeun LH, Domonkos A, et al. 2011. Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Molecular Plant-Microbe Interactions 24, 1345–1358 [DOI] [PubMed] [Google Scholar]
- Journet EP, El Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V. 2001. Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Molecular Plant-Microbe Interactions 14, 737–748 [DOI] [PubMed] [Google Scholar]
- Kalo P, Gleason C, Edwards A, et al. 2005. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308, 1786–1789 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. 2002. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195 [DOI] [PubMed] [Google Scholar]
- Kervestin S, Jacobson A. 2012. NMD: a multifaceted response to premature translational termination. Nature Reviews. Molecular Cell Biology 13, 700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss E, Olah B, Kalo P, et al. 2009. LIN, a novel type of U-box/WD40 protein, controls early infection by rhizobia in legumes. Plant Physiology 151, 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy KT, Endre G, Prabhu R, Penmetsa RV, Veereshlingam H, Cook DR, Dickstein R, VandenBosch KA. 2004. LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections. Plant Physiology 136, 3682–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T, De, Mita S, Games P, Baudin M, Niebel A. 2013. CCAAT-box binding transcription factors in plants: Y so many? Trends in Plant Science 18, 157–166 [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. 2003. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302, 630–633 [DOI] [PubMed] [Google Scholar]
- Limpens E, Mirabella R, Fedorova E, Franken C, Franssen H, Bisseling T, Geurts R. 2005. Formation of organelle-like N-2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proceedings of the National Academy of Sciences, USA 102, 10375–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. 2010. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus . Nature Communications 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 239, 15–27 [DOI] [PubMed] [Google Scholar]
- Marsh J, Rakocevic A, Mitra R, Brocard L, Sun J, Eschstruth A, Long S, Schultze M, Ratet P, Oldroyd G. 2007. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiology 144, 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton P, Jakab J, Penmetsa R, et al. 2007. An ERF transcription factor in Medicago truncatula that is essential for nod factor signal transduction. The Plant Cell 19, 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R, Gleason C, Edwards A, Hadfield J, Downie J, Oldroyd G, Long S. 2004a. A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proceedings of the National Academy of Sciences, USA 101, 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Shaw SL, Long SR. 2004b. Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume–rhizobia symbiosis. Proceedings of the National Academy of Sciences, USA 101, 10217–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Verdenaud M, Ott T, Letort S, de Billy F, Niebel A, Gouzy J, de Carvalho-Niebel F, Gamas P. 2011. Transcription reprogramming during root nodule development in Medicago truncatula . PLoS One 6, e16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J, Muni R, Torres-Jerez I, et al. 2011. Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula . The Plant Journal 65, 244–252 [DOI] [PubMed] [Google Scholar]
- Murray JD. 2011. Invasion by invitation: rhizobial infection in legumes. Molecular Plant-Microbe Interactions 24, 631–639 [DOI] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K. 2007. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315, 101–104 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE. 2013. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nature Reviews. Microbiology 11, 252–263 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Long SR. 2003. Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod factor signaling. Plant Physiology 131, 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR. 1997. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275, 527–530 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Uribe P, Anderson J, et al. 2008. The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. The Plant Journal 55, 580–595 [DOI] [PubMed] [Google Scholar]
- Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt BF, Mantovani R. 2012. The promiscuous life of plant nuclear factor Y transcription factors. The Plant Cell 24, 4777–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae AL, Bonfantefasolo P, Brewin NJ. 1992. Structure and growth of infection threads in the legume symbiosis with Rhizobium leguminosarum . The Plant Journal 2, 385–395 [Google Scholar]
- Ridge RW, Rolfe BG. 1985. Rhizobium sp degradation of legume root hair cell-wall at the site of infection thread origin. Applied and Environmental Microbiology 50, 717–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely BK, Larrainzar E, Haney CH, et al. 2013. Development of tools for the biochemical characterization of the symbiotic receptor-like kinase DMI2. Molecular Plant-Microbe Interactions 26, 216–226 [DOI] [PubMed] [Google Scholar]
- Robledo M, Jimenez-Zurdo JI, Velazquez E, et al. 2008. Rhizobium cellulase CelC2 is essential for primary symbiotic infection of legume host roots. Proceedings of the National Academy of Sciences, USA 105, 7064–7069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. 1999. A plant regulator controlling development of symbiotic root nodules. Nature 402, 191–195 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T. 2007. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiology 145, 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debelle F, Gough C, Bisseling T, Geurts R. 2005. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308, 1789–1791 [DOI] [PubMed] [Google Scholar]
- Soupene E, Foussard M, Boistard P, Truchet G, Batut J. 1995. Oxygen as a key developmental regulator of Rhizobium meliloti N-2-fixation gene-expression within the alfalfa root-nodule. Proceedings of the National Academy of Sciences, USA 92, 3759–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Kouchi H, Hirota A, Hayashi M. 2013. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genetics 9, e1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers ACJ, Auriac MC, Truchet G. 1999. Refined analysis of early symbiotic steps of the Rhizobium–Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126, 3617–3628 [DOI] [PubMed] [Google Scholar]
- Tirichine L, Imaizumi-Anraku H, Yoshida S, et al. 2006. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441, 1153–1156 [DOI] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J. 2007. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315, 104–107 [DOI] [PubMed] [Google Scholar]
- Valoczi A, Varallyay E, Kauppinen S, Burgyan J, Havelda Z. 2006. Spatio-temporal accumulation of microRNAs is highly coordinated in developing plant tissues. The Plant Journal 47, 140–151 [DOI] [PubMed] [Google Scholar]
- van Spronsen P, Bakhuizen R, van Brussel AA, Kijne JW. 1994. Cell wall degradation during infection thread formation by the root nodule bacterium Rhizobium leguminosarum is a two-step process. European Journal of Cell Biology 64, 88–94 [PubMed] [Google Scholar]
- Vasse J, Debilly F, Camut S, Truchet G. 1990. Correlation between ultrastructural differentiation of bacteroids and nitrogen-fixation in alfalfa nodules. Journal of Bacteriology 172, 4295–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voroshilova VA, Demchenko KN, Brewin NJ, Borisov AY, Tikhonovich IA. 2009. Initiation of a legume nodule with an indeterminate meristem involves proliferating host cells that harbour infection threads. New Phytologist 181, 913–923 [DOI] [PubMed] [Google Scholar]
- Xie F, Murray JD, Kim J, Heckmann AB, Edwards A, Oldroyd GED, Downie A. 2012. Legume pectate lyase required for root infection by rhizobia. Proceedings of the National Academy of Sciences, USA 109, 633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Mueller J, et al. 2008. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proceedings of the National Academy of Sciences, USA 105, 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell BS. 1997. Practical data analysis for designed experiments. Chapman & Hall [Google Scholar]
- Zanetti ME, Blanco FA, Beker MP, Battaglia M, Aguilar OM. 2010. A C subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean–Rhizobium etli symbiosis. The Plant Cell 22, 4142–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.