Abstract
Analyzing the chemical space coverage in commercial fragment screening collections revealed the overlap between bioactive medicinal chemistry substructures and rule-of-three compliant fragments is only ∼25%. We recommend including these fragments in fragment screening libraries to maximize confidence in discovering hit matter within known bioactive chemical space, while incorporation of nonoverlapping substructures could offer novel hits in screening libraries. Using principal component analysis, polar and three-dimensional substructures display a higher-than-average enrichment of bioactive compounds, indicating increasing representation of these substructures may be beneficial in fragment screening.
Introduction
Fragment screening has become an increasingly important technique in early stage drug discovery.1 Occupying the chemical space that is commonly described by the ‘rule-of-three’ (Ro3) criteria of physicochemical properties,2 fragment-like molecules are smaller in size than lead-like and drug-like molecules used in conventional high-throughput screening (HTS). Screening of fragment-sized molecules aims to sample a greater coverage of chemical diversity using a smaller collection of compounds,1 at the same time providing better hits as starting points for optimization into chemical leads.3
Typical fragment screening libraries contain compounds that originated either from commercial sources or proprietary synthetic chemical intermediates. While these fragments may mostly conform to the Ro3 physicochemical properties criteria, the chemical space they represent may not entirely project to the lead-like or drug-like chemical spaces that are relevant to medicinal chemistry. To ensure appropriate sampling of chemical space in fragment screening, it is essential to understand the current coverage of known medicinal chemistry space by commercial fragments. Using ChEMBL4 as an open-access medicinal chemistry repository, we investigated the overlap between the fragment-like chemical space of exemplified medicinal chemistry substructures generated from ChEMBL and that of fragments available from commercial vendors listed in the eMolecules database.5 The distribution of biologically active medicinal chemistry compounds was also analyzed, and property trends that differentiate active from inactive compounds were identified. Based upon these results, we make recommendations on the composition of fragment screening libraries to enhance sampling of the chemical space in fragment screening that is enriched in biologically relevant compounds.
Results
Data Collection and Substructure Generation
All compounds annotated as small molecules in ChEMBL4 and with a minimum of one ring (1.01 million unique compounds) were used in this study. Although only 46080 compounds from ChEMBL conformed to the fragment-like Ro3 criteria (considering all physicochemical property criteria,2 apart from topological polar surface area for which no limit was applied in this study),6 all small molecules in the ChEMBL database were fragmented to capture all substructures that are within the sampling space of Ro3-compliant commercial fragments. For that purpose, each compound was subjected to two independent segmentation algorithms to obtain substructures represented by a level 1 scaffold7 and multiple ring segments.8 Level 1 scaffolds generated from the Scaffold Tree algorithm9 represent the majority of molecules using a two-ring substructure, either in the form of a fused ring system or two linked monocycles. The only exception is for substituted monocyclic compounds for which the level 1 scaffold is identical to the entire molecule and therefore contains only one ring.7 The ring segmentation algorithm generates ring systems with directly attached polar functional groups.8 The former method retains linkers in the molecular scaffold, whereas the latter method describes functional groups relevant for molecular recognition. Both methods were applied since the generated substructures are complementary to each other. However, level 1 scaffolds that contained only one substituted monocycle were disregarded because such scaffolds were identical to their parent molecules with all substitutions retained. For example, all compounds comprising substituted phenyl rings would be independently represented, rendering the data set too granular. Nonetheless, substructures containing appropriate functionalities capable of molecular recognition on substituted monocyclic rings were captured using the ring segmentation algorithm. Subsequently, the generated substructures were filtered for compliance with the Ro3 criteria. Although the filtered out substructures are relevant to medicinal chemistry space, they are beyond the sampling space of Ro3-compliant commercial fragments that constitute many typical fragment screening libraries and were hence not included in this study. In total, 72880 level 1 scaffolds and 46163 ring segments were filtered out due to violation of at least one of the Ro3 criteria. In particular, almost 44000 of the filtered level 1 scaffolds were substituted monocycles that contained over three rotatable bonds from various substituents. Finally, substructures with unwanted functionalities such as reactive, metabolically labile groups or toxicophores were also excluded.8 This led to a collection of 13922 unique level 1 scaffolds and 14738 unique ring segments in the ChEMBL data set (Figure 1).
Figure 1.
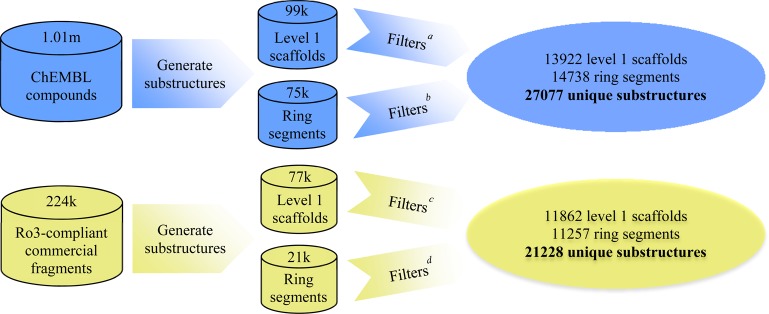
Workflow applied to generate the ChEMBL and commercial fragments data sets. aRule-of-three (Ro3) compliance, unwanted functionalities and substituted monocycle filter; bRo3 compliance and unwanted functionalities filter; cunwanted functionalities and substituted monocycle filter; dunwanted functionalities filter.
The commercial fragments data set was compiled by first applying the Ro3 filter criteria to the eMolecules database containing 5.2 million commercially available compounds5 to include only compounds that represent the chemical space commonly sampled in typical fragment screening libraries. Substructures of 224575 Ro3-compliant commercial fragments were generated using the aforementioned segmentation algorithms. The resultant level 1 scaffold substructures were subjected to both the unwanted functionalities and substituted monocycles filters, whereas only the unwanted functionalities filters were applied to the ring segment substructures. The final commercial fragments data set contained 11862 unique level 1 scaffolds and 11257 unique ring segments (Figure 1).
Substructures and Chemical Space Comparison
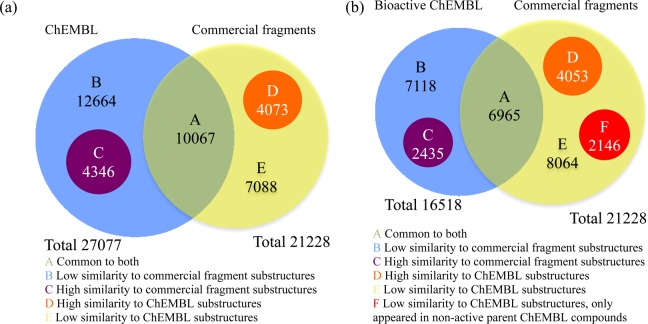
To understand the sampling of exemplified medicinal chemistry space using commercial fragments, the overlap between the substructures generated from both data sets was analyzed. In total, there are 27077 unique substructures derived from ChEMBL and 21228 unique substructures derived from Ro3-compliant commercial fragments (Figure 1). Only 10067 (26% of all the 38238 substructures, the same number of total substructures applies to all subsequent percentage comparisons in this paragraph) substructures are common in both data sets (region A, Figure 2a). The overlap increased when nonidentical but chemically similar substructures were included. Using molecular fingerprints analogous to the Daylight fingerprints (EPFP_7), 4346 substructures (11%, region C) derived from ChEMBL compounds had one of their nearest neighbors in the commercial fragments data set above a 0.85 Tanimoto similarity cutoff, a threshold at which any molecular pairs above this similarity can be considered chemically similar.10 The remaining 12664 substructures from ChEMBL (33%, region B) had low similarity to any commercial fragment substructures. In an analogous comparison, 4073 substructures (11%, region D) derived from Ro3-compliant commercial fragments had a chemically similar substructure in ChEMBL as one of their nearest neighbors. The remaining 7088 substructures (19%, region E) had low similarity to any ChEMBL substructures (Figure 2a).
Figure 2.
Venn diagrams illustrating the overlap between substructures derived from ChEMBL small molecules (blue) and commercial fragments (yellow). The subsets of substructures that have one of the nearest neighbors above a 0.85 Tanimoto similarity cutoff (EPFP_7) in the counterpart data set are represented by purple and orange circles. (a) All ChEMBL substructures versus all commercial fragment substructures; (b) only substructures derived from bioactive parent ChEMBL compounds versus all commercial fragment substructures. The subset of commercial fragment substructures below the 0.85 Tanimoto cutoff that only appeared in nonactive parent ChEMBL compounds is represented by the red circle.
Next, the same analysis was applied to evaluate a subset of the substructures of the ChEMBL data set. This time, only substructures that are derived from biologically active ChEMBL compounds (358240 unique compounds, of which 12018 are Ro3-compliant) were considered. These were defined as the parent compounds annotated with Ki, Kd, IC50, or EC50 ≤ 10 μM.11 This subset, containing 16518 substructures, was compared to those derived from Ro3-compliant commercial fragments (Figure 2b). Only 6965 substructures (23% of all the 30781 substructures, the same number of total substructures applies to all subsequent percentage comparisons in this paragraph) are common to both data sets (region A). When extending the overlap by including chemically similar substructures, 2435 substructures (8%, region C) from bioactive ChEMBL compounds had similar nearest neighbors in commercial fragments. Over 7000 bioactive ChEMBL substructures (23%, region B) had no similar substructures in commercial fragments. Vice versa, 14263 substructures of the commercial fragment set did not overlap with the ChEMBL bioactive set (regions D, E and F). 4053 of these substructures (13%, region D) had a chemically similar substructure in ChEMBL as one of their nearest neighbors. A further 2146 of these substructures (7%, region F) did not resemble Ro3-compliant ChEMBL substructures derived from bioactive compounds, but were present in nonactive parent compounds in ChEMBL (previously in region A of Figure 2a).
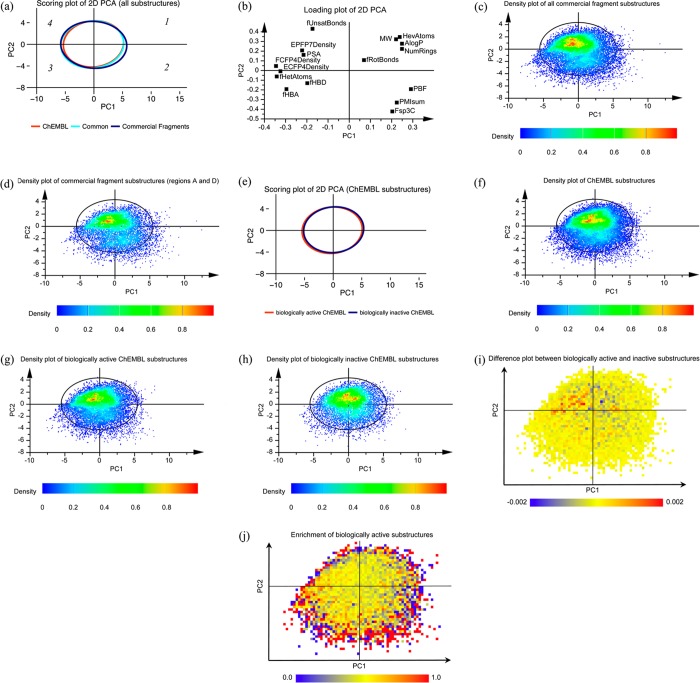
After establishing the overlap between the data sets, the chemical space occupied by the generated substructures was further evaluated. Using 16 descriptors to characterize the physicochemical properties and molecular complexity of the generated substructures (Table 1), principal component analysis (PCA) was performed on the descriptor matrix to visualize the chemical space represented. According to the 2D PCA plot (Figure 3a), the substructures derived from ChEMBL compounds and those derived from Ro3-compliant commercial fragments occupy the same areas of chemical space. Subsequently, the distribution of substructures derived from commercially available fragments within the chemical space was analyzed (Figure 3c). The density plot indicated that the highest representation of these substructures is in quadrants one and four, corresponding to chemical space with increasing molecular size and aromaticity respectively, whereas quadrants two and three, corresponding to chemical space with increasing three-dimensionality and polarity respectively, have lower representation. This distribution remains similar when only the substructures that are identical to, or chemically similar to, bioactive ChEMBL substructures (11018 substructures, regions A and D in Figure 2b) were considered (Figure 3d).
Table 1. Descriptors Used for Describing the Chemical Space Represented by the Substructures Derived from ChEMBL Small Molecules and Ro3-Compliant Commercial Fragmentsc.
| descriptor | abbreviation |
|---|---|
| molecular weight | MW |
| number of heavy atoms | HevAtoms |
| logarithmic octanol/water partition coefficient | AlogP |
| topological polar surface area | PSA |
| number of rings | NumRings |
| fraction ofa | |
| hydrogen-bond acceptors | fHBA |
| hydrogen-bond donors | fHBD |
| heteroatoms | fHetAtoms |
| rotatable bonds | fRotBonds |
| unsaturated bonds | fUnsatBonds |
| sp3-hybridized carbon atomsb | Fsp3C |
| normalizeda | |
| atom type extended connectivity fingerprints | ECFP4Density |
| atom type path fingerprints12 | EPFP7Density |
| functional class extended connectivity fingerprints12 | FCFP4Density |
| sum of normalized principal moments of inertia | PMIsum |
| plane of best fit13 | PBF |
Normalized relative to the number of heavy atoms unless stated otherwise.
Normalized relative to the number of carbon atoms.14
Categorical descriptors with discrete unit values were normalized relative to the number of heavy atoms, unless stated otherwise.
Figure 3.
2D PCA of the chemical space analyzed. Quadrants are counted clockwise with the first quadrant in the top right corner. (a) Scoring plot with ellipses showing the 95% confidence intervals of the distributions of substructures derived from ChEMBL compounds (red), Ro3-compliant commercial fragments (blue), and those common to both data sets (region A in Figure 2a) (cyan). Numbers within the plot in italics illustrate the quadrant numbers. (b) Corresponding loading plot. (c) Density plot of the distribution of all substructures from commercial fragments. The ellipse (same for figures (d), (f), (g), and (h)) corresponds to a confidence level of 95% of Hotelling’s T2 distribution of the entire chemical space analyzed. (d) Density plot of the distribution of commercial fragments that are identical to, or considered chemically similar to, bioactive ChEMBL substructures (regions A and D in Figure 2b). (e) Scoring plot with ellipses showing the 95% confidence intervals of the distributions of substructures derived from biologically active ChEMBL compounds (red) and those only from inactive compounds (blue). (f) Density plot of the distribution in (e). (g) Density plot of the distribution of biologically active ChEMBL substructures in (e). (h) Density plot of the distribution of biologically inactive ChEMBL substructures in (e). (i) Difference plot of (e) between the normalized occupancy of biologically active and inactive substructures within each cell. (j) Enrichment plot of (e) showing the ratio of the normalized occupancy of biologically active substructures to all substructures within each cell.
Finally, the overlap between the ChEMBL substructures derived from bioactive compounds and those that only appeared in inactive compounds was compared (Figures 3e to 3j). The chemical space covered by both subsets is largely similar (Figure 3e). However, the distribution of these substructures is uneven in chemical space (Figures 3f, 3g, and 3h). The density plots indicate that quadrants one and four of the 2D PCA plot, corresponding to chemical space with increasing molecular size and aromaticity, respectively, have the highest representation, whereas quadrants two and three, corresponding to chemical space with increasing three-dimensionality and polarity respectively, have lower occupancy. This distribution remains similar when the substructures that are derived from bioactive compounds and those only from inactive compounds were separately analyzed (Figures 3g and 3h, respectively).
To further elucidate the differences between the biologically active and inactive substructures, the difference and enrichment plots of the 2D PCA plot in Figure 3e were generated by dividing the scoring plot into cells of size 0.25 × 0.25. The difference plot (Figure 3i) shows the difference between the normalized occupancy of biologically active and inactive substructures within each cell. The region of chemical space having the highest positive difference is in quadrant four that corresponds to increasing aromaticity, whereas there are more cells with negative difference observed in quadrant one that corresponds to increasing substructure size. The enrichment plot (Figure 3j) shows the ratio of the normalized occupancy of bioactive substructures to all substructures within each cell, with the average ratio at 0.52. Quadrants two and three, characterizing substructures with increasing three-dimensionality and polarity respectively, appear to have more cells with ratios higher than average compared to quadrants one and four.
Discussion
The application of fragment screening in hit discovery aims to efficiently sample the lead-like and drug-like chemical space using a small collection of compounds. While many typical fragment libraries contain commercial fragments that mostly conform to the Ro3 physicochemical properties criteria, the chemical space these fragments represents may not entirely project to the lead-like and drug-like chemical space that are relevant to medicinal chemistry. By generating substructures from ChEMBL compounds and Ro3-compliant commercial fragments and analyzing the overlap between the two data sets, the chemical space coverage in fragment screening collections derived from commercial fragments was evaluated.
The overlap analyses (Figure 2b) indicate that the number of identical substructures derived from bioactive ChEMBL compounds and commercial fragments (region A) only accounts for less than 25% of all the 30781 substructures present. Even when ChEMBL substructures similar to commercial fragments (region C) are included, more than 7000 substructures (over 40% of the 16518 substructures derived from bioactive ChEMBL compounds, region B) remain unrepresented by commercial fragments. This suggests the overlap between the two data sets is rather low. Almost half of the substructures derived from bioactive medicinal chemistry compounds are not covered using currently available commercial fragments. Chemical syntheses of the substructures that are unique to ChEMBL (region B) would be the most exhaustive way to improve the sampling of known bioactive medicinal chemistry space in fragment screening libraries, even though such compounds may be synthetically challenging (see Figures 4 and S1 for exemplars of substructures in region B of Figure 2b that frequently appear in bioactive parent ChEMBL compounds). Alternatively, we recommend the design of fragment screening libraries should initially and pragmatically focus on exemplifying the substructures that occur in both the ChEMBL set and the commercially available fragment set (region A), or those that occur in commercially available fragments and are similar to substructures derived from bioactive molecules in ChEMBL (region D). These substructures represent compounds that have reported bioactivity in the primary literature or resemble closely bioactive substructures derived from ChEMBL. Hence, including these compounds in fragment screening libraries will ensure appropriate coverage of chemical space relevant to known bioactive medicinal chemistry series in fragment screens. This is useful in particular when financial constraints may limit the number of compounds purchased for a fragment screening library, since these compounds can offer a higher confidence of discovering hit matter given the literature precedence of the represented substructures. However, these substructures may have already been extensively studied and gaining intellectual property rights from these substructures may prove challenging.
Figure 4.
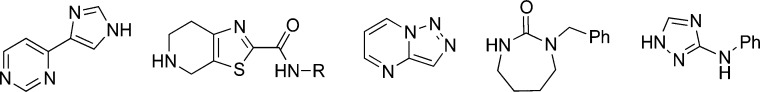
Exemplars of frequently occurring substructures derived from bioactive ChEMBL compounds that have low similarity to commercial fragments (region B of the Venn diagram in Figure 2b).
When considering expanding fragment screening libraries, compounds represented by substructures in commercial fragments that have low similarity to those derived from bioactive ChEMBL compounds (regions E and F, Figure 2b) may become useful to expand the chemical space sampling. Represented by almost 50% of the 21228 substructures derived from commercial fragments, these compounds either have not been subjected to any biochemical or cell assay, or their assay results were not reported in the primary literature (region E), whereas compounds represented by substructures in region F have thus far only resulted in inactive compounds in the primary literature. Although these substructures have not been demonstrated to display any biological activity, they could offer less explored scaffolds that have a better potential in gaining intellectual property rights should hit matter be discovered (see Figure 5 for exemplars of substructures in region E of Figure 2b). In addition, when screening against emerging target classes, the relevant chemical space may not be well-defined and could be different from that which already has extensive coverage in medicinal chemistry literature. Therefore, these compounds with low similarity to currently known medicinal chemistry space can be useful in offering more diverse fragment screening libraries.
Figure 5.
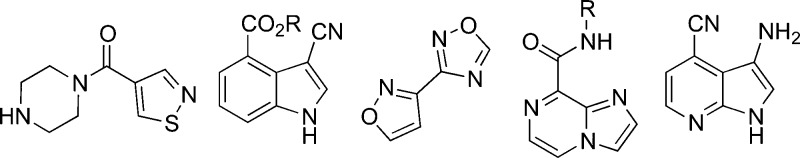
Exemplars of substructures derived from Ro3-compliant commercial fragments that have low similarity to bioactive ChEMBL compounds (region E of the Venn diagram in Figure 2b).
Using 2D PCA, the chemical space described by 16 descriptors illustrates uneven distributions of the substructures generated, even though the substructures derived from ChEMBL compounds and those derived from Ro3-compliant commercial fragments occupy a largely similar chemical space (Figures 3a, 3c, and 3f). In agreement with previous results,11 substructures with increasing aromaticity have the highest representation in commercial fragment substructures (quadrant four, Figures 3c and 3d). The lowest representation in commercial fragment substructures corresponds to chemical space with increasing polarity (quadrant three, Figures 3c and 3d). We speculate that one possible reason for the low representation of polar compounds could be attributed to less robust procedures in the chemical synthesis or purification of polar compounds. This implies that, when designing fragment screening libraries, focusing on the substructures in the commercial fragments set common to, or those have high similarity to, the bioactive ChEMBL substructures (regions A and D, respectively, Figures 2b and 3d) as we recommend here, the coverage of polar substructures relevant to known bioactive medicinal chemistry space could remain low. Figure 6 shows exemplars of polar substructures with low similarity to commercial fragments (region B of the Venn diagram in Figure 2b). Given that among the polar substructures, those derived from active compounds are enriched over those derived from inactive compounds (quadrant three, Figure 3j), it is desirable to increase the sampling of polar chemical space. With limited availability from commercial vendors, we recommend that fragments containing polar substructures should be of a high priority when devoting synthetic resources to create new fragments since this will complement commercial fragment coverage to improve the chemical space sampling of fragment screens.
Figure 6.
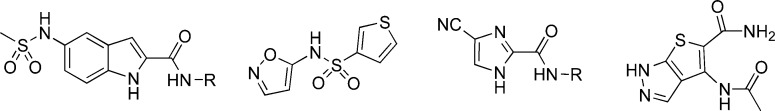
Exemplars of polar substructures derived from bioactive ChEMBL compounds that have low similarity to commercial fragments (region B of the Venn diagram in Figure 2b).
The distributions of substructures derived from bioactive ChEMBL compounds and those derived from inactive compounds are both uneven. Although these two subsets from ChEMBL cover a largely similar chemical space (Figure 3e), the difference and enrichment plots demonstrate the uneven distributions of the two subsets across the chemical space (Figures 3i and 3j). According to the enrichment plot (Figure 3j), the chemical space corresponding to increasing three-dimensionality, and also regions with increasing polarity, display higher-than-average enrichments of biologically active substructures (respectively quadrants two and three, Figure 3j). Such observation suggests that substructures with increasing three-dimensionality and/or increasing polarity could more likely deliver biologically active compounds, supporting recent literature recommendations for improving fragment sampling in three-dimensional space.15−17 However, it should be noted that the population of substructures within many of these enriched cells is evidently lower than the average population (see density plot in Figure 3f). Therefore, the enrichment for bioactive compounds within these cells might be overestimated given the low number of substructures present. Improved sampling of substructures with increasing three-dimensionality and/or increasing polarity will be required to provide a better understanding of the true enrichment of substructures within this chemical space.
Conclusions
The coverage of known medicinal chemistry space by commercial fragments was evaluated to elucidate the sampling of chemical space in fragment screening. This analysis points to strategies which enrich representation of substructures relevant to medicinal chemistry and to promote appropriate sampling of chemical space. The analysis of substructures generated from ChEMBL compounds and Ro3-compliant commercial fragments suggests that the overlap between the two data sets is low. To ensure initial confidence in discovering hit matter using commercial fragments, a subset of the commercial fragment substructures constituting ∼11000 substructures (regions A and D in Figure 2b) that have close similarity to bioactive compounds in known medicinal chemistry space should be used since these substructures have literature precedence to exhibit biological activity. When expanding screening libraries to include more diverse screening compounds, the remaining commercial fragment substructures (regions E and F in Figure 2b) that have low similarity to known medicinal chemistry compounds may offer novel hits that have better potential to gain intellectual property rights in addition to those similar to known bioactive substructures. Applying such strategies to the design of new fragment screening libraries using commercial fragments would maximize the opportunity to discover fragment hits.
When further analyzing the distribution of the substructures in chemical space using 2D PCA, the chemical space characterizing substructures with increasing three-dimensionality and/or increasing polarity apparently display higher-than-average enrichments for bioactive substructures, indicating that compounds with these substructures might be more likely to display biological activity. However, given the limited coverage of polar substructures by commercial fragments, the most effective way to enhance the sampling of polar chemical space in fragment screening is to synthesize new fragments containing these substructures. Together with the improvement of fragment sampling in three-dimensional space that has already been initiated in various academic organizations,15−17 the true enrichment of polar and/or three-dimensional substructures for bioactive compounds can be more thoroughly assessed.
Materials and Methods
Substructure Generation
The level 1 scaffold for individual compound was generated as previously described.7 The ring segments for individual compound were generated by defining substructures as a ring system plus any polar functional groups directly attached to a ring system.8 Polar functional groups were specified as nitrogen, oxygen, or sulfur atoms directly attached to the ring system, carbonyl groups, or double or triple bonded carbon atoms. Polar functional groups linking two ring systems were retained in both ring segment substructures. A mix of pattern matching and rules, implemented in a Python script using the OEChem Toolkit (OpenEye Scientific Software), was used to extract the ring segments.
Descriptor Calculations
The 16 descriptors were calculated using Pipeline Pilot professional client 8.0 (Accelrys, Inc.) applying the definitions in the software, unless stated otherwise. All categorical descriptors with discrete unit values were normalized relative to the number of heavy atoms, unless stated otherwise.
A heteroatom was defined as the elements S, O, or N. An unsaturated bond was defined as a bond with a bond order greater than one. An sp3-hybridized carbon atom was defined as any carbon atom which has an atom hybridization of sp3 according to Pipeline Pilot calculations. The fraction of sp3-hybridized carbon atoms (Fsp3C) was normalized relative to the total number of carbon atoms in the same molecule.14 All fingerprint density descriptors were defined as the ratio between the number of bits in the corresponding fingerprint generated and the number of heavy atoms.
For PMIsum and PBF, one single 3D conformer was generated for each individual substructure using CORINA (Molecular Networks GmbH). The principal moment of inertia (PMI) was then calculated using Pipeline Pilot and the sum obtained after normalizing the PMI to the longest vector. The PBF score for each 3D substructure was calculated as previously described.13
Chemical Space Analysis
The 2D-PCA plots were generated using SIMCA-P+ 13.0.0.0 (Umetrics). The descriptor matrix was normalized to unit variance before carrying out PCA using the PCA-X option under standard settings. The number of principal components was based on automatic cross-validation within the software. The difference and enrichment plots were calculated using Pipeline Pilot 8.0 (Accelrys, Inc.). The difference is defined as that between the normalized occupancy of biologically active substructures (activen) and the normalized occupancy of inactive substructures (inactiven) within each cell. The enrichment is defined as the ratio of activen to the sum of activen and inactiven.
Acknowledgments
N.Y.M. is supported by the Wellcome Trust (WT077705 and WT083481) and Cancer Research U.K. Grant C309/A11566. N.B. is supported by Cancer Research U.K. Grant C309/A11566. We thank OpenEye Scientific Software (Santa Fe, NM, U.S.A.) for a free license of the OEChem Toolkit. We also acknowledge helpful discussions with the 3D Fragment Consortium and Prof. Julian Blagg.
Supporting Information Available
A table listing the most frequently occurring substructures derived from bioactive ChEMBL compounds that have low similarity to commercial fragments. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Supplementary Material
References
- Murray C. W.; Verdonk M. L.; Rees D. C. Experiences in fragment-based drug discovery. Trends Pharmacol. Sci. 2012, 33, 224–232. [DOI] [PubMed] [Google Scholar]
- Congreve M.; Carr R.; Murray C.; Jhoti H. A rule of three for fragment-based lead discovery?. Drug Discovery Today 2003, 8, 876–877. [DOI] [PubMed] [Google Scholar]
- Leeson P. D.; St-Gallay S. A. The influence of the ’organizational factor’ on compound quality in drug discovery. Nat. Rev. Drug Discovery 2011, 10, 749–765. [DOI] [PubMed] [Google Scholar]
- ChEMBL v11. https://www.ebi.ac.uk/chembl/ (accessed Nov 23, 2011).
- eMolecules. http://www.emolecules.com (accessed Mar 1, 2012)
- Koster H.; Craan T.; Brass S.; Herhaus C.; Zentgraf M.; Neumann L.; Heine A.; Klebe G. A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes. J. Med. Chem. 2011, 54, 7784–7796. [DOI] [PubMed] [Google Scholar]
- Langdon S. R.; Brown N.; Blagg J. Scaffold diversity of exemplified medicinal chemistry space. J. Chem. Inf. Model. 2011, 51, 2174–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenk R.; Schipani A.; James D.; Krasowski A.; Gilbert I. H.; Frearson J.; Wyatt P. G. Lessons learnt from assembling screening libraries for drug discovery for neglected diseases. ChemMedChem 2008, 3, 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuffenhauer A.; Ertl P.; Roggo S.; Wetzel S.; Koch M. A.; Waldmann H. The scaffold tree - Visualization of the scaffold universe by hierarchical scaffold classification. J. Chem. Inf. Model. 2007, 47, 47–58. [DOI] [PubMed] [Google Scholar]
- Brown R. D.; Martin Y. C. Use of structure-activity data to compare structure-based clustering methods and descriptors for use in compound selection. J. Chem. Inf. Comput. Sci. 1996, 36, 572–584. [Google Scholar]
- Mok N. Y.; Brenk R. Mining the ChEMBL database: An efficient chemoinformatics workflow for assembling an ion channel-focused screening library. J. Chem. Inf. Model. 2011, 51, 2449–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuffenhauer A.; Brown N.; Selzer P.; Ertl P.; Jacoby E. Relationships between molecular complexity, biological activity, and structural diversity. J. Chem. Inf. Model. 2006, 46, 525–535. [DOI] [PubMed] [Google Scholar]
- Firth N. C.; Brown N.; Blagg J. Plane of Best Fit: A novel method to characterize the three-dimensionality of molecules. J. Chem. Inf. Model. 2012, 52, 2516–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering F.; Bikker J.; Humblet C. Escape from flatland: Increasing saturation as an approach to improving clinical success. J. Med. Chem. 2009, 52, 6752–6756. [DOI] [PubMed] [Google Scholar]
- Hung A. W.; Ramek A.; Wang Y. K.; Kaya T.; Wilson J. A.; Clemons P. A.; Young D. W. Route to three-dimensional fragments using diversity-oriented synthesis. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 6799–6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley A. D.; Pugliese A.; Birchall K.; Bower J.; Brennan P.; Brown N.; Chapman T.; Drysdale M.; Gilbert I. H.; Hoelder S.; Jordan A.; Ley S. V.; Merritt A.; Miller D.; Swarbrick M. E.; Wyatt P. G. Fragment-based hit identification: thinking in 3D. Drug Discovery Today 2013, 18, 1221–1227. [DOI] [PubMed] [Google Scholar]
- Over B.; Wetzel S.; Grutter C.; Nakai Y.; Renner S.; Rauh D.; Waldmann H. Natural-product-derived fragments for fragment-based ligand discovery. Nat. Chem. 2013, 5, 21–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.