Abstract
DICER-like 1 (DCL1) is a major player in microRNA (miRNA) biogenesis and accordingly, its few known loss-of-function mutants are either lethal or display arrested development. Consequently, generation of dcl1 mutants by reverse genetics and functional analysis of DCL1 in late-developing organs are challenging. Here, these challenges were resolved through the unique use of trans-activated RNA interference. Global, as well as organ-specific tomato DCL1 (SlDCL1) silencing was induced by crossing the generated responder line (OP:SlDCL1IR) with the appropriate driver line. Constitutive trans-activation knocked down SlDCL1 levels by ~95%, resulting in severe abnormalities including post-germination growth arrest accompanied by decreased miRNA and 21-nucleotide small RNA levels, but prominently elevated levels of 22-nucleotide small RNAs. The increase in the 22-nucleotide small RNAs was correlated with specific up-regulation of SlDCL2b and SlDCL2d, which are probably involved in their biogenesis. Leaf- and flower-specific OP:SlDCL1IR trans-activation inhibited blade outgrowth, induced premature bud senescence and produced pale petals, respectively, emphasizing the importance of SlDCL1-dependent small RNAs in these processes. Together, these results establish OP:SlDCL1IR as an efficient tool for analysing processes regulated by SlDCL1-mediated gene regulation in tomato.
Key words: Carotenoid, DCL1, leaf, miRNA, mutant, petal, tomato.
Introduction
Small RNAs, ranging from 20 to 24 nucleotides (nt) in size, have many key functions in plants, particularly in genome stability, regulation of gene expression, and defence by directing sequence-specific repression of target RNAs (Chen, 2009; Axtell, 2013a ). Plant microRNAs (miRNAs) constitute a major class of endogenous small RNAs that have emerged as important post-transcriptional regulators of plant development, stress responses, and nutrient homeostasis (Jones-Rhoades et al., 2006; de Lima et al., 2012; Sunkar et al., 2012). In contrast to small interfering RNAs (siRNAs), which are generated via processing of long double-stranded RNA precursors with perfect complementarity, miRNAs are processed from a single-stranded RNA precursor transcript or pri-miRNA that can form imperfect fold-back structures (Reinhart et al., 2002).
The diversification of small RNA pathways in plants can be attributed in part to specific DICER-like (DCL) proteins that specialize in the production of distinct small RNAs. Four types of DCLs are present in all angiosperms (Margis et al., 2006). Among them, the product of DCL1, also termed SHORT INTEGUMENTS1 (SIN1)/SUSPENSOR1 (SUS1)/CARPEL FACTORY (CAF) (Robinson-Beers et al., 1992; Jacobsen et al., 1999; McElver et al., 2001; Schauer et al., 2002), catalyses the formation of predominantly 20- to 22-nt canonical miRNAs (Park et al., 2002; Reinhart et al., 2002; Kurihara and Watanabe, 2004). In addition, certain inverted repeat-derived siRNAs and secondary siRNAs—including trans-acting siRNAs (ta-siRNAs), natural antisense siRNAs, and phased siRNAs—which do not derive from fold-back structures but arise from transcripts targeted by miRNAs, also depend on DCL1 for their accumulation (Allen et al., 2005; Borsani et al., 2005; Henderson et al., 2006; Li et al., 2012b ; Shivaprasad et al., 2012). Moreover, DCL1 was found to contribute to the silencing of a subset of transposons, apparently through an effect on DNA methylation (Laubinger et al., 2010). DCL2 catalyses the formation of 22-nt siRNAs and 22- to 24-nt natural antisense siRNAs (Xie et al., 2004; Borsani et al., 2005; Deleris et al., 2006). DCL3 is responsible for the processing of 24-nt heterochromatin-associated siRNAs (hc-siRNAs) and long miRNAs (Xie et al., 2004; Qi et al., 2005; Wu et al., 2010). DCL4 sequentially processes 21-nt secondary siRNAs including ta-siRNAs and other phased siRNAs, and is required for the production of several Arabidopsis thaliana miRNAs such as miR822 and miR839 (Xie et al., 2004; Gasciolli et al., 2005; Rajagopalan et al., 2006; Howell et al., 2007).
To date, dcl1 loss-of-function mutants have been isolated from Arabidopsis, Oryza sativa, and the moss Physcomitrella patens (Schauer et al., 2002; Liu et al., 2005; Khraiwesh et al., 2010). In Arabidopsis, null dcl1 alleles (sus1) are embryo lethal (Castle et al., 1993). In addition, several hypomorphic dcl1 mutants have been described, such as dcl1-7 (sin 1-1) and dcl1-9 (caf), which show pleiotropic developmental phenotypes due to reduced miRNA levels (Robinson-Beers et al., 1992; Jacobsen et al., 1999; Park et al., 2002). Physcomitrella harbours two DCL1-like proteins, PpDCL1a and PpDCL1b. Deletion of PpDCL1a but not PpDCL1b drastically reduced miRNA levels, indicating its functional equivalence to AtDCL1. ΔPpDCL1a-null mutants showed developmental defects and abnormalities in cell size and shape; their growth was severely retarded and they were developmentally arrested at the filamentous protonema stage (Khraiwesh et al., 2010). In rice, a DCL1 loss-of-function mutant (OsDCL1IR) was generated by RNA interference (RNAi). Silenced plants had reduced levels of miRNAs, developed severe dwarfism, rolled and curly leaves, and tortuous shoots, and were arrested at the young seedling stage (Liu et al., 2005). The lethality and severe growth arrest associated with dcl1 loss-of-function mutants prevents their functional characterization at late developmental stages. It also impedes the generation of dcl1 loss-of-function mutants by reverse genetics in plants whose transformation includes a regeneration phase in tissue culture. For example, attempts to stably silence Nicotiana attenuata DCL1 by RNAi have failed due to lethality of the regenerated transformants (Bozorov et al., 2012).
Solanum lycopersicum (tomato) is one of the most important crops in the fresh vegetable market and the food-processing industry, and is a good model for crop species with fleshy berry fruits (Giovannoni, 2001). Deep sequencing of small RNAs from various tomato tissues has revealed the presence of a complex population of small RNAs, including miRNAs (Moxon et al., 2008; Li et al., 2012b ; Shivaprasad et al., 2012; Axtell, 2013b ). Recently, a tomato dcl4 mutant has been described, demonstrating its function in the biogenesis of ta-siRNAs and their involvement in the regulation of lamina expansion during compound leaf development (Yifhar et al., 2012). The functions of other tomato DCLs, including DCL1, remain unstudied.
In the current study, a trans-activated RNAi approach was employed to silence tomato DCL1 (SlDCL1) globally or in specific organs, and the resulting mutants were utilized to investigate SlDCL1 functions in small RNA biogenesis and development.
Materials and methods
Plant material and growth conditions
The tomato (S. lycopersicum) cv. M82 driver lines 35S:LhG4, FIL:LhG4 (Lifschitz et al., 2006), AP1:LhG4, and AP3:LhG4 (Fernandez et al., 2009; Hendelman et al., 2013a ) are described elsewhere. Tomato plants were grown under greenhouse conditions with temperatures ranging between 15 °C and 30 °C in a tuff–peat mix with nutrients, using 4 litre pots. Germination and seedling growth took place in a growth chamber with a 16h light period and 8h dark period (photosynthetic photon flux density: 50–70 μmol m–2 s–1) at a constant temperature of 24 °C. For crosses, closed flowers were emasculated by removal of the petals and stamens and hand pollinated with the pollen of an appropriate homozygous driver line.
Sequencing of full-length SlDCL1 and SlDCL2a, b, and d 5′-untranslated regions (UTRs)
To determine the full-length sequence of SlDCL1, total RNA was extracted from tomato cv. M82 flowers at anthesis as described below and a 3′ and 5′ RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) procedure was performed on this RNA. The partial Solyc10g005130 sequence was used to design gene-specific primers (all primer sequences are given in Supplementary Table S1 available at JXB online) as follows: for the 5′ end, SlDCL1-5′-RACE and SlDCL1-5′-RACE-nested; for the 3′ end, SlDCL1-3′-RACE and SlDCL1-3′-RACE-nested. The resulting amplified products were gel-purified, cloned, and sequenced. After assembling the full-length sequence of SlDCL1, its 5′ (nt 86–2685) and overlapping 3′ (nt 2481–6168) cDNA sequences were PCR-amplified with SlDCL1_F_XhoI and SlDCL1_R_2685, and SlDCL1_F_2481 and SlDCL1_R_6168, respectively, and sequenced. The 5′ UTR of SlDCL2b (Solyc11g008540) was determined by RLM-RACE of total flower RNA with SlDCL2b-5′-RACE and SlDCL2b-5′-RACE-nested primers. On the basis of their sequence, the 5′ UTRs of SlDCL2a (Solyc06g048960) and SlDCL2d (Solyc11g008530) were determined by PCR on flower cDNA with the primer pairs SlDCL2a_F_28 and qRT-SlDCL2a_R, and SlDCL2d_R_280 and qRT-SlDCL2d_F, respectively. Amplified products were then gel-purified and sequenced directly.
Plasmid construction
For the SlDCL1IR RNAi reporter construct, a 674bp SlDCL1 sequence composed of 264bp and 410bp of its promoter and cDNA, respectively, was amplified by two-step PCR. The primers SlDCL1IRDNA_F, which contains SalI and PstI sites at its 5′ end, and SlDCL1IRDNA_R were used to amplify the promoter sequence. Primers SlDCL1IRRNA_F and SlDCL1IRRNA_R, which contains HindIII/EcoRI sites at its 5′ end, were used to amplify an overlapping fragment that contained the cDNA sequence. Then the 674bp product was assembled by using the amplified fragments as a template for PCR with primer pair SlDCL1IRDNA_F and SlDCL1IRRNA_R. The amplified assembled fragment was restricted with either PstI/EcoRI or SalI/HindIII and cloned in sense and antisense orientation, respectively, around the first intron of the Arabidopsis AKT1 gene to generate max2intpFLAP-SlDCL1IR. Following sequence validation, the max2intpFLAP-SlDCL1IR XhoI fragment was mobilized into the XhoI site of the OP-TATA-BJ36 shuttle vector between an OP array (Moore et al., 1998) and Agrobacterium tumefaciens octopine synthase terminator (OCS) to generate OP:SlDCL1IR. Following orientation validation, the NotI fragment of the OP:SlDCL1IR vector was mobilized into the binary vector pART27 to generate pART27-OP:SlDCL1IR.
Transformation of tomato plants
The binary vector pART27-OP:SlDCL1IR was transformed into tomato cv. M82 as described previously (Hendelman et al., 2013a ). Transgenic progeny were selected by germinating sterilized seeds on selective medium [1× Murashige and Skoog (MS) medium, 3% (w/v) sucrose, 100mg l–1 kanamycin], where only transgenic seedlings developed a branched root system. Further validation was performed by PCR of genomic DNA with the primer pair pFlap_intron_F and OCS_rev to detect the OP:SlDCL1IR transgene.
Total RNA extraction and small RNA gel-blot analysis
Total RNA was isolated from tomato tissues as described previously (Hendelman et al., 2013a ). Blot hybridization analysis was performed according to Stav et al. (2010). Radiolabelled probes were made by 5′-end-labelling DNA oligonucleotides complementary to the corresponding small RNA sequences (probe sequences are listed in Supplementary Table S1 available at JXB online) with [γ-32P]ATP by means of T4 polynucleotide kinase (New England Biolabs, Ipswich, MA, USA). Blots were pre-hybridized and hybridized with EZ-hybridization solution (Biological Industries, Bet-Haemek, Israel). Hybridization was performed at 40 °C overnight. Blots were washed three times at 50 °C with washing buffer (2× SSC, 0.1% SDS) and autoradiographed using a phosphoimager (Fuji).
Real-time quantitative PCR (qRT-PCR) analyses
Total RNA was extracted from the indicated tomato tissues as described above. DNase treatment, concentration determination, preparation of cDNA from 1 μg of total RNA, and real-time quantitative PCR were performed as described in Hendelman et al. (2013b ). The relative expression levels of all transcripts were calculated using the two standard curve method normalized to TIP41 or CAC as reference genes.
Small RNA libraries constructions, sequencing, and bioinformatic analyses
Total RNA was extracted from shoots of control (35S:LhG4) and SlDCL1-silenced (35S>>SlDCL1) seedlings 14 d after sowing (two biological replicates per genotype combined from 12 seedlings each) using Bio-TRI RNA reagent (Bio-Lab, Jerusalem, Israel) according to the manufacturer’s protocol. Extracted RNA samples were used to construct two control and two SlDCL1-silenced small RNA libraries (Supplementary Table S2 available at JXB online) using the TruSeq small RNA kit (Illumina, San Diego, CA, USA) as described by the manufacturer, and submitted to sequencing on an Illumina HiSeq 2000 analyser. Sequencing was performed by Illumina. The data for this article have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE51959. The small RNA sequences from all sequenced libraries were parsed, mapped, quantified, and managed using The Cache ASsisted Hash Search with Xor logic (CASHX) Pipeline (release 2.3) (Fahlgren et al., 2009). All reads were mapped to the tomato genome (SGN release version SL2.40; http://solgenomics.net/organism/Solanum_lycopersicum/genome) and perfectly matching reads in the size range of 18–26 nt were retained. The small RNA CASHX database was annotated with the Repeats Master version 5.0 (ftp://ftp.solgenomics.net/tomato_genome/repeats/), Plant snoRNAs Database (version 1.2; http://bioinf.scri.sari.ac.uk/cgi-bin/plant_snorna/home), SILVA rRNA database (http://www.arb-silva.de/), miRbase (release 19, http://www.mirbase.org/), and the tomato genome annotation (SGN release version ITAG2.3; ftp://ftp.solgenomics.net/tomato_genome/annotation/ITAG2.3_release/). After annotation, only ‘clean’ reads not matching tRNAs, rRNAs, small nuclear RNAs (snRNAs), or small nucleolar RNAs (snoRNAs) were used for further analysis. For all small RNA data analyses, clean reads from both biological replicates were used. The differential expression between control and SlDCL1-silenced clean small RNAs (two biological replicates per genotype) was performed by the analysis of digital gene expression data in the R (EdgeR) package (version 3.07) (Robinson et al., 2010). By using this package, first low count reads were removed in each library by retaining reads with at least 1 count per million in at least three samples. Then normalization factors were calculated, effective library sizes were estimated by using normalization factors, fitted according to the Negative Binomial model, estimates of common dispersions were made, and differential expression tests were performed. The resultant P-values were adjusted for false discovery rate (FDR) and only adjusted P-values <0.05 were considered statistically significant. Cluster analysis of clean reads against the tomato genome was performed using the ShortStack software tool (version 0.4.1) (Axtell, 2013b ) with options -mindepth=10 and -inv. The Inv file was generated from the tomato SL2.40 genome by einverted (EMBOSS 6.3.1) with option -maxrepeat 10000. Clean small RNAs from both biological replicates of each genotype were used as input to the ShortStack pipeline.
Carotenoid determination in petals
Total pigments from petals at anthesis (~100mg) were extracted with acetone. Acetone samples were dried under a stream of N2 and dissolved in 450 μl of ethanol. Then, 50 μl of 60% KOH (w/v) were added and samples were incubated in the dark for 16h at 4 °C for saponification. Saponified samples were mixed gently with 0.5ml of diethyl ether and 0.1ml of 12% (w/v) NaCl/H2O. The upper phase was collected and dried under a stream of N2 and dissolved in acetone for further analysis. Total carotenoid content was determined spectroscopically at 440nm using an extinction coefficient of 2400 (Schiedt and Liaaen-Jensen, 1995). For identification, carotenoids were separated by reverse-phase high performance liquid chromatography (RP-HPLC) using a Waters 600 system (Waters, Milford, MA, USA) and a Spherisorb ODS2 C18 column (silica 5 μm, 3.2 mm×250mm) (Phenomenex, Torrance, CA, USA). The mobile phase consisted of acetonitrile:water (9:1; A) and ethylacetate (B), at a constant flow rate of 1.6ml min–1. The gradient was: 100% to 80% A during 8min; 80% to 65% A during 4min; followed by 65% to 45% A during 14min; and a final segment of 7min at 100% B. Spectra at the wavelength range of 250–700nm of eluting HPLC solvent were recorded and absorption peaks were detected. Carotenoids were identified by their absorption spectra and retention time.
Results
Generation of a reporter line that is able to silence the tomato DCL1 homologue upon trans-activation
The full-length sequence of the tomato DCL1 homologue was determined (SlDCL1; GenBank accession no. JX962774). Sequence analysis suggested that it is targeted by sly-miR162, similar to AtDCL1 mRNA (Xie et al., 2003), and encodes a DCL1-like protein (Supplementary Fig. S1A available at JXB online). Analysis of published RNA sequencing data indicated that SlDCL1 is expressed at similar levels in vegetative and reproductive organs, with maximum expression in the root and minimum expression in breaking fruit (Tomato Genome Consortium, 2012) (Supplementary Fig. S1B available at JXB online).
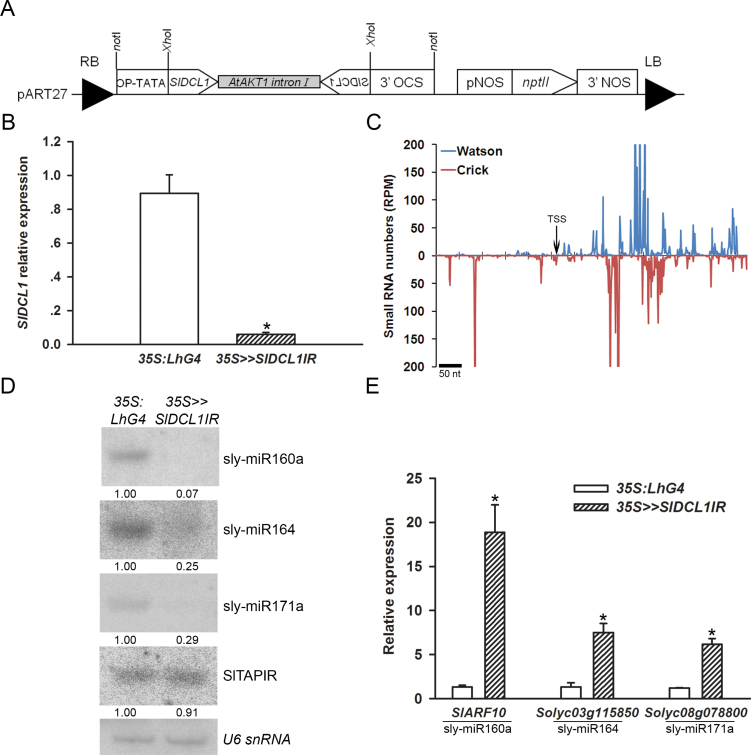
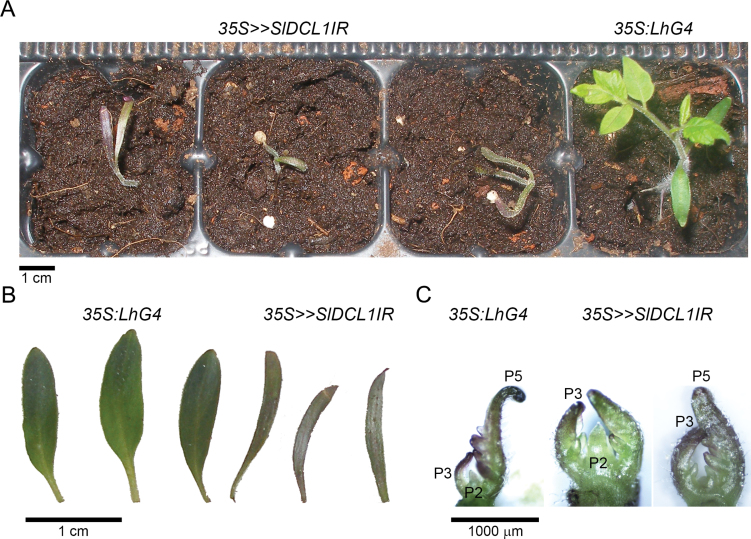
In the absence of a tomato dcl1 mutant, its function was addressed by silencing it in transgenic tomato. However, plant dcl1 loss-of-function mutants are likely to be either lethal or arrested during early development (Errampalli et al., 1991; Castle et al., 1993; Liu et al., 2005). To overcome this problem, a trans-activated RNAi was employed via the OP/LhG4 transcription factor system. A responder RNAi construct (OP:SlDCL1IR) that spanned the transcriptional start site of SlDCL1 was constructed with the intention of improving knockdown efficiency by invoking post-transcriptional as well as transcriptional silencing of SlDCL1 (Fig. 1A; Supplementary Fig. S1A available at JXB online). Following OP:SlDCL1IR transformation into M82 tomato and characterization of regenerated transgenic responder plants, the OP:SlDCL1IR-13 responder line (hereafter OP:SlDCL1IR) was selected for further analysis because SlDCL1 was almost completely silenced (~5% of control plants) in its trans-activated F1 progeny (35S>>SlDCL1IR) (Fig. 1B). The strong silencing was associated with the accumulation of siRNAs that matched SlDCL1 mRNA as well as its putative promoter (Fig. 1C), demonstrating the functionality of the RNAi construct and the usefulness of trans-activation for DCL1 silencing. In agreement with DCL1 being the primary producer of canonical miRNAs (Park et al., 2002; Kurihara and Watanabe, 2004), northern blots of 35S>>SlDCL1IR seedlings revealed decreased levels of several known tomato miRNAs, but no significant change in the levels of type III fold-back transposon anionic peroxidase inverted repeat (TAPIR)-derived heterochromatic siRNA (Moxon et al., 2008) (Fig. 1D). Consistent with the down-regulation of miRNAs, corresponding target mRNA levels increased significantly (Fig. 1E). In accordance with the perturbation of their miRNA pathway, the 35S>>SlDCL1IR seedlings could only germinate in tissue culture, had narrow cotyledons, and their development was arrested at the first to third true leaf primordium stage (Fig. 2).
Fig. 1.
Molecular characterization of 35S>>SlDCL1IR plants. (A) Schematic representation of the binary responder SlDCL1 RNAi (pART27-OP:SlDCL1IR) construct. (B) Quantitative RT-PCR analysis of SlDCL1 transcript in 35S>>SlDCL1IR seedlings 28 d after sowing. Primers were designed around the miR162 complementary site, and TIP41 expression values were used for normalization. Data are means ±SD of three independent biological repeats, each measured in triplicate. An asterisk indicates a significant difference as determined by Student’s t-test (P ≤ 0.01). (C) Schematic of siRNA accumulation at the SlDCL1IR RNAi sequence. The siRNA sequences were determined by deep sequencing of the seedling RNA used for qRT-PCR analysis in B. The normalized abundances of siRNAs were plotted relative to the sequence used for RNAi (Supplementary Table S1 available at JXB online) as a function of the positions of their 5′ ends. Maximum normalized reads per million (RPM) were set at 200/–200 to enable the visualization of relatively low abundance siRNAs. TSS, transcription start site as determined by RACE. (D) RNA gel-blot analysis of small RNAs in the RNA samples analysed in B. The total RNA (2.5 μg) was hybridized with the indicated small RNA antisense probe. An antisense probe for U6 snRNA served as a loading control. Small RNA expression was normalized to U6 snRNA, and levels are indicated below each panel. (E) qRT-PCR analysis of miRNA target transcripts in the RNA samples analysed in B. The corresponding miRNA is shown below each target mRNA. TIP41 expression values were used for normalization. Error bars indicate ±SD of three biological replicates, each measured in triplicate. Asterisks indicate significant difference as determined by Student’s t-test (P ≤ 0.01).
Fig. 2.
Phenotypic characterization of 35S>>SlDCL1IR seedlings. (A–C) Phenotypes of control (35S:LhG4) and 35S>>SlDCL1IR plants. (A) Seedlings at 28 d after sowing. (B) A magnified view of cotyledons from the seedlings in A. (C) A magnified view of shoot apices from the seedlings in A.
Small RNAs that depend on SlDCL1 for their biogenesis
To determine the effect of SlDCL1 silencing on global small RNA metabolism in tomato, the small RNA populations of control and 35S>>SlDCL1IR seedlings were compared using Illumina sequencing by synthesis technology. Two small RNA libraries, representing biological replicates, were prepared and sequenced from each genotype, generating >19 million raw reads in total (Supplementary Table S2 available at JXB online). For the analyses presented here, only sequences that were 18–26 nt in length were used. Depending on the library, read representation was between 6 and 4 million, from which ~70% matched the published tomato genome. Around 30% of the matched reads originated from tRNA, rRNA, snRNA, and snoRNA sequences. The remaining genome-matched reads (referred to as clean reads hereafter) were used for further analysis and represented ~1 million distinct tomato small RNA sequences (Supplementary Table S2 available at JXB online). The abundance of small RNAs was calculated as reads per million (RPM). After processing, clean small RNAs were annotated as described in the Materials and methods (Supplementary Table S3 available at JXB online).
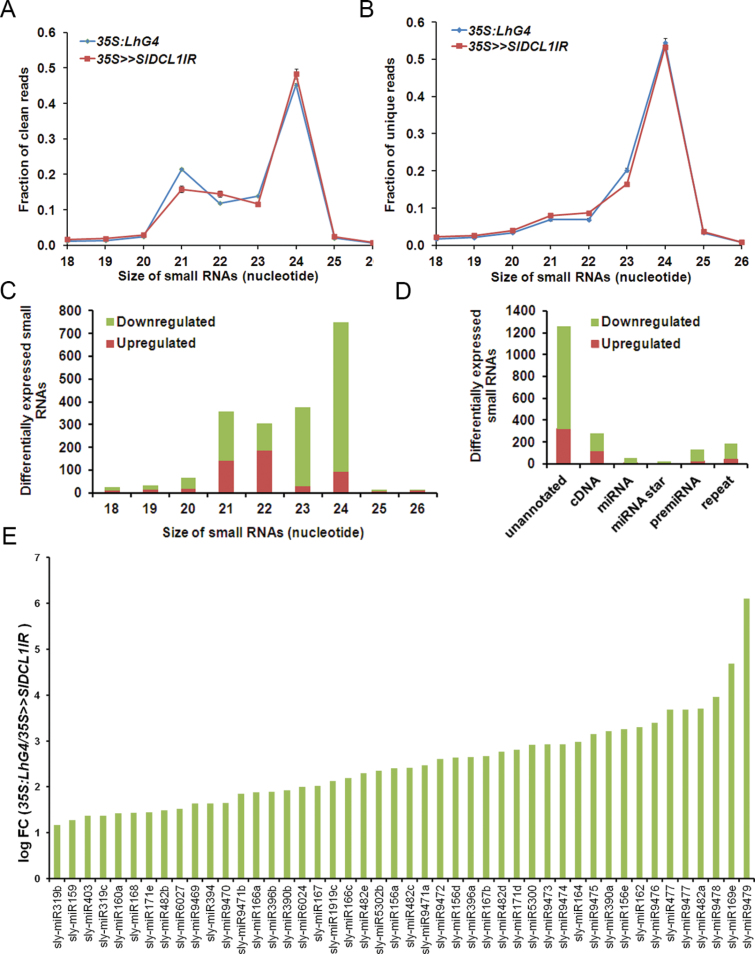
Consistent with the distribution of small RNA sizes that is typically observed in rice and Arabidopsis (Kasschau et al., 2007; Wu et al., 2010), in 35S:LhG4, the 24-nt small RNAs accounted for 45% of the population, making them the most abundant size class, with 21-nt small RNAs forming a secondary peak (Fig. 3A). Compared with 35S:LhG4 control seedlings, in 35S>>SlDCL1IR, the 21-nt size class was under-represented, whereas the 22-nt size class was over-represented (Fig. 3A), but a significant change in diversity was not observed (Fig. 3B). The question then was which tomato small RNAs require SlDCL1 for their biogenesis. EdgeR and ShortStack analyses were employed to identify the small RNAs and their producing loci, respectively, that show significantly modified expression on SlDCL1 down-regulation (Robinson et al., 2010; Axtell, 2013b ). First, to identify and annotate the wild-type tomato small RNA clusters, the 35S:LhG4 clean small RNAs were subjected to ShortStack analysis. Then, the expression of 35S>>SlDCL1IR clean small RNAs from identified wild-type clusters was quantified and the clusters were re-annotated accordingly. ShortStack analysis of control clean small RNAs identified 128 621 small RNA-producing clusters, of which 92 were annotated as MIRNA-derived clusters (Supplementary Table S4 available at JXB online). Since DCL1 is known to catalyse the formation of predominantly 21-nt RNAs (Park et al., 2002; Reinhart et al., 2002; Qi et al., 2005), the analysis was focused on the clusters that contained significant numbers (≥25 RPM) of 21-nt small RNAs and are therefore more likely to involve SlDCL1 in their biogenesis. Only 995 clusters fulfilled this criterion, and 132 of these showed at least a 2-fold change in their 21-nt small RNA numbers upon SlDCL1 silencing (hereafter termed DEclusters). Eighty-nine DEclusters were underexpressed in 35S>>SlDCL1IR relative to controls (Supplementary Table S5 available at JXB online). Cluster_61425, which derived from the MIR395a locus and contained ~30-fold fewer 21-nt small RNAs in 35S>>SlDCL1IR, showed the strongest decrease. As expected, MIRNA-derived small RNAs were almost exclusively down-regulated in 35S>>SlDCL1IR seedlings, and MIRNA-encoding loci comprised the largest group (61%, 32/89) of underexpressed DEclusters (Supplementary Table S5 available at JXB online). The results of the ShortStack analysis also indicated that several miRBase tomato miRNAs were not the most abundant small RNAs from their hairpin precursors and corresponded either to the predicted miRNA* [sly-miR6027 (Cluster_134922), sly-miR1919a-c (1892963), sly-miR5301 (Cluster_93957)] or to an undefined pre-miRNA position [sly-miR5302 (Cluster_93957)]. In the case of sly-miR5301, the predicted miRNA was found to be a close homologue of miR482. EdgeR analysis identified 2.5% (1421 of 56 052) and 0.9% (494 of 56 052) of the relatively abundant clean small RNAs (at least 1 RPM in three libraries), that had at least 2-fold fewer or more reads (FDR ≤0.05), respectively, in 35S>>SlDCL1IR relative to the control (Supplementary Table S6 available at JXB online). Among the down-regulated small RNAs, many were 21–22 nt in length originating from MIRNA genes, but also small RNAs 23–24 nt in length, which derived from non-annotated genomic loci and not likely to be produced by SlDCL1 (Qi et al., 2005), were present (Fig. 3C, D). In contrast, among the up-regulated small RNAs, most were 21–22 nt in length, derived from non-annotated genomic loci (Fig. 3C, D). In agreement with the ShortStack analysis, edgeR analysis indicated the significant down-regulation of 47 miRNAs in 35S>>SlDCL1IR relative to controls (Supplementary Table S6 available at JXB online). Annotation of these miRNAs indicated that 14 were identical to miRBase, 16 were homologues of known miRNAs, and 17 represented novel miRNAs. The degree of miRNA down-regulation was 4-fold on average, but varied considerably between miRNAs, indicating differential SlDCL1 requirement for their biogenesis (Fig. 3E).
Fig. 3.
Characterization of small RNAs from control (35S:LhG4) and 35S>>SlDCL1IR seedlings. (A, B) Size distribution of small RNAs in the indicated genotypes. The distributions of total (A) and unique (B) small RNAs are shown. Data are means ±SD of two biological replicates. (C–E) Characterization of differentially expressed small RNAs. (C) Size distribution of differentially expressed small RNAs. (D) Features of genomic loci that generate the small RNAs shown in C. (E) Change in miRNA accumulation upon SlDCL1 silencing.
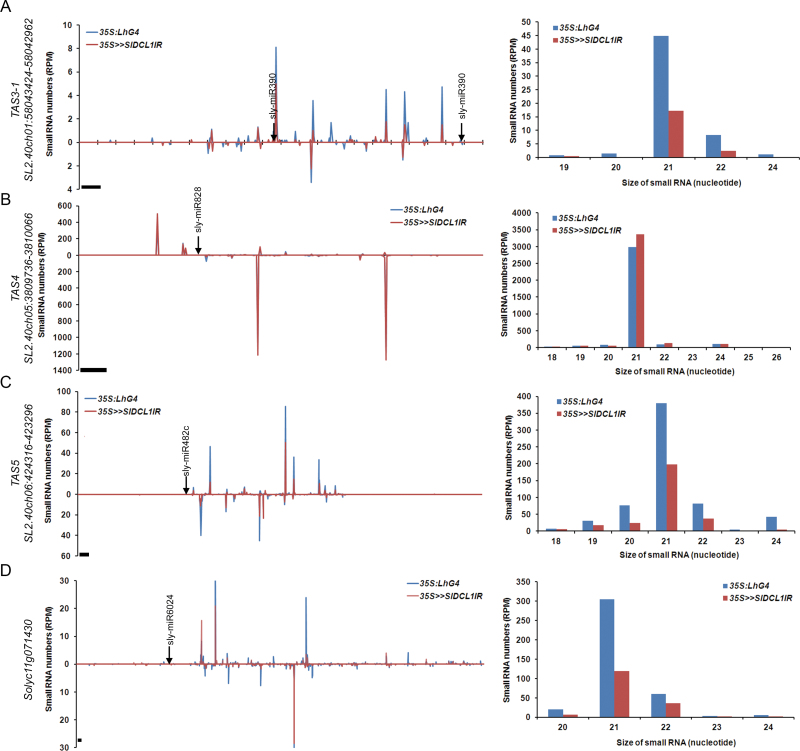
In tomato, sly-miR390- and sly-miR482c-guided cleavage sets the phase for the biogenesis of trans-acting siRNA gene 3 (TAS3)-derived and TAS5-derived ta-siRNAs, respectively (Li et al., 2012a ). EdgeR analysis suggested that sly-miR390 and sly-miR482c are significantly down-regulated, by 3.9- and 5.4-fold, respectively, in 35S>>SlDCL1IR seedlings (Fig. 3E). Consistent with this, the present analysis revealed significantly fewer TAS3-1 and TAS5 ta-siRNAs in 35S>>SlDCL1IR seedlings, thus confirming the requirement of SlDCL1 for ta-siRNA biogenesis (Fig. 4A, C). In contrast, a slight increase, rather than the expected decrease, was observed in the numbers of TAS4-derived (Cluster 99854) ta-siRNAs (Fig. 4B). Since the expression of TAS4 is induced by different stresses (Hsieh et al., 2009), a possible explanation is that an increase in TAS4 transcription occurred in the stressed 35S>>SlDCL1IR seedlings.
Fig. 4.
Effect of SlDCL1 silencing on phased siRNA accumulation. Schematic of 35S>>SlDCL1IR and control siRNA accumulation at indicated TAS (A–C) and R genes (D). The normalized abundances of siRNAs were plotted relative to their mRNA sequence as a function of the positions of their 5′ ends. Values above and below zero indicate siRNA matching of the plus and minus strand, respectively. The corresponding miRNA cleavage site is indicated. Bars=21 nt. Size distribution of total gene-derived siRNAs is shown on the right. Data are means of two biological replicates.
Except for ta-siRNAs, which depend on initial cleavage by miRNA for their biogenesis (Allen et al., 2005), cleavage by 22-nt miRNAs was found to set up the production of secondary 21-nt siRNAs from various target RNAs (Chen et al., 2010; Cuperus et al., 2010). In tomato and Medicago, such a mechanism was suggested to trigger the production of phased secondary 21-nt siRNAs from nucleotide-binding site (NBS)–leucine-rich repeat (LRR) R genes, thus preventing their unregulated expression (Li et al., 2010; Zhai et al., 2011; Shivaprasad et al., 2012). Accordingly, most (78%, 14/18) of the gene-derived DEclusters contained 21-nt siRNAs that matched NBS–LRR-encoding loci (Supplementary Table S5 available at JXB online). The 21-nt small RNA numbers from this DEcluster set were reduced by 2- to 5.3-fold upon SlDCL1 silencing, probably as a result of lower levels of the miRNA that triggers their biogenesis. An example of an NBS–LRR gene-derived DEcluster (Cluster_146573), which is predicted to be triggered by sly-miR6024-mediated cleavage, is shown in Fig. 4D. Indeed, in 35S>>SlDCL1IR seedlings, sly-miR6024 was down-regulated 4-fold and Solyc11g071430-derived 21-nt phased siRNA numbers decreased by 2.6-fold. The rest of the down-regulated gene-derived DEclusters were generated from genes with unknown functions. In addition, the analysis identified 13 repeat-associated DEclusters that were down-regulated upon SlDCL1 silencing, suggesting its involvement in the biogenesis of siRNAs from these loci (Supplementary Table S5 available at JXB online).
SlDCL1 silencing induces the up-regulation of certain SlDCL2 genes
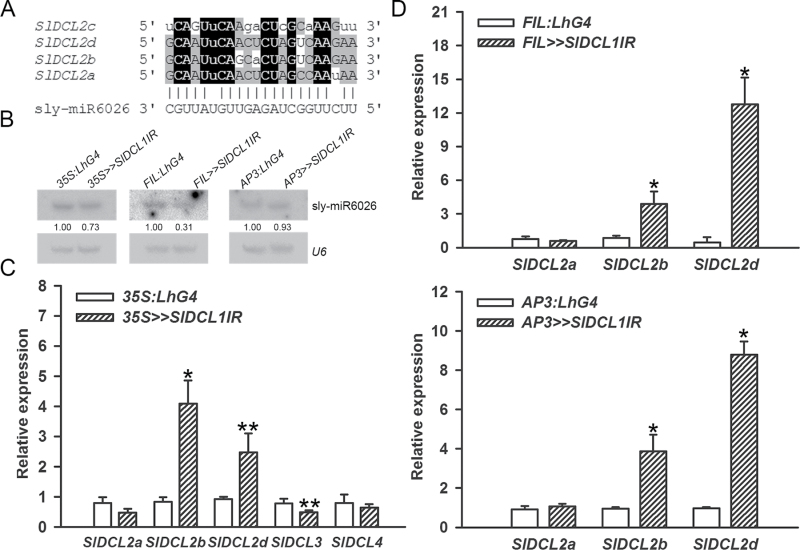
It has been suggested that soybean and Medicago DCL2 mRNAs are targeted by the 22-nt miR1515 and miR1507, respectively, triggering the generation of 21-nt secondary phased siRNAs (Li et al., 2010; Zhai et al., 2011). Tomato has four DCL2 genes (SlDCL2a–d), three of which (SlDCL2b–d) are clustered on chromosome 11 (SL2.40ch11:2690601–2724508) (Bai et al., 2012). Similar to Medicago DCL2, ShortStack analysis identified small RNA clusters that were derived from SlDCL2a (clusters 78419–78430), SlDCL2b (clusters 136462–136472), SlDCL2c (clusters 136447–136454), and SlDCL2d (clusters 136455–136460). These clusters were almost all (34/35) dominated by 21-nt small RNAs (DicerCall=21) that in a few cases were significantly phased (Supplementary Table S5, Fig. S2 available at JXB online). In addition, a legitimate target site for the 22-nt sly-miR6026 was predicted in the 5′ UTRs of SlDCL2a, b, and d but not in SlDCL2c, which contains a different sequence instead (Fig. 5A). Cleavage by 22-nt miRNAs has been shown to induce the biogenesis of 21-nt secondary siRNAs (Cuperus et al., 2010), raising the possibility that sly-miR6026 acts as a trigger for siRNA biogenesis from SlDCL2a, b, and d. In Medicago, miR1507 has been suggested to target DCL2 as well as NBS–LRR genes (Zhai et al., 2011). Consistent with this, sly-miR6026 has been predicted to target the Tobacco mosaic virus resistance gene of tomato (TM2, Solyc09g018220) (Li et al., 2012b ), and ShortStack analysis identified 21-nt small RNA clusters that derived from it (clusters 113162 and 113163) (Supplementary Table S5 available at JXB online), suggesting that similar to miR1507 in Medicago, in tomato, sly-miR6026-guided cleavage of SlDCL2a, b, and d as well as the NBS–LRR TM2 gene triggers secondary siRNA production. RNA gel blots detected a slight decrease in sly-miR6026 upon global and tissue-specific (see below) SlDCL1 silencing (Fig. 5B). However, the present analysis indicated that a significant increase, rather than a decrease, in the numbers of SlDCL2a, b, and d-derived small RNAs had occurred in 35S>>SlDCL1IR seedlings (Supplementary Table S5, Fig. S2 available at JXB online). The most significant increase was in SlDCL2b (DEclusters 136464–136470, average of ~4.73±0.97-fold) and SlDCL2d (DEclusters 136455–136458, average of ~3.13±1.02-fold). Possible explanations include an increase in the expression of either SlDCL4, which is required for 21-nt phased siRNA biogenesis (Yifhar et al., 2012), or SlDCL2b and d, whose transcripts may serve as substrates for sly-miR6026-mediated cleavage. qRT-PCR analysis of these genes in different SlDCL1-depleted tissues confirmed the significant increase in SlDCL2b and d accumulation, whereas accumulation of SlDCL2a, SlDCL3, and SlDCL4 was not significantly changed (Fig. 5C, D). Since DCL2 has been shown to be required for the formation of 22-nt siRNAs (Xie et al., 2004; Gasciolli et al., 2005; Deleris et al., 2006), the up-regulation of SlDCL2b and d in 35S>>SlDCL1IR seedlings might explain the global increase in 22-nt numbers (Fig. 3A) and the majority (43%) of the 22-nt up-regulated DEclusters (Supplementary Table S5 available at JXB online).
Fig. 5.
Certain SlDCL2 paralogues are up-regulated upon SlDCL1 silencing. Sequence alignment between SlDCL2 genes (A) predicted to serve as sly-miR6026 targets. Identical nucleotides in all or at least three sequences are shaded in black and grey, respectively. Watson–Crick pairing with sly-miR6026 is shown below. Mismatched nucleotides are in lower case. (B) RNA gel-blot analysis of sly-miR6026 in the indicated genotypes from seedlings 28 d after sowing, leaf P6-7 primordia, and flower petals in 10mm buds (left to right). Total RNA (5 μg) from the indicated tissues was hybridized with sly-miR6026 antisense probe. Sly-miR6026 expression was normalized to U6 snRNA and levels are indicated below each panel. (C, D) qRT-PCR analysis of the expression levels of the indicated SlDCL genes in the RNA samples analysed in B: seedlings 28 d after sowing (C), leaf P6-7 primordia (D top), and flower petals in 10mm buds (D bottom). Error bars indicate ±SD of three biological replicates, each measured in triplicate. Single (P ≤ 0.01) and double (P ≤ 0.05) asterisks indicate significant difference as determined by Student’s t-test.
Depletion of SlDCL1 from developing leaf primordium inhibits blade expansion
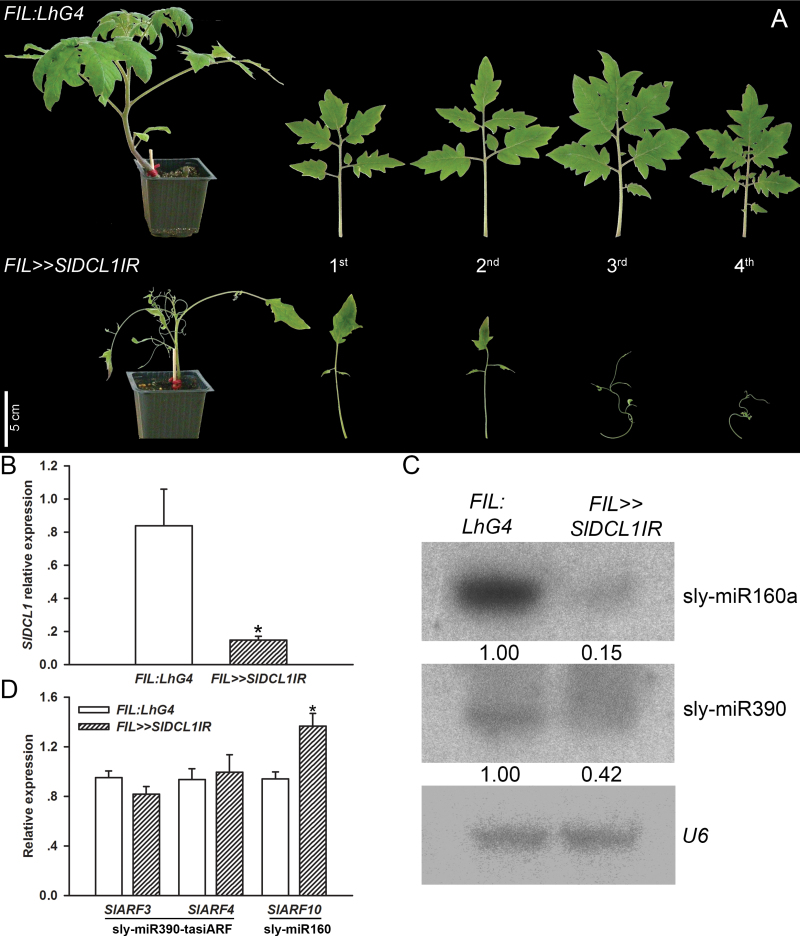
Constitutive SlDCL1 silencing induced seedling lethality before lateral organs could develop (Fig. 2). However, tissue-specific driver lines allowed investigation of the requirement of SlDCL1 of leaf and flower development. In tomato, the Filamentous Flower (FIL) promoter drives strong expression throughout young leaf primordia soon after they initiate from the shoot apical meristem (SAM), but not while they are in the SAM (Lifschitz et al., 2006; Shani et al., 2009). Thus, leaf-specific SlDCL1 silencing was induced by crossing the OP:SlDCL1IR responder line with the FIL:LhG4 driver line. As expected, germination of FIL>>SlDCL1IR progeny seedlings was normal (Supplementary Fig. S3 available at JXB online). In addition, relatively subtle phenotypes were observed in the first two leaves. These leaves contained one instead of two pairs of primary leaflets, which were smaller than controls, and had a morphologically distinct terminal leaflet blade (Fig. 6A). More severe phenotypes were observed in later formed leaves. Compared with FIL:LhG4 control leaves, these leaves were much smaller, and had wavy rachises that supported almost bladeless primary and terminal leaflets (Fig. 6A). As the plants matured, their cotyledons became abnormally elongated. In addition, precocious outgrowth of cotyledonary as well as juvenile axillary meristems was observed, suggesting loss of apical dominance in these plants (Supplementary Fig. S3 available at JXB online). The bladeless FIL>>SlDCL1IR leaf phenotype was reminiscent of the later formed wiry leaves of TAS3 pathway mutants that overaccumulate Auxin Response Factor 3 (SlARF3) and SlARF4, due to reduced levels of ta-siARF (Yifhar et al., 2012). Reduced tomato leaf lamina outgrowth has also been associated with ectopic accumulation of miR160-resistant SlARF10 (Hendelman et al., 2012). Molecular analysis of young leaf primordia confirmed the silencing of SlDCL1 (Fig. 6B) and, accordingly, revealed reduced accumulation of sly-miR160a and sly-miR390, which is required for ta-siARF biogenesis (Fig. 6C) (Yifhar et al., 2012). Nevertheless, significant up-regulation of SlARF3, SlARF4, and SlARF10 was not detected in these primordia, most probably due to insufficient down-regulation of sly-miR390 and sly-miR160a, respectively (Fig. 6D).
Fig. 6.
Characterization of SlDCL1-depleted leaves. (A) Comparison of whole M82 tomato plants 42 d after sowing and representative leaves (from left to right) between control (FIL:LhG4) and FIL>>SlDCL1IR. Scale bar=5cm. (B) qRT-PCR analysis of SlDCL1 transcript levels in FIL:LhG4 and FIL>>SlDCL1IR P5 leaf primordia. (C) RNA gel-blot analysis of miRNAs in the RNA samples analysed in B. Total RNA (2 μg) was hybridized with the indicated miRNA antisense probe. Expression levels were determined relative to the control after normalization to U6 and are indicated below each panel. (D) qRT-PCR analysis of miRNA target transcripts in the RNA samples analysed in B. The corresponding miRNA is shown below each target mRNA. (B, D) CAC expression values were used for normalization. Error bars indicate ±SD of three biological replicates, each measured in triplicate. Asterisks indicate significant difference as determined by Student’s t-test (P ≤ 0.01).
Partial depletion of SlDCL1 from petals results in lower accumulation of carotenoids
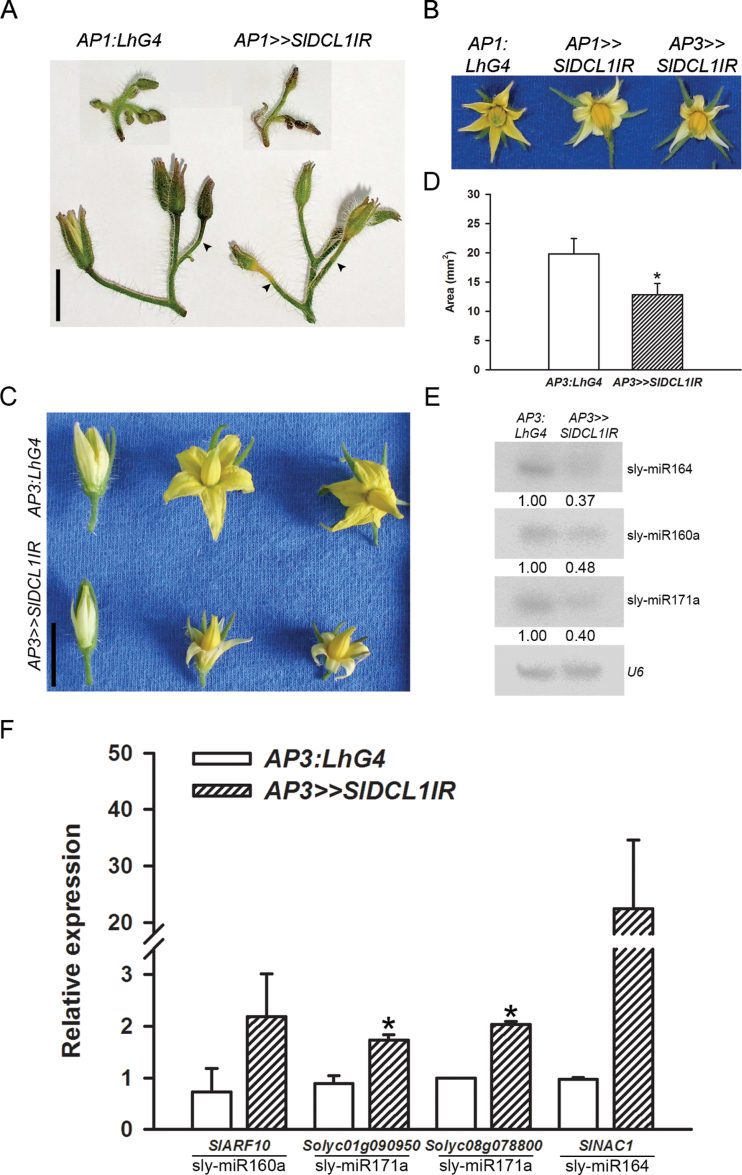
The flower-specific AP1:LhG4 driver line drives expression throughout the young floral primordia and, as the buds mature, its activity is mainly confined to developing sepals and petals. The AP3:LhG4 driver line drives expression throughout the developing petals, in the stamen vasculature, and on the abaxial side of anthers (Hendelman et al., 2013a ). Hence, flower-specific SlDCL1 silencing was induced by crossing the OP:SlDCL1IR responder line to these driver lines. Both AP1>>SlDCL1IR and AP3>>SlDCL1IR plants showed a wild-type phenotype during vegetative growth (data not shown). Moreover, young AP1>>SlDCL1IR buds did not show any developmental aberrations despite containing only 40% of the wild-type SlDCL1 levels (Supplementary Fig. S4A available at JXB online). However, most of the buds underwent premature senescence at 4–5mm in size and could not develop into mature flowers. The senescence initiated as yellowing of the receptacle area in one or more buds from the same inflorescence. This yellowing spread downward along the stack to the abscission zone and upward to the sepals, ultimately leading to complete drying of the bud (Fig. 7A). Only ~10% of the buds developed into mature flowers that produced pale petals (Fig. 7B). Interestingly, the AP3>>SlDCL1IR flowers produced similarly pale and slightly smaller petals (Fig. 7B–D). The composition of carotenoids in AP3>>SlDCL1IR petals was analysed to determine whether the pale phenotype is associated with quantitative or qualitative differences. While the carotenoid profile in AP3>>SlDCL1IR petals is relatively similar to that of the control, consisting of mainly neoxanthin and violaxanthin, the total level of carotenoids was reduced >2-fold (Table 1). Additional visual abnormalities were not observed in the AP3>>SLDCL1IR flowers (Fig. 7C). qRT-PCR analysis of SlDCL1 levels in isolated petals, at the start of yellowing, revealed that it accumulated to only 25% of controls, indicating partial silencing of SlDCL1 in the petals (Supplementary Fig. S4B available at JXB online). Accordingly, miRNA levels were down-regulated and corresponding target mRNAs were up-regulated in the silenced petals (Fig. 7E, F).
Fig. 7.
Characterization of SlDCL1-depleted flowers. (A) Phenotype of representative control (AP1:LhG4) and AP1>>SlDCL1IR inflorescences bearing young (top) and mature (bottom) buds. Arrowheads mark the abscission zone. Scale bars=1cm. (B) Representative flowers of the indicated genotypes at anthesis. (C) Representative flowers of the indicated genotypes at different developmental stages: 2 d before anthesis, anthesis, and 1 d post-anthesis (left to right). (D) Quantitation of petals area at anthesis in control and AP3>>SlDCL1IR. Error bars represent ± SD (n=54). (E) RNA gel-blot analysis of miRNAs in petals from 10mm buds of the indicated genotypes. Total RNA (5 μg) was hybridized with the corresponding miRNA antisense probe. Expression levels were determined relative to the control after normalization to U6 and are indicated below each panel. (F) qRT-PCR analysis of miRNA target transcripts in the RNA samples analysed in E. The corresponding miRNA is shown below each target mRNA. CAC expression values were used for normalization. Error bars indicate ±SD of three biological replicates, each measured in triplicate. (D, F) Asterisks indicate significant difference as determined by Student’s t-test (P ≤ 0.01).
Table 1.
Carotenoid composition in control and AP3>>SlDCL1IR petals
| AP3:LhG4 a | AP3>>SlDCL1lR a | |
|---|---|---|
| Neoxanthin | 71.6±2.2 | 66.4±4.2 |
| Violaxanthin | 24.3±1.9 | 23.0±2.4 |
| Antheraxanthin | 1.2±0.5 | 1.4±0.4 |
| Lutein | 1.8±0.4 | 5.6±1.9 |
| β-Carotene | 1.1±0.4 | 3.7±0.5 |
| Totalb (μg g fresh weight–1) | 620.6±112.8 | 246.4±28.8 |
a Numbers correspond to percentage of total carotenoids (n=4).
b All isomers.
Discussion
As part of the effort to unveil the roles of miRNA regulation in tomato development, transgenic tomato responder lines were previously generated that, by trans-activation, can express the viral silencing suppressors Tomato bushy stunt virus P19 and Beet western yellows virus P0, thereby perturbing the miRNA pathway in the activated plants (Stav et al., 2010; Hendelman et al., 2013a ). However, because P19 binds siRNA duplexes and P0 can lead to the decay of various ARGONAUTES, their transgenic expression may interfere with endogenous siRNA pathways as well (Dunoyer et al., 2004; Baumberger et al., 2007). In the current study, the OP:SlDCL1IR tomato responder line was used; this line is a more specific miRNA-inhibiting tool that enables the silencing of SlDCL1, the tomato homologue of DCL1, by trans-activation. DCL1 has been found to be the predominant DCL involved in the biogenesis of canonical miRNAs, with no other DCL acting as its surrogate (Gasciolli et al., 2005). Accordingly, trans-silencing of SlDCL1 to 5% of its normal levels was associated with diminished miRNA levels, demonstrating that in tomato, like in Arabidopsis and rice, DCL1 is central for miRNA production.
The involvement of DCL1 in the biogenesis of certain siRNAs renders the criterion of DCL1 dependence for accumulation insufficient to warrant annotation of a small RNA as an miRNA (Meyers et al., 2008). ShortStack is a recently published stand-alone application that can predict and annotate plant miRNAs based on small RNA sequencing data (Axtell, 2013b ). By combining ShortStack predictions and the SlDCL1 dependence criterion, 33 new SlDCL1-dependent miRNAs were confidently discovered, 17 of which represent novel plant miRNAs. This method is suggested for reliable annotation of canonical miRNA in tomato. A total of 47 miRNAs that depend on SlDCL1 for their biogenesis were identified in this study. These miRNAs were shown to be differentially reduced by SlDCL1 silencing. This differential response ranged from a 2-fold to a 64-fold decrease and might indicate the in vivo cleavage efficiency and specificity of SlDCL1 for various miRNA precursors. Interestingly, sly-miR159a, which is encoded by a single precursor (SL2.40ch03:61786005–61786301), was reduced by only 2.4-fold in 35S>>SlDCL1IR plants, indicating relatively efficient processing and low sensitivity to SlDCL1 silencing. Indeed, the sly-miR159a precursor has been successfully used as a backbone for the expression of an artificial miRNA (amiR) precursor in transgenic tomato and it was more efficiently processed than sly-miR168a-based amiR (reduced by 2.7-fold in 35S>>SlDCL1IR) (Vu et al., 2013).
The present data revealed that SlDCL1 silencing causes two significant but contrasting changes in the small RNA population of tomato seedlings. The 35S>>SlDCL1IR seedlings exhibited a lower proportion of 21-nt small RNAs and a higher proportion of 22-nt small RNAs. The former was also observed in other dcl1 mutants and can be attributed to a substantial reduction in 21-nt miRNAs which depend on DCL1 for their biogenesis, and phased siRNAs, which depend on initial miRNA cleavage for their production (Kasschau et al., 2007; Shivaprasad et al., 2012). Nevertheless, the increase in 22-nt small RNAs has not been observed in other dcl1 mutants (Kasschau et al., 2007; Wu et al., 2010). In Arabidopsis, DCL2 has been shown to be required for the formation of 22-nt long siRNAs (Xie et al., 2004; Gasciolli et al., 2005; Deleris et al., 2006). The tomato genome encodes four DCL2 paralogues, three of which (SlDCL2a, b, and d) are clustered on chromosome 11 (Bai et al., 2012). The results of the present study indicate that SlDCl2b and SlDCl2d are up-regulated upon SlDCL1 silencing, suggesting that these paralogues are responsible for the observed increase in 22-nt small RNAs. Sly-miR6026, which is predicted to target SlDCL2a, b, and d, was not significantly down-regulated upon SlDCL1 silencing. Consistent with this, SlDCL2b- and SlDCL2d-derived siRNAs increased rather than decreased and SlDCl2a was not up-regulated in 35S>>SlDCL1IR seedlings. Therefore, the up-regulation of SlDCl2b and SlDCl2d is probably due to an increase in transcription rather than reduced targeting by sly-miR6026. One appealing explanation for this is the existence of a specific SlDCl2b and SlDCl2d miRNA-regulated transcriptional activator that is up-regulated upon SlDCL1 silencing.
Consistent with the severe developmental phenotypes of known dcl1 mutants (Schauer et al., 2002; Liu et al., 2005; Khraiwesh et al., 2010), the 35S>>SlDCL1IR seedlings were growth arrested at the first to third leaf stage, emphasizing the importance of miRNAs for tomato’s post-germination development. Nevertheless, the trans-activated RNAi strategy enabled, for the first time, SlDCL1 silencing in a specific organ, without disturbing prior development. This enabled a direct unbiased assessment of SlDCL1 functions in the silenced tissue. In addition, activation of the OP:SlDCL1IR construct by different promoters generated variable silencing strengths that were manifested by hypomorphic phenotypes. Reducing SlDCL1 levels by 85% in the leaf primordia led to strong inhibition of blade outgrowth, but did not modify its abaxial–adaxial polarity, regulated in part by miR166 and miR390 (Chitwood et al., 2007). Similarly, the hypomorphic Arabidopsis mutant dcl1-9 (caf) produced polar leaves that were thinner than the wild type and ~10% of them did not develop any blade (Jacobsen et al., 1999), together indicating that blade outgrowth is dependent on DCL1-mediated silencing in simple as in compound leaf species. Indeed, perturbation of the post-transcriptional regulation of SlARF10, and SlARF3 and SlARF4, which are normally cleaved by sly-miR160a and tasiARF, respectively, has been shown to reduce blade outgrowth dramatically in tomato leaves (Hendelman et al., 2012; Yifhar et al., 2012). Surprisingly, none of these target genes was significantly up-regulated in FIL>>SlDCL1IR leaf primordia, suggesting that they are not the main cause for the bladeless phenotype. In addition, FIL>>SlDCL1IR plants also demonstrated loss of apical dominance. Since both blade outgrowth and apical dominance are regulated by auxin (Cline, 1997; Koenig et al., 2009), disruption of its signalling pathway by SlDCL1 silencing is likely to be a major cause for the observed FIL>>SlDCL1IR phenotypes.
The distinct yellow colour of tomato flowers is a result of accumulation of carotenoids, mainly the xanthophylls neoxanthin and violaxanthin (Hirschberg, 2001). A novel pale phenotype was observed in AP1>>SlDCL1IR and AP3>>SlDCL1IR petals with partially silenced SlDCL1. This phenotype was associated with a reduction of >2-fold in the total rather than an alteration in the ratio between the individual carotenoids. The exact reason for this is currently unknown. Nevertheless, by profiling Arabidopsis dcl1 mutant embryos, it was shown that the two most up-regulated miRNA targets are responsible for a large part of the mutant embryo phenotype, thus proving the usefulness of dcl1 mutants for revealing specific miRNA regulatory modules involved in development (Nodine and Bartel, 2010; Willmann et al., 2011). The availability of a tomato dcl1 mutant, as presented in the current study, now enables the use of a similar approach to elucidate the molecular bases of the observed phenotypes.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Sequence analysis of the tomato DCL1 homologue.
Figure S2. Effect of SlDCL1 silencing on SlDCL2-derived siRNAs accumulation.
Figure S3. Additional phenotypes of FIL>>SlDCL1IR plants.
Figure S4. Quantitative analysis of SlDCL1 levels in trans-activated flowers.
Table S1. List of primers and probes used in this study.
Table S2. Summary of small RNA profiling.
Table S3. Summary of small RNA annotations.
Table S4. Summary of ShortStack-predicted miRNAs.
Table S5. List of ShortStack small RNA clusters.
Table S6. EdgeR results and differentially expressed miRNAs.
Acknowledgements
We thank members of the lab for critical reading of the manuscript. This work was supported by a grant from the Chief Scientist of the Israel Ministry of Agriculture and Rural Development no. 256-0828-10 to TA. RS acknowledges support from the USDA (2007-35100-18318) and OAES. This is contribution no. 112/2013 series, from the Agricultural Research Organization, the Volcani Center, Bet Dagan 50250, Israel.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221 [DOI] [PubMed] [Google Scholar]
- Axtell MJ. 2013a. Classification and comparison of small RNAs from plants. Annual Review of Plant Biology 64, 137–159 [DOI] [PubMed] [Google Scholar]
- Axtell MJ. 2013b. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 19, 740–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Yang GS, Chen WT, Mao ZC, Kang HX, Chen GH, Yang YH, Xie BY. 2012. Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum . Gene 501, 52–62 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC. 2007. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Current Biology 17, 1609–1614 [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. 2005. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123, 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozorov TA, Pandey SP, Dinh ST, Kim SG, Heinrich M, Gase K, Baldwin IT. 2012. DICER-like proteins and their role in plant–herbivore interactions in Nicotiana attenuata. Journal of Integrative Plant Biology 54, 189–206 [DOI] [PubMed] [Google Scholar]
- Castle LA, Errampalli D, Atherton TL, Franzmann LH, Yoon ES, Meinke DW. 1993. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Molecular and General Genetics 241, 504–514 [DOI] [PubMed] [Google Scholar]
- Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH. 2010. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proceedings of the National Academy of Sciences, USA 107, 15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. 2009. Small RNAs and their roles in plant development. Annual Review of Cell and Developmental Biology 25, 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Guo M, Nogueira FT, Timmermans MC. 2007. Establishing leaf polarity: the role of small RNAs and positional signals in the shoot apex. Development 134, 813–823 [DOI] [PubMed] [Google Scholar]
- Cline M. 1997. Concepts and terminology of apical dominance. American Journal of Botany 84, 1064. [PubMed] [Google Scholar]
- Cuperus JT, Carbonell A, Fahlgren N, et al. 2010. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nature Structural and Molecular Biology 17, 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313, 68–71 [DOI] [PubMed] [Google Scholar]
- de Lima JC, Loss-Morais G, Margis R. 2012. MicroRNAs play critical roles during plant development and in response to abiotic stresses. Genetics and Molecular Biology 35, 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O. 2004. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. The Plant Cell 16, 1235–1250 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Errampalli D, Patton D, Castle L, Mickelson L, Hansen K, Schnall J, Feldmann K, Meinke D. 1991. Embryonic lethals and T-DNA insertional mutagenesis in Arabidopsis. The Plant Cell 3, 149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Sullivan CM, Kasschau KD, et al. 2009. Computational and analytical framework for small RNA profiling by high-throughput sequencing. RNA 15, 992–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AI, Viron N, Alhagdow M, et al. 2009. Flexible tools for gene expression and silencing in tomato. Plant Physiology 151, 1729–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. 2005. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Current Biology 15, 1494–1500 [DOI] [PubMed] [Google Scholar]
- Giovannoni J. 2001. Molecular biology of fruit maturation and ripening. Annual Review of Plant Physiology and Plant Molecular Biology 52, 725–749 [DOI] [PubMed] [Google Scholar]
- Hendelman A, Buxdorf K, Stav R, Kravchik M, Arazi T. 2012. Inhibition of lamina outgrowth following Solanum lycopersicum AUXIN RESPONSE FACTOR 10 (SlARF10) derepression. Plant Molecular Biology 78, 561–576 [DOI] [PubMed] [Google Scholar]
- Hendelman A, Kravchik M, Stav R, Zik M, Lugassi N, Arazi T. 2013a. The developmental outcomes of P0-mediated ARGONAUTE destabilization in tomato. Planta 237, 363–377 [DOI] [PubMed] [Google Scholar]
- Hendelman A, Stav R, Zemach H, Arazi T. 2013b. The tomato NAC transcription factor SlNAM2 is involved in flower-boundary morphogenesis. Journal of Experimental Botany 64, 5497 5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE. 2006. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38, 721–725 [DOI] [PubMed] [Google Scholar]
- Hirschberg J. 2001. Carotenoid biosynthesis in flowering plants. Current Opinion in Plant Biology 4, 210–218 [DOI] [PubMed] [Google Scholar]
- Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC. 2007. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. The Plant Cell 19, 926–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ. 2009. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiology 151, 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz EM. 1999. Disruption of an RNA helicase/RNase III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126, 5231–5243 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. 2006. MicroRNAS and their regulatory roles in plants. Annual Review of Plant Biology 57, 19–53 [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. 2007. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biology 5, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W. 2010. Transcriptional control of gene expression by microRNAs. Cell 140, 111–122 [DOI] [PubMed] [Google Scholar]
- Koenig D, Bayer E, Kang J, Kuhlemeier C, Sinha N. 2009. Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 136m 2997–3006 [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y. 2004. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proceedings of the National Academy of Sciences, USA 101, 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Zeller G, Henz SR, Buechel S, Sachsenberg T, Wang JW, Ratsch G, Weigel D. 2010. Global effects of the small RNA biogenesis machinery on the Arabidopsis thaliana transcriptome. Proceedings of the National Academy of Sciences, USA 107, 17466–17473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Orban R, Baker B. 2012a. SoMART: a web server for plant miRNA, tasiRNA and target gene analysis. The Plant Journal 70, 891–901 [DOI] [PubMed] [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. 2012b. MicroRNA regulation of plant innate immune receptors. Proceedings of the National Academy of Sciences, USA 109, 1790–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Deng Y, Wu T, Subramanian S, Yu O. 2010. Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiology 153, 1759–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y. 2006. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proceedings of the National Academy of Sciences, USA 103, 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li P, Li X, Liu C, Cao S, Chu C, Cao X. 2005. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiology 139, 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margis R, Fusaro AF, Smith NA, Curtin SJ, Watson JM, Finnegan EJ, Waterhouse PM. 2006. The evolution and diversification of Dicers in plants. FEBS Letters 580, 2442–2450 [DOI] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, et al. 2001. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159, 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, et al. 2008. Criteria for annotation of plant MicroRNAs. The Plant Cell 20, 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore I, Galweiler L, Grosskopf D, Schell J, Palme K. 1998. A transcription activation system for regulated gene expression in transgenic plants. Proceedings of the National Academy of Sciences, USA 95, 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T. 2008. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Research 18, 1602–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine MD, Bartel DP. 2010. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes and Development 24, 2678–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Current Biology 12, 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ. 2005. Biochemical specialization within Arabidopsis RNA silencing pathways. Molecular Cell 19, 421–428 [DOI] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. 2006. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes and Development 20, 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. 2002. MicroRNAs in plants. Genes and Development 16, 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Beers K, Pruitt RE, Gasser CS. 1992. Ovule development in wild-type Arabidopsis and two female-sterile mutants. The Plant Cell 4, 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A. 2002. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends in Plant Science 7, 487–491 [DOI] [PubMed] [Google Scholar]
- Schiedt K, Liaaen-Jensen S. 1995. Carotenoids: isolation and analysis. Basel: Birkhäuser Verlag [Google Scholar]
- Shani E, Burko Y, Ben-Yaakov L, Berger Y, Amsellem Z, Goldshmidt A, Sharon E, Ori N. 2009. Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. The Plant Cell 21, 3078–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC. 2012. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. The Plant Cell 24, 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stav R, Hendelman A, Buxdorf K, Arazi T. 2010. Transgenic expression of tomato bushy stunt virus silencing suppressor P19 via the pOp/LhG4 transactivation system induces viral-like symptoms in tomato. Virus Genes 40, 119–129 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Li YF, Jagadeeswaran G. 2012. Functions of microRNAs in plant stress responses. Trends in Plant Science 17, 196–203 [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu TV, Choudhury NR, Mukherjee SK. 2013. Transgenic tomato plants expressing artificial microRNAs for silencing the pre-coat and coat proteins of a begomovirus, Tomato leaf curl New Delhi virus, show tolerance to virus infection. Virus Research 172, 35–45 [DOI] [PubMed] [Google Scholar]
- Willmann MR, Mehalick AJ, Packer RL, Jenik PD. 2011. MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiology 155, 1871–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y. 2010. DNA methylation mediated by a microRNA pathway. Molecular Cell 38, 465–475 [DOI] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biology 2, E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Kasschau KD, Carrington JC. 2003. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Current Biology 13, 784–789 [DOI] [PubMed] [Google Scholar]
- Yifhar T, Pekker I, Peled D, et al. 2012. Failure of the tomato trans-acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome. The Plant Cell 24, 3575–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, De Paoli E, et al. 2011. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes and Development 25, 2540–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.