Abstract
SUMMARY
Rhabdomyosarcoma is a soft-tissue sarcoma with molecular and cellular features of developing skeletal muscle. Rhabdomyosarcoma has two major histological subtypes, embryonal and alveolar, each with distinct clinical, molecular, and genetic features. Genomic analysis show that embryonal tumors have more structural and copy number variations than alveolar tumors. Mutations in the RAS/NF1 pathway are significantly associated with intermediate- and high-risk embryonal rhabdomyosarcomas (ERMS). In contrast, alveolar rhabdomyosarcoma (ARMS) have fewer genetic lesions overall and no known recurrently mutated cancer consensus genes. To identify therapeutics for ERMS, we developed and characterized orthotopic xenografts of tumors that were sequenced in our study. High throughput screening of primary cultures derived from those xenografts identified oxidative stress as a pathway of therapeutic relevance for ERMS.
INTRODUCTION
Rhabdomyosarcoma is the most common soft-tissue sarcoma of childhood and adolescence (Reis et al., 1999). Using contemporary multimodal therapies, more than 75% of patients with localized disease are cured (Crist et al., 2001). However, despite the availability of new agents and intensification of therapy, patients with recurrent or metastatic rhabdomyosarcoma experience 5-year survival rates of only 17% or 30%, respectively (Pappo et al., 1999). Rhabdomyosarcoma can be divided into two broad histopathologic subtypes: embryonal rhabdomyosarcoma (ERMS), which accounts for about 60% of all rhabdomyosarcomas, and alveolar rhabdomyosarcoma (ARMS), which accounts for about 25%(Newton et al., 1988). The remaining cases are classified as mixed, unspecified or undifferentiated sarcomas(Rudzinski et al., 2013). Patients with ERMS have a relatively good prognosis, and the tumors are characterized by the loss of heterozygosity at the 11p15 locus(Ognjanovic et al., 2009; Scrable et al., 1989). In contrast, patients with ARMS have an inferior clinical outcome, and their tumors are often characterized by a translocation between the FOXO1 gene on chromosome 13q14 and either PAX3 on chromosome 2q35 or PAX7 on chromosome 1p36(Barr, 1997; Raney et al., 2001).
Current treatment protocols for rhabdomyosarcoma are designed to deliver risk-based therapy (low, intermediate, or high) based exclusively on the clinical and pathologic features at the time of initial presentation(Malempati and Hawkins, 2012). However, the outcome for an individual child, particularly one with intermediate-risk disease features, is still difficult to predict. This indicates that clinical and pathologic features alone are imprecise, and biologic signatures within clinical subgroups might offer a more reliable method for predicting outcome and assigning risk-based or targeted therapies(Davicioni et al., 2010).
In this study, we characterized the genomic, epigenomic, molecular and cellular features of rhabdomyosarcoma and integrated those data with drug sensitivity data to identify druggable pathways for this devastating pediatric cancer.
RESULTS
Sequencing of Rhabdomyosarcoma
We performed whole-genome sequencing (WGS) analysis on 16 rhabdomyosarcoma tumors from 13 patients and on matched normal tissue. Nine of the patients had ERMS, and 4 had ARMS. The distribution of patient age, sex, race, primary site, and stage reflected the clinical heterogeneity of the disease. We also sequenced 3 recurrent tumor samples from 2 ERMS patients (Table S1).
Using a paired-end–sequencing approach, we generated 4,529 Gb of sequence data for the samples described; 4,260 Gb (94%) were successfully mapped to the reference genome (Table S2). The average genome coverage was 42×, and the average exon coverage was 37×; 99% of SNPs detected across all 29 genomes showed concordance with their corresponding SNP array genotype calls at the same genomic positions (Table S2). We also performed RNA sequencing on 15 of the 16 tumors used for WGS (Table S2).
All somatic alterations including sequence mutations and structural variations (SVs) were experimentally validated by custom-capture technology and Illumina sequencing. We identified 22,123 validated somatic sequence mutations and 1,275 validated SVs across the 16 tumors (Table S2). These included 409 nonsilent tier-1 mutations in genes, 1,980 tier-2 mutations in regulatory regions or evolutionarily conserved regions of the genome, 19,202 tier-3 mutations in nonrepetitive regions of the genome that are not part of tiers 1 or 2 (Table S2). The average number of sequence mutations was 1,382.7 per case (range, 290-3,135), with 25.6 mutations per case (range, 5-65) resulting in amino acid changes (Table S2). The average number of validated SVs was 79.7 per case (range, 2-299). The estimated mean background mutation rate was 1.06×10−6 per base (range, 2.35×10−7–2.42×10−6). We also identified and validated 22 SVs that are predicted to produce an in-frame fusion protein in our discovery cohort (Table S2). RNA sequencing revealed that 8 of those predicted in-frame fusions were expressed in the tumors (Table S2). The majority (5/8) of the expressed in-frame fusion transcripts were the result of the PAX3/7-FOXO1 translocations in ARMS tumors. The other 3 transcripts were FGFR1- WHSC1L1 in SJRHB001, NSD1-ZNF346 in SJRHB011, and a 2.7 kb intra-genic deletion in COL4A2 in SJRHB002 (Table S2 and Figure S1A-S1C). These results show that despite an overall high number of SVs in ERMS cases, very few resulted in functional fusion genes.
Copy Number Variations and Structural Variations in Rhabdomyosarcoma
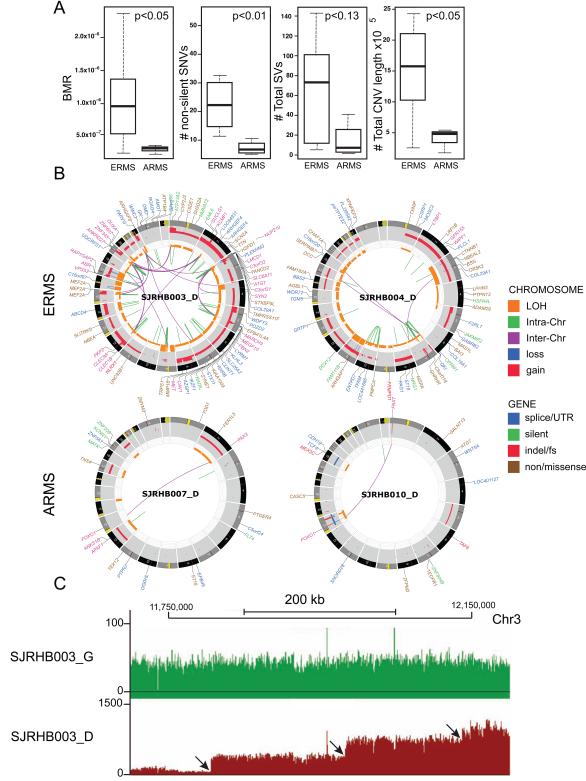
Previous aCGH studies have shown that ARMS tumors tend to have fewer copy number variants (CNVs) than do ERMS tumors(Williamson et al., 2010). We found a trend toward increased SVs and CNVs in ERMS, compared to those in ARMS, in our discovery cohort, but SV data did not achieve statistical significance because of the small number of ARMS tumors (Figures 1A and 1B). SVs and CNVs can be caused by the gradual, progressive accumulation of chromosomal and regional lesions during each round of cell division, or they may be caused by a single acute event such as chromothripsis (Stephens et al., 2011). We found no evidence of chromothripsis in the 16 rhabdomyosarcoma tumors in our discovery cohort (Data not shown, see Supplemental Experimental Procedures for details). Instead, the multi-level CNVs in genomic regions with multiple complex SVs suggested accumulation of sequential chromosomal lesions rather than a single acute event (Figure 1C).
Figure 1. The genomic landscape of embryonal rhabdomyosarcoma is distinct from that of alveolar rhabdomyosarcoma. (A).
Boxplots of validated BMR, number of non-silent SNVs, total SVs and number of total CNVs in the ERMS and ARMS tumors in the discovery cohort. (B) Representative CIRCOS plots of validated mutations and chromosomal lesions in two ERMS and two ARMS tumors in the discovery cohort. Loss of heterozygosity (orange), gain (red), and losses (blue) are shown. Intrachromosomal translocations (green lines) and interchromosomal translocations (purple lines) are indicated. Sequence mutations in Refseq genes included silent single nucleotide variants (SNVs, green), nonsense and missense SNVs (brown), splice-site and UTR mutations (dark blue), and insertion/deletion mutations (red). (C) Representative plot of sequence reads on chromosome 3 for the matched germline (green) and tumor (red) sample. Distinct regions of copy number change are indicated by arrows. For clarity, some of the gene names have been removed from the CIRCOS plots. In SJRHB003 and SJRHB004, the labels for gene disrupting SVs have been removed. See also Tables S1,2 and Figure S1.
The background mutation rate (BMR) and overall number of single-nucleotide variants (SNVs) was significantly higher in ERMS tumors than in ARMS tumors (Figure 1C). Mutation spectrum estimated using validated SNVs throughout the whole genome (Figures S1D and S1E) indicated that ERMS tumors had significantly more G→T SNVs than did ARMS tumors (26.7% vs 19.9%, p = 0.031), and the combined proportion of G→T transversions in rhabdomyosarcoma (24.9%) was higher than that found in T-cell acute lymphoblastic leukemia (T-ALL) and medulloblastoma (MB) (T-ALL + MB: 17.8%, p = 0.0003)(Robinson et al., 2012; Zhang et al., 2012) but lower than that found in lung cancer (32.7%, p=0.00014)(2012). Oxidative stress stimulated p38 MAPKs and consistent with this observation, MAKP12 expression was significantly higher in ERMS compared to ARMS (FDR = 0.02, 2.1-fold change). In addition, MAP2K6 was overexpressed in ERMS (FDR = 0.0003, 3.3-fold change). Taken together, these data are consistent with the hypothesis that oxidative stress contributes to the SNVs identified in rhabdomyosarcomas.
Intratumoral Heterogeneity and Clonal Evolution in ERMS Recurrence
As a first step to analyze intratumor heterogeneity in the discovery cohort, the tumor purity (ratio of tumor cells to all cells) was estimated from the WGS data. For each tumor, regions of the genome that had copy number alterations (CNAs) and corresponding changes in their loss of heterozygosity (LOH) were surveyed to identify the maximum proportional representation of a somatic lesion in the tumor. We were able to estimate the tumor purity in 15 of the 16 tumors (ranged from 67% to 98% in our discovery cohort, Figure S2C). Next, we analyzed intratumor heterogeneity using the purity adjusted mutant allele frequency (MAF) derived from deep sequencing of all SNVs by capture enrichment and Illumina sequencing. We excluded SJRHB013 in this analysis because of low tumor purity. The majority of tumors (13/15) had tumor purity adjusted MAF peaks corresponding to 0.5 indicating that the tumor purity estimates from CNAs and LOH analysis is accurate (Figure S2). Using somatic SNVs found in the diploid chromosomal regions, we found that there was a major MAF peak around 0.25 for 3 of the samples (SJRHB001, SJRHB009 and SJRHB004). These data suggest that there are relatively few mutations in the founding clone and a subclone representing ~50% of the tumor has the majority of SNVs. Overall, 10 of the 15 tumors had evidence of significant intratumor heterogeneity and two of the tumors (SJRHB004 and SJRHB011_D) had more than 2 subclones (Figure S2).
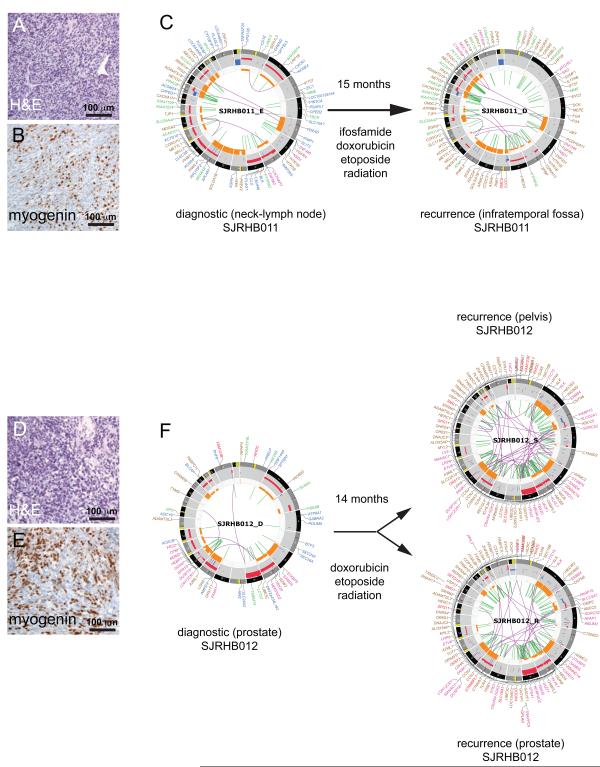
To characterize the clonal evolution(Nowell, 1976) of ERMS following treatment, we analyzed tumors from 2 patients who had diagnostic and recurrent tumors sequenced along with their matching normal germline genomes. In the case SJRHB011, a recurrent tumor sample (SJRHB011_D) was compared to the diagnostic tumor isolated 15 months earlier (SJRHB011_E) (Figure 2A-2C). In the case SJRHB012, tumor samples from 2 recurrent sites (SJRHB012_R and SJRHB012_S) that were collected at the same time were compared to the primary tumor (SJRHB012_D) isolated 14 months earlier (Figure 2D-2F). In both cases, the patients received chemotherapy and radiation prior to surgical resection of the recurrent tumors. We designed a single capture chip for each patient that included all tier 1-4 predicted mutations in primary and recurrent tumors from their corresponding WGS analysis. Deep sequencing of the captured DNA allowed us to calculate the MAFs for all SNVs across each of these 5 tumor samples. This approach allowed us to distinguish de novo SNVs that arose in the recurrent samples from mutations that were present at low frequencies in the primary tumor and were missed due to intratumor heterogeneity and/or limited coverage of the WGS.
Figure 2. Recurrent embryonal rhabdomyosarcomas acquire new mutations. (A, B).
Hematoxylin and Eosin (H&E) staining (A) and myogenin immunohistochemistry (IHC) (B) of a section of the diagnostic tumor from patient SJRHB011. (C) CIRCOS plots of validated sequence mutations and chromosomal lesion in the diagnostic tumor and the recurrent specimen. (D,E) H&E staining (D) and myogenin IHC (E) of a section of the diagnostic tumor from patient SJRHB012. CIRCOS plot of validated sequence mutations and chromosomal lesion in the diagnostic tumor and the recurrent specimen. Tumor from two different sites were collected at recurrence for this patient. CIRCOS plots are presented as in Figure 1B. In SJRHB011_D, the labels for gene disrupting SVs have been removed. In SJRHB012_R,S the labels for gene disrupting SVs, non-coding mutations and silent mutations have been removed. See also Fig. S2.
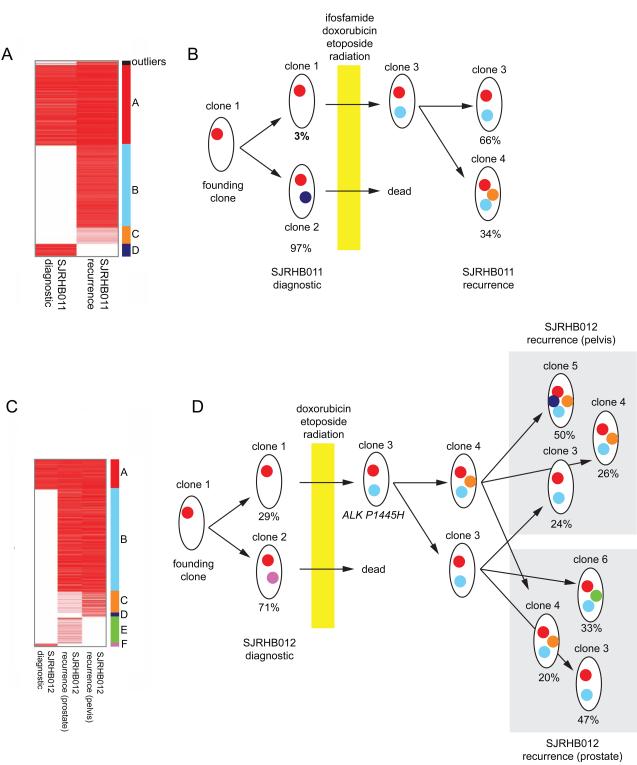
In SJRHB011, there were 3,524 SNVs with sufficient coverage for our analysis and we focused on 841 heterozygous mutations in diploid regions of the genome without LOH. There were 4 major clusters (A-D) (Figure 3A). Cluster A SNVs are found in cells that were present in the major clone in both primary (MAF=0.49) and recurrent (MAF=0.53) tumors. Cluster B SNVs are found in cells in the major clone of the recurrent tumor but not the primary tumor. Cluster C SNVs are present in the recurrent tumor in a subclone and Cluster D SNVs were present in the majority of cells in the primary tumor but were lost in the recurrent sample after chemotherapy (Figure 3A). At diagnosis, there was a major clone (clone 2) and a minor clone (clone 1) contributing 97% and 3% to the tumor, respectively (Figure 3B). Following treatment, the major clone (clone 2) was eliminated and some of the cells from the minor clone (clone 1) acquired additional SNVs (clone 3) to seed the recurrent tumor. This recurrent tumor then further evolved into two major clones (clone 3 and clone 4) contributing 66% and 34% to the tumor, respectively (Figure 3B).
Figure 3. Embryonal rhabdomyosarcomas clonally evolve following treatment. (A).
Heatmap of the 841 SNVs (MAF represented by the red intensity) used in the analysis of clonal evolution for SJRHB011. The 4 distinct clusters (A-D) are labeled with different colors on the right side of the heatmap. (B) Model of clonal evolution of SJRHB011. Clusters of SNVs are displayed as dots in colors corresponding to those shown in (A). (C) Heatmap of 1,049 SNVs (MAF represented by the red color intensity) used in the evolutionary analysis of SJRHB012. The 6 distinct clusters (A-F) are labeled with different colors on the right of the heatmap. (D) Clusters of SNVs are displayed as dots in colors corresponding to those shown in (A).
A similar analysis was performed for SJRHB012 using 1,049 validated SNVs in diploid regions of the genome without LOH. For this tumor, there were 6 clusters of SNVs (Figure 3C). Cluster A SNVs are present in all 3 tumor samples (diagnostic and two recurrent sites) from this patient (Figure 3C,3D). Cluster B SNVs were present in virtually all tumor cells at both recurrent sites but absent from the primary tumor (Figure 3D). These mutations were likely acquired early during progression prior to spreading to the secondary sites. An ALK mutation (P1445H) is one of the mutations in this cluster. Cluster C SNVs were absent from the diagnostic tumor but were present at one site of recurrence (pelvis) as the dominant clone and at the other site (prostate) as a minor clone (Figure 3D). Cluster D and Cluster E SNVs were unique to the pelvic or prostate recurrent sites, respectively. Cluster F SNVs were found in the dominant clone in the primary tumor but were lost following treatment (Figure 3D).
Similar to SJRHB011, the recurrent tumors in SJRHB012 were derived from the founding minor clone (clone 1) after acquiring additional mutations, including the ALKP1445H mutation (clone 3). A fraction of the clone 3 cells acquired additional mutations and became clone 4. Both clones 3 and 4 seeded the 2 recurrent tumor sites and continued to evolve (clone 5 and 6). Taken together, these data highlight the complex genetic changes and clonal evolution that occur in ERMS tumors following treatment.
SJRHB011, 012 and 013 Xenografts
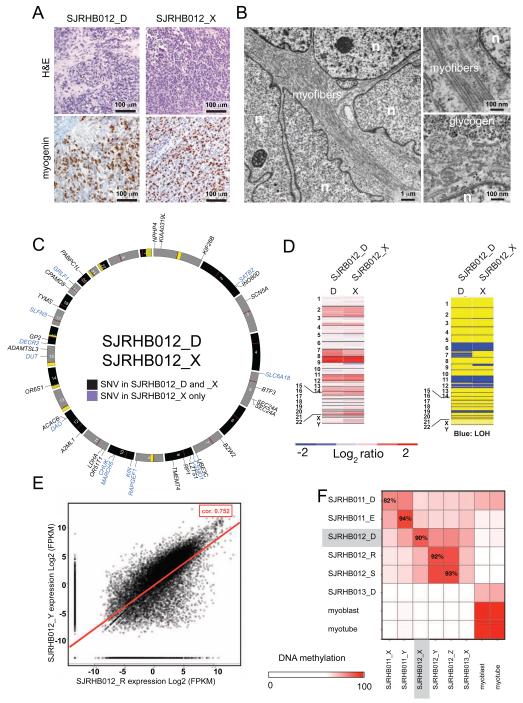
To begin to study ALK and other signaling pathways that may be perturbed in primary and recurrent ERMS, we developed and characterized 6 orthotopic xenografts derived from the diagnostic and recurrent samples for SJRHB011 and SJRHB012 and from the post-treatment SJRHB013 tumor specimen. Immunohistochemistry and transmission electron microscopy confirmed that the cellular features of the patient’s tumors were preserved in the xenografts in immunocompromised mice (Figure 4A,4B and Figure S3A-S3F). SNP 6.0 analysis, exome sequencing, RNA-seq and DNA methylation analysis confirmed that these xenografts closely recapitulate the molecular features of their primary tumors (Figure 4C-4F and Figure S3A-S3F). There was no correlation between acquisition of the ALKP1445H mutation and ALK protein levels in SJRHB012 (Figure S3F, S3G). To test the sensitivity of these ERMS xenograft tumor cells to ALK inhibitors and other therapeutics, we developed and validated a short-term culture protocol that was amenable to high-throughput screening. There was no relationship between ALK mutation status and sensitivity to ALK inhibitors for SJRHB012 or the other ERMS xenografts (SJRHB011 and SJRHB013) (Figure S3H).
Figure 4. Orthotopic xenografts retain molecular and cellular features of the patient’s tumor. (A).
Representative H&E and myogenin immunohistochemistry from a primary tumor (SJRHB012_D) and the corresponding xenograft (SJRHB012_X). (B) Transmission electron micrographs of SJRHB012_X showing features of rhabdomyosarcoma including myofibers and glycogen. Nuclei (n) are indicated. (C) Circos plot of exonic SNVs for the SJRHB012_D/SJRHB012_X pair. Gene names in black contain SNVs found in the primary and xenograft samples and those shown in blue are unique to the xenograft. (D) SNP 6.0 analysis of copy number changes (left) and LOH (right) for the matched primary and xenograft samples with red showing gain and blue showing loss for copy number and blue showing LOH for the lower panel. (E) Correlation analysis of the RNA-seq data for a representative primary tumor and xenograft pair with a coefficient of 0.752 for this pair (red line). (F) Heatmap of DNA methylation analysis for the matched diagnostic and xenograft pairs. See also Fig. S3.
Recurrent Mutations in Genes Implicated in Muscle Development and Homeostasis
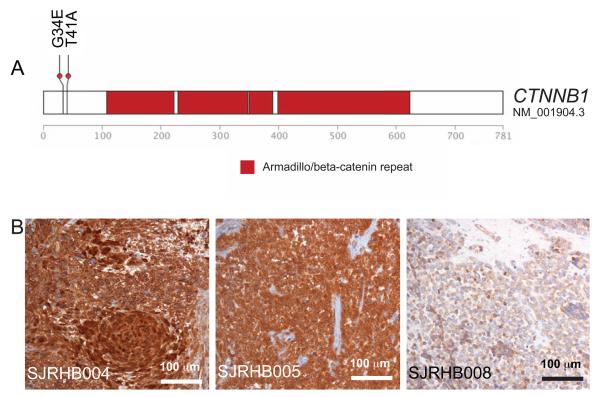
It has been proposed that rhabdomyosarcomas may arise from multipotent mesenchymal progenitor cells, muscle progenitor cells, or muscle stem cells (satellite cells) (Hettmer and Wagers, 2010). These possibilities are not mutually exclusive, and it is possible that ERMS and ARMS may have distinct cellular origins. To explore muscle-differentiation pathways in ERMS and ARMS, we performed network analysis, as previously described (Hu et al., 2012; Zhang and Horvath, 2005), using published gene expression data for ERMS and ARMS tumors (Davicioni et al., 2010; Williamson et al., 2010). The WNT and sonic hedgehog (SHH) pathways differed significantly between the two rhabdomyosarcoma subtypes. To validate and extend those data, we extended our RNA-Seq analysis to 32 tumors including the 15 samples from the discovery cohort described above. We also performed DNA methylation analysis using the Illumina 450k BeadChip on 32 tumors (20 ERMS and 12 ARMS) (Figure S4A and Table S3). Several genes in the WNT and SHH pathways were differentially expressed and a subset of those were differentially methylated (Table S4). We did not identify any recurrent gene mutations in the SHH pathway in our discovery cohort, but we did identify β-catenin–activating mutations in the WNT pathway in SJRHB004 and SJRHB005 (Figure 5A). Tumors with β-catenin activating mutations accumulated nuclear β-catenin protein in contrast to those that expressed wild-type β-catenin (Figure 5B). We extended our immunohistochemical analysis of β-catenin and found that 20% (6/30) of ERMS had some evidence of nuclear localization and 0% (0/17) of ARMS had evidence of nuclear β-catenin (Table S1). The nuclear β-catenin expression was heterogeneous, ranging from rare cells to extensive (3+) staining within the tumor tissue (Table S1).
Figure 5. Embryonal rhabdomyosarcomas have WNT mutations. (A).
Activating mutations in the β-catenin gene in SJRHB004 and SJRHB005 in the discovery cohort. (B) Immunohistochemistry of β-catenin for SJRHB004, SJRHB005 and an alveolar rhabdomyosarcoma, SJRHB008. See also Fig. S4 and Tables S3-S6.
To estimate the frequency of mutations identified in our discovery cohort, we performed targeted resequencing of all exons for 139 of those genes with mutations in an additional cohort comprising 21 ERMS, 13 ARMS, and 3 unspecified tumors (Table S5). Approximately half of the tumors had no mutations in any of the 139 genes, so we performed exome sequencing on a subset (28/36) of the tumors (Table S5). All mutations identified in this subset were validated using nextgen amplicon sequencing.
To determine if any of the muscle-specification or -differentiation pathways were mutated in rhabdomyosarcomas, we analyzed the distribution of mutations in genes that are enriched in developing muscle compared to other human tissues. We selected all genes that were enriched by at least 4 fold in 1 of the 3 cell populations (myoblast, myotube, or skeletal muscle) for a total of 1,295 genes. We identified 130 mutations across 101 genes from our group of muscle-enriched genes in the discovery and validation cohorts. Nine of those genes (ANKRD10, COL4A3, DMD, KLHL4, LTBP1, MIB1, MYOD1, P2RX6, and SYNE2) were mutated in 2 independent tumor samples, and TTN was mutated in 3 samples (Table S6). Four genes (CTNNA3, MACF1, MAP3K5, and MEF2A) had 2 different mutations in the same patient’s tumor (Table S6). Overall, 70% (19/27) of the ERMS and 44% (8/18) of the ARMS had a mutation in at least 1 of the 1,295 muscle-enriched genes in our analysis, but we detected no statistically significant enrichment in mutations in the muscle-development/differentiation pathways. Taken together, these data on the mutational landscape of ERMS and ARMS do not provide any additional evidence about cellular origins for these tumors.
The most common chromosomal alteration in rhabdomyosarcoma is the gain of chromosome 8 in ERMS tumors(Williamson et al., 2010). We analyzed the gene expression in ERMS tumors with chromosome 8 gains in comparison to those without chromosome 8 gains. Overall, there was a slight increase in expression of genes on chromosome 8 in the ERMS tumors with chromosomal gains (Figure S4B). However, none of the individual genes on chromosome 8 were significantly upregualted in the tumors with chromosome 8 gain. There were 7 genes on other chromosomes with significantly different expression (FDR≤ 0.05) in the ERMS tumors with chromosome 8 gains compared to those without chromosome 8 gains. Among those genes, MICU1, MCU, MAMDC2 and ISL1 were altered by ≥ 2-fold (Figure S4C). MCU is a calcium uniporter in mitochrondria and MICU1 is an EF-hand protein that is the key regulator of MCU. Both MCU and MICU1 are downregulated in ERMS tumors with chromosome 8 gains. Previous studies have suggested that perturbations in MCU/MICU1 expression may lead to changes in oxidative metabolism and sensitization to calcium overload (Csordas et al., 2013). These data may particularly relevant to rhabdomyosarcoma because of the features of muscle differentiation in this tumor and the role of calcium in muscle contraction.
Recurrent Gene Mutations in Cancer Consensus Pathways
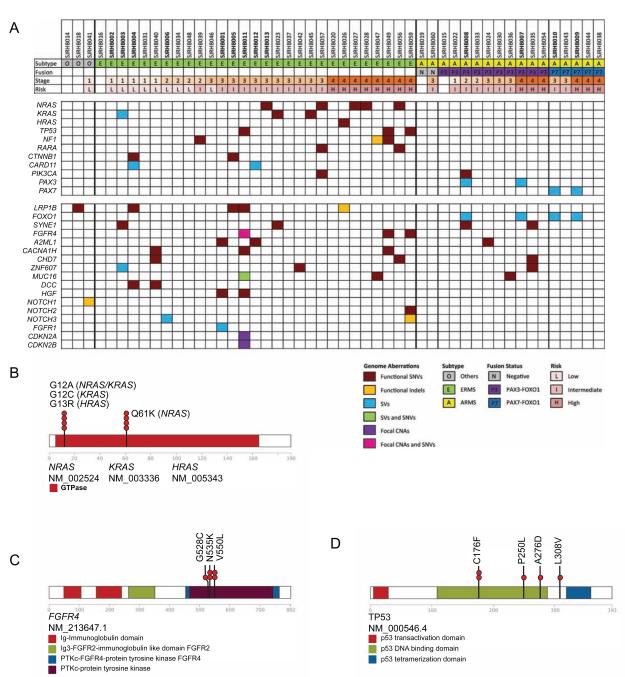
Combined sequence analyses demonstrated recurrent genetic lesions (SNVs, indels, and SVs) in 8 cancer consensus genes (NRAS, KRAS, TP53, NF1, RARA, CTNNB1, CARD11, and PIK3CA), as well as the expected PAX3/7-FOXO1 translocations in ARMS tumors (Figure 6A). One ERMS tumor (SJRHB026) also had an oncogenic HRAS mutation. All but one of the cancer consensus gene mutations were found in ERMS, and 15% (4/27) of ERMS had multiple cancer consensus gene mutations (Figure 6A). Cancer consensus gene mutations were validated in 88% (7/8) of the high-risk ERMS tumors, 73% (7/11) of the intermediate-risk, and 20% (2/10) of the low-risk. The most common cancer consensus gene mutations were in the RAS pathway (NRAS, KRAS, HRAS, and NF1) (Figure 6A,6B and Table 1). When we combined the discovery and validation cohorts, 75% (6/8) of high-risk ERMS tumors had RAS pathway mutations, 45% (5/11) of intermediate-risk, and 0% (0/10) of low-risk (Table 1); thus, RAS pathway mutations had a significant association with ERMS risk group classification (p=0.00015). KRAS, HRAS, NRAS, and NF1 gene mutations were rarely found in combination with other cancer consensus gene mutations, and they were never found in ARMS.
Figure 6. The RAS and p53 pathways are recurrently mutated in embryonal rhabdomyosarcomas. (A).
Summary of mutations in cancer consensus genes and recurrent mutations in non-cancer consensus genes in the discovery (bold) and validation cohorts. Tumor samples are organized by histological subtype and stage. (B) Distribution of oncogenic mutations in NRAS, KRAS and HRAS. (C,D) Distribution of missense mutations in FGFR4 (C) and TP53 (D). See also Fig. S5 and Tables S7,S8.
Table 1.
RAS, NF1, TP53 and FGFR4 mutations in embryonal rhabdomyosarcoma.
| Sample # | 1Risk | NRAS | KRAS | HRAS | NF1 | TP53 | FGFR4 |
|---|---|---|---|---|---|---|---|
| SJRHB013 | I | Q61K/WT | |||||
| SJRHB027 | H | Q61K/WT | |||||
| SJRHB028 | H | Q61K/WT | |||||
| SJRHB056 | H | G12A/WT | |||||
| SJRHB057 | I | Q61K/WT | |||||
| SJRHB023 | I | G12C/WT | |||||
| SJRHB045 | I | G12A/WT | |||||
| SJRHB026 | H | G13R/WT | |||||
| SJRHB047 | H | C1939_N1942fs/WT | |||||
| SJRHB039 | I | L1855I/WT | |||||
| SJRHB049 | H | 4W784C | 4A276D | 4V550L | |||
| SJRHB011 | I | C176F/ Δ | N535K/WT | ||||
| SJRHB059 | H |
2P250L+ L308V |
3G528C+ V550L |
I, intermediate risk, H, high risk group.
Data from exome capture, there were relatively few reads for the corresponding wild type nucleotide suggesting deletion or copy neutral LOH.
Data from exome capture, there were relatively few reads for the corresponding wild type nucleotide suggesting deletion or copy neutral LOH.
Data from exome capture, the wild type allele was not present possibly reflecting deletion or copy neutral LOH.
In our cohort, TP53 gene mutations were accompanied by FGFR4 mutations (Figure 6A, 6C, 6D and Table 1). In addition to TP53 gene mutations, we also performed detailed analysis of CNVs for TP53, MDM2, MDM4 and CDKN2A using the WGS and SNP 6.0 data and combined those data with RNA-sequence data, p53 IHC and fluorescence in situ hybridization (FISH) analysis of all 4 loci (Table S7 and Figure S5A). There were 4 ERMS samples (SJRHB003, SJRHB011, SJRHB049 and SJRHB059) with SNVs in TP53, and all 4 had elevated nuclear accumulation of p53 protein (Figure S5B). One ERMS tumors (SJRHB016) had a homozygous deletion of TP53 with reduced gene expression (Table S7). One of the ERMS tumors (SJRHB012) and one of the ARMS tumors (SJRHB054) had focal MDM2 gains (>100 copies and 14 copies, respectively) with increased MDM2 gene expression and one of the ERMS tumors (SJRHB020) had focal homozygous deletion of CDKN2A with reduced gene expression (Table S7 and Fig. S5). There was one ERMS sample (SJRHB002) with a gain of MDM4 (3-5 copies in 33% of the cells) (Figure S5C) but there was no increase in gene expression (Table S7). Overall, by combining WGS, exome sequencing, SNP 6.0 analysis, transcriptome analysis, p53 IHC and FISH analysis, mutations in the p53 pathway were more common in ERMS (8/31) than ARMS (1/14). There were no mutations in TP53 or CDKN2A in our ARMS cohort.
Previous gene expression array analysis of ERMS and ARMS tumors led to the identification of a 34 metagene expression signature that is predictive of overall survival for rhabdomyosarcoma(Davicioni et al., 2010). We used RNA sequence data for 32 tumors from the discovery and validation cohorts to rank the tumors based on their 34 metagene signature (Table S8). There was no significant association of RAS pathway mutations with 34-metagene rank in our cohort and a larger study will be required to establish the prognostic significance of RAS pathway mutations in intermediate risk ERMS patients.
Targeting Oxidative Stress and Ras Pathways in ERMS
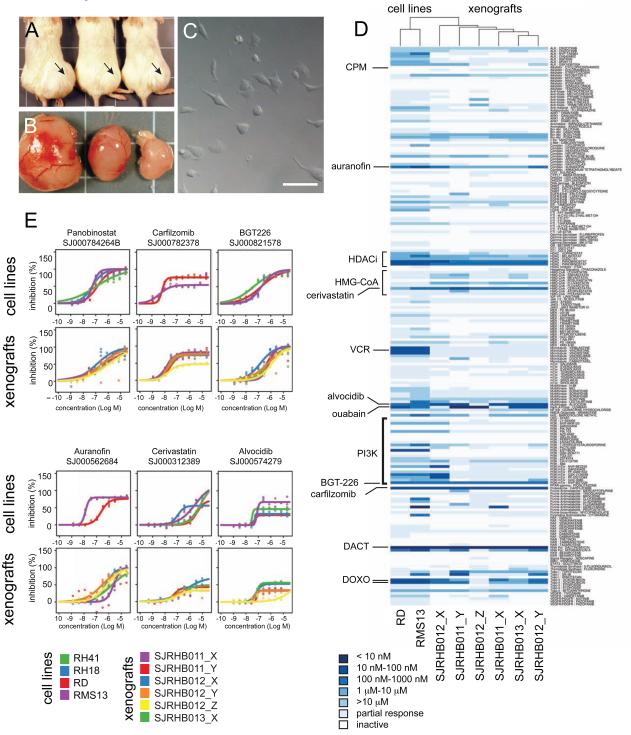
Skeletal muscle cells have unique energy metabolism because of their aerobic capacity and ability to rapidly adapt for short-term anaerobic activity. As a result of their unique metabolic properties, muscle cells also have a robust antioxidant defense system to protect the DNA, lipids and proteins from the deleterious effects of excess reactive oxygen species (ROS). Cancer cells also have elevated ROS due to their increased metabolic activity, oncogenic stimulation (i.e. RAS) and mitochondrial dysfunction. Therefore, we reasoned that rhabdomyosarcomas may be particularly susceptible to therapeutics that increase ROS or target the cells ability to protect against oxidative stress. In addition, the association of RAS pathway mutations with ERMS risk group provided an additional pathway for interventions using molecular targeted therapy. To test the efficacy of therapeutics that target these two pathways, we developed a method to perform acute short-term cultures with the primary rhabdomyosarcoma orthotopic xenografts described above. Briefly, the tumor cells are injected into the flank muscle of immunocompromised mice in matrigel and several weeks later when the tumor mass is palpable, they are isolated and dispersed into a single cell suspension (Figure 7A-7C). The cells can be grown for up to 96 hours in muscle differentiation medium in 384 well dishes with reproducible survival and growth kinetics. Next, we developed a custom compound library with 207 compounds including FDA approved drugs, molecules in clinical development and well-characterized small molecule with biological activity (Table S9). The library included chemotherapeutics used to treat pediatric cancer, agents that perturb the oxidative stress pathway and molecules that target the Ras pathway. The library was screened in dose response in triplicate against each of the 6 xenografts and several rhabdomyosarcoma cell lines (Table S9). As a positive control, we used staurosporine and as a negative control we used DMSO at the same concentration in the drug treated wells. There was robust separation between the positive and negative controls and the mean z’ was between 0.48 and 0.81 across the experiments (Table S10 and Figure S6A, S6B). Overall, the xenografts were less sensitive to the agents in the targeted library than the cell lines (Figure 7D). None of the drugs that target the Ras pathway had significant activity against the xenografts including the tumor that had an NRAS mutation (SJRHB013). We also included molecules that target the PI3K pathway because of the interplay between the Ras and PI3K pathways(Gysin et al., 2011). None of these agents had significant activity except for the dual PI3K and mTOR inhibitor BGT-226 (Figure 7D,7E). There are now several clinical trials testing the combination of PI3K inhibitors and Ras pathway inhibitors so we tested the combination of BGT-226 with all of the drugs in our library. Even these combinations failed to show significant increase in cytotoxicity for our ERMS xenografts (Table S9). These data are consistent with phosphoprotein analysis for the Ras and PI3K pathways (Figure S6C, S6D) showing little if any deregulation of those pathways in our xenografts.
Figure 7. Embryonal rhabdomyosarcoma xenografts are sensitive to drugs that target oxidative stress. (A).
Pictures of orthotopic xenograft of SJRHB012_X in the muscle of NSG immunocompromised mice. (B) Tumors isolated from the corresponding mice shown in (a). (C) Differential interference contrast micrograph of primary SJRHB012_X cells in a 384 well dish for drug screening. (D) Heatmap and unsupervised clustering of drug sensitivity for two rhabdomyosarcoma cell lines (RD and RMS13) and the 6 xenografts characterized in this study. (E) Dose response curves of cell lines and xenografts to some of the compounds investigated. Abbreviations: CPM, cyclophosphamide; HDACi, histone deacetylase inhibitors; VCR, (E) vincristine; DACT, actinomycin-D; DOXO, doxorubicin. Scale bar in c, 10 μm. See also Fig. S6 and Tables S9, S10.
In contrast to compounds targeting Ras and PI3K pathways, the agents that targeted oxidative stress were more active. The HDAC inhibitors were active as a class with panobinostat showing the most activity (Figure 7D, 7E). In addition, carfilzomib, auranofin, cerivastatin, alvocidib and ouabain showed significant activity against the ERMS xenografts. Each of these drugs can increase oxidative stress and several have been shown to be synergistic by targeting oxidative stress and inducing mitochondrial cell death (discussed below). Taken together, these data suggest that drugs that increase oxidative stress and ROS production in ERMS cells may be effective for the treatment of diagnostic and recurrent ERMS tumors.
DISCUSSION
Genetic Landscape of Rhabdomyosarcoma
It has been previously shown that chromosome 8 was gained in 74% of ERMS tumors(Williamson et al., 2010). We identified chromosome 8 gains in 92% (11/12) of the ERMS tumors and we found that two genes on chromosome 10 that control calcium uptake in mitochondria (MCU, MICU1) are downregulated in the tumors with chromosome 8 gain. This may indicate that there is a negative regulator of MCU and MICU1 on chromosome 8. In a separate aCGH study of 26 ERMS tumors, CDKN2A-homozygous deletions were found in 23% (6/26) of tumors and heterozygous deletion through loss of chromosome 9p in most of the remaining tumors (Paulson et al., 2011). In our cohort, we identified 1 ERMS tumor showing CDKN2A-homozygous loss (8%), and 1 other ERMS tumor showing a heterozygous deletion. We did not find any CDKN2A mutations in ARMS tumors. We also analyzed the other key genes in the p53 pathway including TP53, MDM4 and MDM2. In total, mutations in the p53 pathway were identified in 19% of the ERMS tumors (6/31) and 7% (1/14) of ARMS tumors. This difference between the 2 studies may be due, in part, to the larger number of low-risk ERMS tumors in our study.
Paulson et al. also reported patterns of activation of the RAS pathway by oncogenic mutation in HRAS, KRAS, or NRAS (42%, 12/26) or by homozygous deletions of NF1 (15%, 4/26)15. These data are consistent with that from 3 other studies showing RAS pathway mutations in 35% (5/14), 22% (7/31)(Martinelli et al., 2009; Stratton et al., 1989) and 11.7%(Shukla et al., 2012) of ERMS tumors. In our study, 58% (11/19) of the high-risk and intermediate-risk ERMS tumors had RAS pathway mutations, and RAS pathway mutations were significantly associated with risk group assignment (p=0.0015). None of the ARMS tumors in our discovery or validation cohorts (17 specimens) had RAS pathway mutations or TP53/FGFR4 gene mutations. A much larger cohort of ERMS tumors will be needed to determine the incidence and significance of RAS pathway mutations with respect to overall survival.
The only cancer consensus gene that was mutated in our ARMS cohort was PIKC3A in 1 sample (SJRHB008). There was also one ERMS tumor with a PIKC3A mutation (SJRHB057). One of the mutations (H1047R) is a hotspot mutation and the other (N345K) is a rare mutation that has been shown to be oncogenic in cell culture(Gymnopoulos et al., 2007). These data suggest that multiple cancer consensus pathways are mutated in ERMS and the PAX3/7-FOXO1 translocation is the major oncogenic driver in ARMS.
In a previous study comparing genetically engineered mouse models of rhabdomyosarcoma to primary human tumors, Keller and colleagues analyzed the p53, SHH, RB and RAS pathways by comparing their gene expression signatures (Rubin et al., 2011). They concluded that fusion negative rhabdomyosarcomas (ERMS) with a RAS signature always had perturbations in at least one other pathway (SHH, RB or P53). No other genetic or histological data was presented in that study to confirm the findings. This is important because signaling through these pathways is complex and using a metagene approach may not accurately identify pathway perturbations. This is particularly true for the Ras pathway that is regulated primarily at the level of protein phosphorylation rather than gene expression.
In our characterization of the genomic landscape of rhabdomyosarcoma, we did not find significant overlap of mutations in the p53, Shh, Rb and Ras pathways. However, we cannot rule out the possibility of non-genetic factors contributing to pathway perturbations and more comprehensive integration of genetic and epigenetic profiles will help to resolve this discrepancy. Indeed, our data suggest that several genes that regulate the RAS pathway or similar receptor tyrosine kinase pathways are differentially methylated in rhabdomyosarcomas including THEM4, DAB2, KSR1, ELMO1, SH3D19, NCK2 and JAK1.
Developmental Pathways in Rhabdomyosarcoma
Rhabdomyosarcomas express multiple genes and pathways characteristic of muscle cell differentiation, and electron microscopic analysis of ERMS and ARMS tumors has demonstrated that tumor cells have features of skeletal muscle including myofibers (Skalli et al., 1988). Our analysis of previously published gene-expression array data revealed that expression of genes in the SHH and WNT pathways differ significantly between ERMS and ARMS tumors and our RNA-Seq and DNA methylation analysis provided additional validation. No mutations in the hedgehog pathway were found in our cohort but we identified β-catenin–activating mutations in 7% (2/29) of ERMS tumors, and nuclear accumulation of β-catenin in 20% (6/30). In a recent study(Shukla et al., 2012), CTNNB1 mutations were identified in 2/60 ERMS tumors but there was no validation of nuclear accumulation of β-catenin. The nuclear accumulation of β-catenin was heterogeneous and in tumor sample SJRHB004, the β-catenin–activating mutation was in a minor clone, and in SJRHB005, it was present in the major clone. There was no evidence of activation of WNT signaling in ARMS tumors in our cohort. Previous studies in mice have shown that β-catenin plays an important developmental stage–specific role in muscle cell development, and activation of β-catenin can lead to ectopic proliferation(Hutcheson et al., 2009). Activation of β-catenin is not sufficient to induce tumorigenesis of muscle progenitors or differentiation of muscle cells in mice, but in combination with other mutations, it may contribute to tumorigenesis in human rhabdomyosarcoma. Indeed, in our cohort, β-catenin– activating mutations were found in combination with LRP1B mutations.
Beyond SHH and WNT signaling we identified 101 muscle-enriched genes mutated in rhabdomyosarcoma. Among those 101 genes, only 8 were recurrently mutated in at least 2 tumor samples, and 1 (TTN) was recurrently mutated in 3 samples. The TTN gene is one of the largest genes in the genome, and it is mutated in most pediatric solid tumors; thus, it is difficult to assess the functional significance of TTN mutations in rhabdomyosarcoma. A much larger study will be required to elucidate the significance of mutations in genes implicated in muscle development or differentiation. Our DNA methylation data also provided additional insight into muscle development pathways that may be deregulated in rhabdomyosarcoma including LBX1, FOXK1, ZFHX3, MEF2D, HOXD3, ZFP42, DLK2, MYF5 and FHL3.
Intratumor heterogeneity and Clonal Evolution of Rhabdomyosarcoma
We identified significant intratumor heterogeneity in 10/15 of the tumors and 2 of them had multiple clones. Interestingly, 3 of the tumors had a founding clone with relatively few SNVs and the majority of SNVs in the tumor were present in a clone that grew out of that clone. There was no difference between ARMS and ERMS tumors with respect to tumor heterogeneity.
We included two patients in our discovery cohort that had diagnostic and recurrent tumors to begin to explore the clonal evolution(Nowell, 1976) of ERMS after treatment. This is of particular interest because the overall survival for patients with recurrent rhabdomyosarcoma is less than 30% and nothing is known about how the tumors evolve in the context of current standard of care therapy. Our analysis suggests that chemotherapy can completely eliminate the major clone in a diagnostic tumor and a minor subclone can then seed the recurrent tumor following therapy and continue to acquire mutations. Thus, not only are the two ERMS tumors in our analysis complex with respect to tumor heterogeneity at diagnosis, but they are significantly different at the time of recurrence and at individual recurrent sites. These data highlight the importance of performing comprehensive genomic analyses of diagnostic and recurrent tissue specimens from multiple sites for rhabdomyosarcoma in order to identify the genetic lesions that contribute to progression and resistance to therapy and to more effectively identify therapeutic approaches for those recurrent rhabdomyosarcoma patients. As more targeted agents become available, biopsy of the recurrent tumor may become an important intervention for selection of therapy since the mutations present at recurrence may differ from those present at initial diagnosis.
Implications for Preclinical Models of ERMS and ARMS
To model ARMS in the mouse, Keller et al. generated a conditional Pax3-Foxo1 knock-in mouse line that could be induced to express the Pax3-Foxo1 fusion protein with cell type–specific Cre expression (Keller et al., 2004). However, expression of the fusion protein was not sufficient to promote ARMS formation, and only when combined with conditional inactivation of Trp53 or Cdkn2a did the mice develop ARMS. In our study, none of the ARMS tumors had TP53 or CDKN2A mutations. Indeed, only one of the 14 ARMS samples in our cohort had a lesion in the p53 pathway (MDM2 amplification). It is not known if this species difference reflects a divergence in the cellular origins of the tumor, differences in the epigenetic landscape of the cell of origin for ARMS across species, or some other cell autonomous or non–cell autonomous difference that affects tumor initiation or progression. Our data suggest that conditional activation of the RAS pathway will prove useful for modeling intermediate- or high-risk ERMS in the mouse, and inactivation of the Trp53 gene in combination with an activating mutation in Fgfr4 may provide another model of intermediate- or high-risk ERMS.
Targeting the Oxidative Stress Pathway in ERMS
Skeletal muscle cells and cancer cells have high levels of ROS because of their unique metabolic demands. Therefore, we reasoned that a cancer with features of skeletal muscle (rhabdomyosarcoma) may have even higher levels of ROS than other cancer cells and be particularly sensitive to therapeutics that increase oxidative stress. Indeed, one of the most active agents for the treatment of rhabdomyosarcoma (actinomycin-D) increases oxidative stress(Minai et al., 2013; Tsuruga et al., 2003). The genomic data presented here suggests that ERMS tumors have elevated ROS because they have higher rates of G→T transversions and some increase in expression of genes in the p38 MAPK pathway(Wagner and Nebreda, 2009). Also, the majority of ERMS tumors have chromosome 8 gains (92% in our cohort) and show deregulation of MCU/MICU1 expression which in turn can lead to mitochondrial dysfunction and oxidative stress(Block and Gorin, 2012). Several genes implicated in regulation of metabolism, mitochondrial function and oxidative stress were differentially methylated in rhabdomyosarcomas including PTK2, COX7A1, NOSIP, NOS1, ATP2A3, DDAH1, GLRX and TXNDC12. In our study, the ERMS xenografts are sensitive to the thioredoxin reductase inhibitor auranofin (Liu et al., 2012). The thioredoxin pathway is regulated by epigenetic processes controlled in part by HDACs and one of the cytotoxic mechanisms of HDAC inhibitors is perturbations in the expression of the thioredoxin pathway(Butler et al., 2002). Our data on the activity of HDAC inhibitors for the ERMS xenografts are consistent with this mechanism of action and the susceptibility of the oxidative stress pathway in these tumors. Two other drugs that show activity against the ERMS xenografts (carfilzomib and alvocidib) act synergistically with HDAC inhibitors by targeting mitochondrial function and increasing oxidative stress in cancer cells (Dasmahapatra et al., 2006; Dasmahapatra et al., 2010; Dasmahapatra et al., 2011; Huang et al., 2010). Indeed, one of the major mechanisms of action of the HDAC inhibitors is through changes in expression of thethoredoxin reductase pathway and our data on HDAC inhibitors as a class is consistent with this hypothesis. Cervistatin is a synthetic statin used to lower cholesterol and prevent cardiovascular disease. In cardiac myocytes, it is believed that statins reduce oxidative stress. However, it has been shown that this effect is cell-type specific and in skeletal muscle, statins can have the opposite effect and increase oxidative stress contributing to rhabdomyolysis which is a major side effect of statins(Bouitbir et al., 2012). Cervistatin was withdrawn from the market due to the high rate of deaths and other side effects related to rhabdomyolysis. Finally, ouabain is a Na+/K+ ATPase inhibitor that can have pleiotropic effects in cells. In myocytes, ouabain can lead to opening of ATP-sensitive mitochondrial potassium channels and concomitant increase in ROS (Tian et al., 2003). Taken together, these genomic, molecular and cellular data suggest that therapeutics that increase reactive oxygen in rhabdomyosarcoma may be particularly effective and may contribute synergistically to current standard of care using vincristine, actinomycin and cyclophosphamide (VAC). The synergistic effects of carfilzomib or alvocidib with HDAC inhibitors is particularly promising.
EXPERIMENTAL PROCEDURES
Full details of sample acquisition, molecular and biochemical procedures, informatics and whole genome sequencing are provided in the Supplementary Information. 48 of the tumors in this study were from St. Jude Children’s Research Hospital (SJCRH) patients and 5 were obtained from the Nationwide biorepository. The SJCRH Institutional Review Board approved experiments involving human subjects and informed consent was obtained from all subjects. The SJCRH Institutional Animal Care and Use Committee approved all experiments involving animals.
Rhabdomyosarcoma Xenografts
Excess, deidentified tumor material was collected from patients with rhabdomyosarcoma at St. Jude Children’s Research Hospital in agreement with local institutional ethical regulations and IRB approval. Tumor tissue was initially implanted in the flank location of NSG mice. After sufficient tumor growth, orthotopic xenografts were created by processing the flank tumor tissue into a single cell suspension by enzymatic dissociation and injection into the hind leg muscle of CD-1 nude mice. Tumor growth was monitored by manual palpation. After tumor development, the tumor was harvested and processed using the same dissociation technique to isolate cells for drug screening.
Orthotopic Xenograft Intramuscular Injection
Rhabdomyosarcoma cells were suspended in matrigel (BD Worldwide Cat#354234) at a concentration of 1×104 cells per microliter and placed on ice. Recipient CD-1 nude mice were manually restrained and injected with 100 μl of cell suspension intramuscularly into the lateral thigh region of the hind leg.
Rhabdomyosarcoma Tumor Dissociation
Rhabdomyosarcoma tumor tissue was harvested from CD-1 nude orthotopic xenografts. Tumor was placed through a tumor press and then rinsed with Dulbecco’s modified eagle media (DMEM, Lonza Cat#12-604F). Tumor suspension was transferred to a 50 ml conical tube and filled with DMEM. Dissociation was done by adding 600 μl of trypsin (10 mg/ml, Sigma Cat#T9935) and 50 mg of type II collagenase (275 u/mg,Worthington Cat#4177) and tube placed in 37 degree water bath for 10 minutes. Dissociation was stopped by adding 600 μl of soybean trypsin inhibitor (10 mg/ml, Sigma Cat#T6522). Deoxyribonuclease I (2 mg/ml, Sigma Cat#D4513) and magnesium chloride (1 M) were added in equal amounts in 60 μl increments until tumor fragments easily settled at the bottom of the tube. Tumor suspension is filtered with a 40 μm cell strainer and centrifuged at 450g (G=RCF) for 5 minutes. Supernatant is discarded and 10 ml of red blood cell lysis solution (5 Prime Cat#2301310) added and allowed to incubate at room temperature for 10 minutes. Solution of phosphate buffered saline without calcium or magnesium (PBS-minus, Lonza Cat#17-516F) /10% Fetal Bovine Serum (FBS, Biowest Cat#SO1520) was added to fill 50 ml conical tube and cell suspension centrifuged at 450g (G=RCF) for 5 minutes. Supernatant is discarded and cell pellet was resuspended in PBS-minus/10%FBS for counting.
Supplementary Material
Highlights.
There are higher rates of mutation in ERMS than in ARMS tumors.
RAS pathway mutations are associated with intermediate- and high-risk ERMS.
ERMS tumor cells have elevated oxidative stress.
ERMS tumors are sensitive to drugs that target oxidative stress.
SIGNIFICANCE.
Our data show that the genetic landscape of ERMS and ARMS are distinct. ARMS have fewer mutations overall and no recurrent cancer consensus gene mutations. In contrast, ERMS have a high rate of recurrent mutations in the RAS pathway. We selected six ERMS patient tumors to generate orthotopic xenografts and optimized a culture system for high-throughput screening of primary xenograft tumor cells to test their drug sensitivity. None of the molecular targeted therapeutics for the RAS pathway had any significant activity but the oxidative stress pathway is a promising cellular target for primary and recurrent ERMS. These data suggest that cell biology studies may be combined with genomic analyses to identify druggable pathways for rhabdomyosarcoma.
ACKNOWLEDGMENTS
We would like to thank S. Frase for assistance with electron microscopy, B. Vadodaria for technical assistance and A. McArthur for editing the manuscript. This work was supported, in part, by Cancer Center Support (CA21765) from the NCI, grants to M.A.D from the NIH (EY014867 and EY018599 and CA168875), and the American Lebanese Syrian Associated Charities (ALSAC). M.A.D. is a Howard Hughes Medical Institute Early Career Scientist. The whole-genome sequencing was supported as part of the St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project. Finally, we thank John and Andra Tully and the Tully Family Foundation for generous support of Pediatric Solid Tumor Research at St. Jude Children’s Research Hospital.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBER
The European Bioinformatics Institute accession number for all sequence data, RNA-Seq data and DNA methylation data is EGAS00001000256.
REFERENCES
- Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FG. Chromosomal translocations involving paired box transcription factors in human cancer. The international journal of biochemistry & cell biology. 1997;29:1449–1461. doi: 10.1016/s1357-2725(97)00095-2. [DOI] [PubMed] [Google Scholar]
- Block K, Gorin Y. Aiding and abetting roles of NOX oxidases in cellular transformation. Nature reviews Cancer. 2012;12:627–637. doi: 10.1038/nrc3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouitbir J, Charles AL, Echaniz-Laguna A, Kindo M, Daussin F, Auwerx J, Piquard F, Geny B, Zoll J. Opposite effects of statins on mitochondria of cardiac and skeletal muscles: a 'mitohormesis' mechanism involving reactive oxygen species and PGC-1. European heart journal. 2012;33:1397–1407. doi: 10.1093/eurheartj/ehr224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler LM, Zhou X, Xu WS, Scher HI, Rifkind RA, Marks PA, Richon VM. The histone deacetylase inhibitor SAHA arrests cancer cell growth, up-regulates thioredoxin-binding protein-2, and down-regulates thioredoxin. Proceedings of the National Academy of Sciences of the United States of America; 2002. pp. 11700–11705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist WM, Anderson JR, Meza JL, Fryer C, Raney RB, Ruymann FB, Breneman J, Qualman SJ, Wiener E, Wharam M, et al. Intergroup rhabdomyosarcoma study-IV: results for patients with nonmetastatic disease. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2001;19:3091–3102. doi: 10.1200/JCO.2001.19.12.3091. [DOI] [PubMed] [Google Scholar]
- Csordas G, Golenar T, Seifert EL, Kamer KJ, Sancak Y, Perocchi F, Moffat C, Weaver D, de la Fuente Perez S, Bogorad R, et al. MICU1 Controls Both the Threshold and Cooperative Activation of the Mitochondrial Ca(2+) Uniporter. Cell metabolism. 2013;17:976–987. doi: 10.1016/j.cmet.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmahapatra G, Almenara JA, Grant S. Flavopiridol and histone deacetylase inhibitors promote mitochondrial injury and cell death in human leukemia cells that overexpress Bcl-2. Molecular pharmacology. 2006;69:288–298. doi: 10.1124/mol.105.016154. [DOI] [PubMed] [Google Scholar]
- Dasmahapatra G, Lembersky D, Kramer L, Fisher RI, Friedberg J, Dent P, Grant S. The pan-HDAC inhibitor vorinostat potentiates the activity of the proteasome inhibitor carfilzomib in human DLBCL cells in vitro and in vivo. Blood. 2010;115:4478–4487. doi: 10.1182/blood-2009-12-257261. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dasmahapatra G, Lembersky D, Son MP, Attkisson E, Dent P, Fisher RI, Friedberg JW, Grant S. Carfilzomib interacts synergistically with histone deacetylase inhibitors in mantle cell lymphoma cells in vitro and in vivo. Molecular cancer therapeutics. 2011;10:1686–1697. doi: 10.1158/1535-7163.MCT-10-1108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Davicioni E, Anderson JR, Buckley JD, Meyer WH, Triche TJ. Gene expression profiling for survival prediction in pediatric rhabdomyosarcomas: a report from the children's oncology group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:1240–1246. doi: 10.1200/JCO.2008.21.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proceedings of the National Academy of Sciences of the United States of America; 2007. pp. 5569–5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gysin S, Salt M, Young A, McCormick F. Therapeutic strategies for targeting ras proteins. Genes & cancer. 2011;2:359–372. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettmer S, Wagers AJ. Muscling in: Uncovering the origins of rhabdomyosarcoma. Nature medicine. 2010;16:171–173. doi: 10.1038/nm0210-171. [DOI] [PubMed] [Google Scholar]
- Hu Y, Wu G, Rusch M, Lukes L, Buetow KH, Zhang J, Hunter KW. Integrated cross-species transcriptional network analysis of metastatic susceptibility. Proceedings of the National Academy of Sciences of the United States of America; 2012. pp. 3184–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JM, Sheard MA, Ji L, Sposto R, Keshelava N. Combination of vorinostat and flavopiridol is selectively cytotoxic to multidrug-resistant neuroblastoma cell lines with mutant TP53. Molecular cancer therapeutics. 2010;9:3289–3301. doi: 10.1158/1535-7163.MCT-10-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcheson DA, Zhao J, Merrell A, Haldar M, Kardon G. Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for beta-catenin. Genes & development. 2009;23:997–1013. doi: 10.1101/gad.1769009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes & development. 2004;18:2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li Y, Yu S, Zhao G. Recent advances in the development of thioredoxin reductase inhibitors as anticancer agents. Current drug targets. 2012;13:1432–1444. doi: 10.2174/138945012803530224. [DOI] [PubMed] [Google Scholar]
- Malempati S, Hawkins DS. Rhabdomyosarcoma: review of the Children's Oncology Group (COG) Soft-Tissue Sarcoma Committee experience and rationale for current COG studies. Pediatric blood & cancer. 2012;59:5–10. doi: 10.1002/pbc.24118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli S, McDowell HP, Vigne SD, Kokai G, Uccini S, Tartaglia M, Dominici C. RAS signaling dysregulation in human embryonal Rhabdomyosarcoma. Genes, chromosomes & cancer. 2009;48:975–982. doi: 10.1002/gcc.20702. [DOI] [PubMed] [Google Scholar]
- Minai L, Yeheskely-Hayon D, Yelin D. High levels of reactive oxygen species in gold nanoparticle-targeted cancer cells following femtosecond pulse irradiation. Scientific reports. 2013;3:2146. doi: 10.1038/srep02146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton WA, Jr., Soule EH, Hamoudi AB, Reiman HM, Shimada H, Beltangady M, Maurer H. Histopathology of childhood sarcomas, Intergroup Rhabdomyosarcoma Studies I and II: clinicopathologic correlation. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1988;6:67–75. doi: 10.1200/JCO.1988.6.1.67. [DOI] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Ognjanovic S, Linabery AM, Charbonneau B, Ross JA. Trends in childhood rhabdomyosarcoma incidence and survival in the United States, 1975-2005. Cancer. 2009;115:4218–4226. doi: 10.1002/cncr.24465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappo AS, Anderson JR, Crist WM, Wharam MD, Breitfeld PP, Hawkins D, Raney RB, Womer RB, Parham DM, Qualman SJ, Grier HE. Survival after relapse in children and adolescents with rhabdomyosarcoma: A report from the Intergroup Rhabdomyosarcoma Study Group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1999;17:3487–3493. doi: 10.1200/JCO.1999.17.11.3487. [DOI] [PubMed] [Google Scholar]
- Paulson V, Chandler G, Rakheja D, Galindo RL, Wilson K, Amatruda JF, Cameron S. High-resolution array CGH identifies common mechanisms that drive embryonal rhabdomyosarcoma pathogenesis. Genes, chromosomes & cancer. 2011;50:397–408. doi: 10.1002/gcc.20864. [DOI] [PubMed] [Google Scholar]
- Raney RB, Anderson JR, Barr FG, Donaldson SS, Pappo AS, Qualman SJ, Wiener ES, Maurer HM, Crist WM. Rhabdomyosarcoma and undifferentiated sarcoma in the first two decades of life: a selective review of intergroup rhabdomyosarcoma study group experience and rationale for Intergroup Rhabdomyosarcoma Study V. Journal of pediatric hematology/oncology. 2001;23:215–220. doi: 10.1097/00043426-200105000-00008. [DOI] [PubMed] [Google Scholar]
- Reis SM, Gurney JG, Linet M, Tamra M, Young JL, Bunin GR. National Cancer Institute SEER Program. NIH Pub; Bethesda, MD: 1999. Cancer incidence and survival among children and adolescents: United States SEER Program 1975-1995; pp. 99–4649. [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BP, Nishijo K, Chen HI, Yi X, Schuetze DP, Pal R, Prajapati SI, Abraham J, Arenkiel BR, Chen QR, et al. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell. 2011;19:177–191. doi: 10.1016/j.ccr.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudzinski ER, Teot LA, Anderson JR, Moore J, Bridge JA, Barr FG, Gastier-Foster JM, Skapek SX, Hawkins DS, Parham DM. Dense pattern of embryonal rhabdomyosarcoma, a lesion easily confused with alveolar rhabdomyosarcoma: a report from the Soft Tissue Sarcoma Committee of the Children's Oncology Group. American journal of clinical pathology. 2013;140:82–90. doi: 10.1309/AJCPA1WN7ARPCMKQ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrable H, Cavenee W, Ghavimi F, Lovell M, Morgan K, Sapienza C. A model for embryonal rhabdomyosarcoma tumorigenesis that involves genome imprinting. Proceedings of the National Academy of Sciences of the United States of America; 1989. pp. 7480–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla N, Ameur N, Yilmaz I, Nafa K, Lau CY, Marchetti A, Borsu L, Barr FG, Ladanyi M. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:748–757. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalli O, Gabbiani G, Babai F, Seemayer TA, Pizzolato G, Schurch W. Intermediate filament proteins and actin isoforms as markers for soft tissue tumor differentiation and origin. II. Rhabdomyosarcomas. The American journal of pathology. 1988;130:515–531. [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Fisher C, Gusterson BA, Cooper CS. Detection of point mutations in N-ras and K-ras genes of human embryonal rhabdomyosarcomas using oligonucleotide probes and the polymerase chain reaction. Cancer research. 1989;49:6324–6327. [PubMed] [Google Scholar]
- Tian J, Liu J, Garlid KD, Shapiro JI, Xie Z. Involvement of mitogen-activated protein kinases and reactive oxygen species in the inotropic action of ouabain on cardiac myocytes. A potential role for mitochondrial K(ATP) channels. Molecular and cellular biochemistry. 2003;242:181–187. [PubMed] [Google Scholar]
- Tsuruga M, Dang Y, Shiono Y, Oka S, Yamazaki Y. Differential effects of vitamin E and three hydrophilic antioxidants on the actinomycin D-induced and colcemid-accelerated apoptosis in human leukemia CMK-7 cell line. Molecular and cellular biochemistry. 2003;250:131–137. doi: 10.1023/a:1024912806686. [DOI] [PubMed] [Google Scholar]
- Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nature reviews Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- Williamson D, Missiaglia E, de Reynies A, Pierron G, Thuille B, Palenzuela G, Thway K, Orbach D, Lae M, Freneaux P, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:2151–2158. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Statistical applications in genetics and molecular biology. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.