Abstract
Large scale mapping of transcriptomes has revealed significant levels of transcriptional activity within both unannotated and annotated regions of the genome. Interestingly, many of the novel transcripts demonstrate tissue-specific expression and some level of sequence conservation across species, but most have low protein-coding potential. Here we describe progress in identifying and characterizing long noncoding RNAs and review how these transcripts interact with other biological molecules to regulate diverse cellular processes. We also preview emerging techniques that will help advance the discovery and characterization of novel transcripts. Finally, we discuss the role of long non-coding RNAs in disease and therapeutics.
Keywords: Long noncoding RNA, next generation sequencing, epigenetics, therapeutics
Pervasive transcription in mammalian genomes
A major advance in molecular biology over the past 25 years has been the discovery of and demonstration of function for long noncoding RNAs (lncRNAs). The maturation of high throughput genomic tools such as next generation sequencing and large scale tiling arrays has helped accelerate the pace of discovery. A series of projects involving either cDNA library sequencing [1, 2] or their hybridization to genomic arrays [3–7] provided comprehensive maps of the transcriptional landscapes within cells. The recent ENCODE project [8–10], the most comprehensive effort yet for surveying transcription in human cells, confirmed earlier reports of pervasive transcription in mammalian genomes. ENCODE performed RNA-seq across 15 cell lines and detected primary transcripts from a cumulative 75% of the human genome [9]. There appear to be over 9000 genomic loci which give rise to lncRNAs in human cells [10]. The noncoding RNA Expression Database (NRED) for mouse lncRNAs reported expression of about 3000 transcripts in six different biological contexts [11]. Their noncoding status is deduced from a lack of sequence homologies to known proteins, the absence of substantial open reading frames, codon substitution frequencies which deviate from that for protein coding regions [12, 13], and tandem mass spectrometry analysis [9, 14] which determine whether peptides corresponding to the RNA sequences of interest are represented. Noncoding transcripts seem to be concentrated within the cell nucleus, and are often expressed at significantly lower levels than coding RNA [9]. In addition, an increasing number of studies have begun to analyze the proportion of polyadenylated to non-polyadenylated transcripts making up the noncoding transcriptome. A significant component of the noncoding transcriptome, at least in human cells, seem to be non-polyadenylated [9, 15, 16] – an observation that could partly be explained by the discovery of polyA- transcripts emanating from many active enhancer elements in the human genome (enhancer RNAs, or eRNAs)[17–20].
The complex transcriptional landscape in mammalian cells, confounded by the lack of functional annotations for the majority of lncRNAs, has made it particularly challenging to classify novel noncoding transcripts. An arbitrarily selected cutoff of 100–200 nt is commonly used to broadly distinguish lncRNAs from 21–35 nt “small RNAs” such as microRNAs, Piwi-interacting RNAs (piRNAs), and small-interfering RNAs (siRNAs). However, such oversimplifications inevitably create difficulties in some cases, such as for the class of short RNAs which are transcribed very close to transcription start sites [6, 21, 22]. These transcripts usually fall below the 200 nt threshold but are biologically distinct from Ago-associating short transcripts, small-nuclear RNAs (snRNAs) or small-nucleolar RNAs (snoRNAs). LncRNA above the 200 nt threshold may be classified to reflect their locations relative to genomic elements. Transcriptional loci may (a) overlap with annotated gene bodies with transcription initiating from either exons or introns from the sense or antisense strands, (b) lie within cis-regulatory regions of genes as in the case of eRNAs or (c) lie in intergenic regions, giving rise to long intergenic noncoding RNAs (lincRNAs). These diverse classes of lncRNAs and their biological roles in the cell are the focus of this review. In any case, more biologically meaningful ways for classifying noncoding transcripts should be possible in the near future as we glean more insight into lncRNA mechanisms.
lncRNAs: functional transcripts vs. biological noise
The biological relevance of pervasive transcription and their associated lncRNAs is a current topic of debate [23–26]. Their low sequence conservation across model organisms and low expression levels have led some to postulate that many lncRNAs could arise from low fidelity RNA polymerase (RNAP) activity [27] and that this spurious activity is of little significance. However, in-depth analyses of lncRNA sequences may suggest the contrary. First, promoter regions and splice sites of lncRNAs have a degree of sequence conservation comparable to that for protein-coding genes [10, 28]. Second, while sequence conservation along the length of lncRNAs may be lower than that for mRNA [10, 29], lncRNA function may not necessarily depend on strict sequence conservation, especially if only small segments of the lncRNA are in contact with proteins, or if conservation of secondary structures takes precedence over that of primary sequences [29, 30]. For example, the well-characterized Xist RNA harbors only short segments of conserved sequence, but is known to play a critical role in dosage compensation [31].
Information from RNA-seq performed by the ENCODE consortium also reported that the boundaries of many human genes need to be expanded to account for extensive transcriptional activities. This has correspondingly resulted in a more than three-fold decrease in length of intergenic regions. Annotated genes can express between 10 to 12 isoforms simultaneously, of which a significant portion are novel elements that are non protein-coding [9]. There is concern that such lncRNAs are merely extensions of the nearby coding transcripts. The distinction can be made by coupling RNA-seq or tiling array data to end capture assays such as CAGE (cap analysis of gene expression) for 5’ ends [8], 3P-seq for 3’ ends [30] or RNA paired end ditag (PET) sequencing to mark both ends. Intersection of CAGE and PET libraries with the lncRNA database revealed that a significant portion of lncRNAs has unique start and stop sites, suggesting that lncRNA transcription can occur independently [10]. Novel elements also covered a large majority of intronic sequences – raising the question of how distinctions between novel lncRNA and unprocessed nascent transcripts may be made. Apart from the presence of unique 5’ and 3’ ends, the fact that some intronic lncRNAs can be detected in cytoplasmic fractions of cell extracts also argues against their being nascent mRNA [5]. In fact, the prevalence of lncRNAs emanating within close vicinities of coding genes is likely to be a reflection of the cis-acting nature of some transcripts (as reviewed below), or of gene regulation elicited from transcriptional overlap (such as imprinting of Igf2r by the lncRNA Airn [32]).
The idea of regulation by transcriptional interference, such as that proposed for Airn, is a reminder that the function of some lncRNAs may hinge on the transcriptional process, rather than the RNA product. Experiments to distinguish between the two, such as that done for imprinted lncRNA Kcnq1ot1 [33], have not been systematically performed. These would be crucial experiments in many cases, such as for eRNAs, which are noncoding, predominantly non-polyadenylated transcripts originating from a subset of putative enhancer elements [18, 19, 34]. eRNA levels demonstrate strong correlation with transcriptional activities of corresponding coding genes, yet it is still unclear in many cases whether eRNA synthesis is important for enhancer/promoter activation and the eventual activation of target genes, or if eRNAs are merely by-products of active enhancers in close association with gene promoters and the basal transcriptional machinery. Recent work by Kraus and colleagues showed that inhibition of eRNA transcription via flavopiridol, an inhibitor of transcription elongation, has little impact on the establishment of epigenetic marks (e.g. H3K4me1) or loading of RNA polymerase II (RNAPII) and other coactivators (e.g. E1A binding protein p300 (EP300) and CREB binding protein (CREBBP)) at enhancers [20]. In addition, enhancer/promoter loopings were also largely unaffected in the absence of eRNAs [20]. This suggests that molecular features usually associated with enhancers can occur independently of eRNA synthesis. It is important to note that further experiments are needed to determine whether eRNAs contribute to other aspects of enhancer function and target gene expression since flavopiridol have effects beyond transcription elongation [20].
As we begin to appreciate the complexities of transcriptional activity in the genome, it is clear that the traditional concept of a gene needs to be redefined. Fundamental differences between mRNA and lncRNAs point to the inadequacies of applying rules used to assess mRNA function on other transcripts whose functions lie outside the realm of protein production. In addition, coding and noncoding transcripts emanating from overlapping genomic loci blurs the distinction between regulatory and protein-coding sequences. Future work in unraveling lncRNA function and how underlying genomic sequences contribute to function will be key to understanding the true nature of the genome.
Mechanisms of lncRNA function
LncRNAs have been implicated in the regulation of a diverse array of biological processes including dosage compensation [35], imprinting [33, 36], cell cycle control [37–39], development [30, 40], and gametogenesis [41]. The function of lncRNAs cannot currently be predicted from sequence information alone, unlike proteins which often have well-defined modular domains and whose functions may be deduced from those of related proteins. An emerging theme, however, is the capacity of lncRNAs to modulate gene expression, either through action in cis on neighboring genes [33, 35, 36, 42, 43] or action in trans regardless of gene location [20, 44].
Chromatin modification by lncRNAs
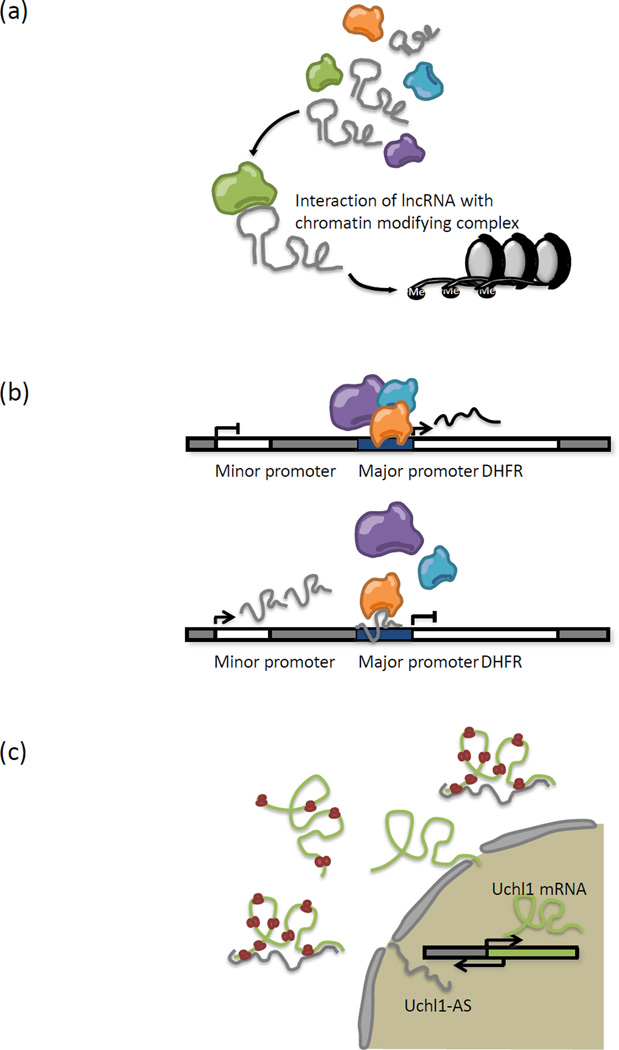
A classic example of lncRNA-mediated chromatin modification comes from eutherian dosage compensation, a whole-chromosome silencing mechanism that depends on expression of Xist RNA [35]. Synthesis of Xist RNA from the future inactive X chromosome (Xi) during early development triggers large scale recruitment of Polycomb repressive complex 2 (PRC2) in cis to the chromosome, establishing facultative heterochromatin extensively marked by the repressive H3K27me3 modification [45]. Native RNA immunoprecipitation (RIP) of Enhancer of Zeste 2 (EZH2), the catalytic subunit of PRC2, has shown that Xist RNA interacts with PRC2 during X-chromosome inactivation (XCI) to initiate and spread chromosomal silencing [42]. This RNA-protein interaction is believed to involve the repeat A region within Xist [46]. In line with RIP data, another study reported that ectopic expression of Xist from an autosomal locus is sufficient for the deposition of H3K27me3 around the site of transgene integration, providing support for a direct role of Xist in PRC2 recruitment and H3K27me3 deposition [47]. Apart from Xist, PRC2 is found to complex with other lncRNAs such as Kcnq1ot1, antisense noncoding RNA in the INK4 locus (ANRIL), and HOX transcript antisense RNA (HOTAIR) [33, 39, 44]. In the case of HOTAIR, action occurs in trans [48, 49]. Together, these observations lend credibility to the hypothesis that lncRNAs play crucial roles in recruitment of chromatin-modifying complexes to appropriate genomic loci both in cis and in trans (Figure 1a). The dependency on lncRNAs (and their secondary structures) to target PRC2 may explain the many as yet unsuccessful searches for DNA-based polycomb responsive elements (PREs) in mammalian systems [50].
Figure 1. Mechanisms for long noncoding RNA (lncRNA) function.
Characterization of lncRNA function has revealed the ability of these transcripts to regulate gene expression through chromatin remodeling, control of transcription initiation and post-transcriptional processing. (a) lncRNAs such as Xist, Kcnq1ot1, Airn and HOTAIR have been found to interact with chromatin remodeling proteins such as polycomb repressive complex 2 and G9a (represented in green) to mediate deposition of repressive chromatin marks. (b) lncRNAs can directly regulate messenger RNA synthesis at genomic loci by interacting with transcription factors (see text) or components of the basal transcriptional machinery. In the case of DHFR regulation, an upstream lncRNA transcribed from the minor promoter has been shown to bind both the major DHFR promoter as well as TFIIB, leading to displacement of TFIIB from the major promoter. (c) lncRNAs can regulate co-transcriptional processes such as RNA splicing (see text) and translation. The Uchl1-AS RNA is transcribed in times of cellular stress and acts to speed up translation of Uchl1 mRNA by enhancing polysome loading onto the mRNA in the cytoplasm. The mechanisms for this activity is not well understood, although it has been proposed that the SINEB2 repeat element could play a crucial role.
LncRNA partners for other chromatin-associated proteins have also been found. The H3K9 methyltransferase G9a, which has been implicated in imprinting, associates with the lncRNA Airn to mediate silencing of the Igf2r/Slc 22a2/Slc22a3 gene cluster on the paternal allele in the murine placenta [36] and with Kcnq1ot1 at the imprinted Kcnq1 domain [33]. In budding yeast, the IRT1 transcript recruits both Set2 (a histone methyltransferase) and Set3 (a histone acetylase) to the IME1 locus for regulation of gametogenesis [41]. Interactions of lncRNAs with proteins are by no means exclusive to repressive chromatin modifiers; Mixed-lineage leukemia (MLL), a component of the Trithorax complex which deposits activating methylation marks at H3K4, has been shown to be recruited by lncRNAs associated with homeotic genes such as Hoxb5/6as [51], Evx1as [51] and Mistral [52], gene activation of Xist has been shown to involve Jpx RNA-mediated eviction of a CCCTC-binding factor (CTCF) repressor [53], and promoter associated transcripts at rRNA genes associate with DNA methyltransferase 3b to mediate gene silencing [54].
These findings prompt the question of how lncRNAs achieve targeting to genomic loci with high specificities. Here, several hypotheses have been proposed [48]. Theoretically, lncRNAs may form triplexes with genomic DNA containing complementary sequences. Alternatively, targeting specificity may be achieved with favorable chromatin architectures. For example, lncRNAs could demonstrate preferential loading of chromatin modifiers to segments of the genome which are in close proximity. Chromosome conformation capture experiments have suggested this mechanism [55, 56]. The third possibility calls upon additional DNA binding factors to bridge the gap between lncRNAs and chromatin. In the case of Xist RNA, work by Jeon and Lee has revealed the role of the transcription factor Yin Yang 1 (YY1) in tethering Xist RNA to the X inactivation center (Xic) on Xi [47]. YY1 was found to bind both Xist DNA and RNA, and its depletion resulted in a loss of Xist loading on the Xi. These observations suggested that YY1 is the docking factor responsible for the cis-acting nature of Xist RNA.
Regulation of transcription initiation
The classical noncoding U1 snRNA, a component of the spliceosome, interacts with transcriptional initiation factor TFIIH to boost initiation rates of the basal transcriptional complex [57]. Novel lncRNAs have demonstrated similar capabilities, bypassing chromatin-modifying complexes to communicate directly with gene promoters, the basal transcriptional machinery, and transcription factors. These lncRNAs are usually synthesized from regulatory loci such as enhancers and promoters and act in cis to mediate rapid, sensitive, and localized transcriptional regulation. For example, the Evf2 lncRNA is transcribed from an ultraconserved enhancer at the Dlx5-6 gene cluster and forms a complex with the transcription factor Dlx2 to elicit activation of the Dlx gene cluster [58]. Depletion of Evf2 resulted in the decrease in GABAergic interneurons in the early postnatal hippocampus and dentate gyrus of mice [59]. Recent studies have uncovered more lncRNAs that function as transcriptional activators in both mice and humans [17, 18, 60]. Many of these transcripts are synthesized at enhancers and the majority influences the activity of enhancers, or help with the recruitment of protein factors to enhancers. For example, two lncRNAs highly expressed in aggressive prostate cancers bind to the androgen receptor (AR) to enhance AR loading at gene enhancers, even in the absence of AR ligands [61]. Activating lncRNAs were also found in association with Mediator, acting as cofactors which help to model chromatin architecture and enhance kinase activity [62]. The transcription of the noncoding transcripts at enhancers is also proposed to play a role in enhancer activation by mediating the deposition of H3K4 mono- and di-methylation [63].
The ability of lncRNAs to interact with both the basal transcriptional machinery and key regulatory sequences on the chromatin, possibly through RNA-DNA interactions, is demonstrated by the lncRNA transcribed from the minor promoter of the human dihydrofolate reductase (DHFR) gene [64] (Figure 1b). The noncoding transcript is proposed to form a triplex with the major DHFR promoter and bind to TFIIB to displace the pre-initiation complex from the DHFR locus, thereby blocking gene expression. The DHFR study presents a case where lncRNAs mediate gene repression by interfering with the activity of the basal transcriptional machinery. Similarly, murine B2 RNA and human Alu RNA, both of which are transcribed from short interspersed elements (SINEs), mediate repression of heat shock genes by binding to and deactivating RNAPII [65, 66]. Although these RNAs all bind the transcription initiation complex, they bear little resemblance to each other in sequence or structure [66]. Primary and/or secondary structures relevant for these interactions are currently of significant interest. Identification of more noncoding RNAs operating in the same manner should shed light on this question.
Co- and Post-transcriptional regulation
Co- and post-transcriptional processes such as splicing, transport, translation of mRNA, and subcellular localization of proteins may also be controlled by lncRNAs. Interaction of lncRNAs with primary coding transcripts can occlude splice junctions and result in production of alternative isoforms. Studies which look at the regulation of the transcription factor Zeb2, which has been implicated in epithelial-mesenchymal transitions (EMT) during embryogenesis and cancer transformation, have revealed that Zeb2 is regulated post-transcriptionally by its natural antisense transcript (NAT). The noncoding NAT, synthesized from the antisense strand of the Zeb2 promoter, shields an internal ribosome entry site (IRES) within the 5’ UTR of Zeb2 from mRNA splicing, thereby allowing for increased rates of Zeb2 translation and driving EMT [67]. In the absence of antisense expression, the loss of the IRES results in significantly lower levels of Zeb2 protein (Figure 1c).
The expression of ubiquitin carboxy-terminal hydrolase L1 (Uchl1), a gene implicated in brain function and neurodegenerative diseases such as Parkinson’s disease and Alzheimer’s disease [68], has also been found to be regulated by a lncRNA. The Uchl1 antisense transcript (Uchl1-AS), which partially overlaps with the 5’ end of Uchl1 mRNA, is initially concentrated in the nucleus but translocates to the cytoplasm under conditions of cellular stress [69]. Once there, Uchl1-AS promotes translation of Uchl1 mRNA by enhancing polysome loading on the mRNA. mRNA levels remain unperturbed in the presence of increasing amounts of the antisense transcript, attesting to the participation of Uchl1-AS in regulating post-transcriptional processes. Characterization of Uchl1-AS revealed that the 5’ overlap region, as well as a short interspersed nuclear element (SINE) B2 repeat element harbored within the transcript, are critical for lncRNA function [69]. How the two elements work together to increase polysome loading onto mRNA remains unknown. It is likely that some form of RNA duplex formation occurs at the overlapping region, perhaps inducing changes in the architecture of mRNA to allow for efficient translation. The requirement of a SINEB2 repeat element is intriguing because Xist RNA also consists of repeat regions that play critical roles in chromosome silencing [70]. A search through the mouse cDNA FANTOM3 database for antisense transcripts with similar properties (5’ overlap and presence of SINEB2 repeat) identified another lncRNA at the Uxt gene that is also capable of eliciting increases in protein levels post-transcriptionally [69]. In addition, a survey of human lincRNAs also revealed an enrichment for transposable elements [71]. More in-depth analyses of RNA structure for a larger number of lncRNAs are needed to explore possible links between the presence of particular genomic elements (e.g. repeat sequences) and lncRNA function (discussed below).
Challenges in the lncRNA field
While sequencing technologies have allowed for rapid discovery of lncRNAs, elucidating the biological roles of lncRNA in vivo remain challenging. The problem arises partly because novel lncRNAs seem to be governed by a set of rules distinct from that used by proteins. Numerous strategies have been employed by different groups to address the challenges, such that the set of functionally annotated lncRNAs is expanding very rapidly (well beyond the examples this review has been able to highlight) as evidenced by the establishment of several lncRNA databases to provide comprehensive documentation of sequence information, evolutionary conservation, expression profiles and functional evidence [11, 72]. Many groups turned to clues including the level and specificity of expression, or the chromatin state around a locus of transcription to focus on lncRNAs with potentially higher chances of biological relevance from the large pool of transcripts [73–75]. It is expected that biologically significant lncRNAs will be tightly regulated and be highly expressed only in appropriate contexts. Alternatively, the strategy of identifying lncRNAs which can perturb expression of specific genes makes follow up studies much more manageable, as demonstrated by the process through which the evolutionarily conserved lncRNA NRON was identified [76]. Here, work by Schultz and colleagues utilized RNAi strategies to screen for highly conserved noncoding transcripts which regulate the expression of NFAT, a transcription factor implicated in initiation of T-cell receptor mediated immune responses. NRON is believed to modulate the nuclear trafficking of NFAT by associating with well known nuclear import proteins, and depletion of NRON correlates with increased activity of NFAT [76]. Other strategies, such as narrowing down the search to specific lineage differentiation pathways, cell types, or to those regulated by proteins of interest simplify the search for biological function. These methodologies have enabled identification of lncRNAs with roles in adipocyte differentiation [77] and in cancer [37, 78, 79]
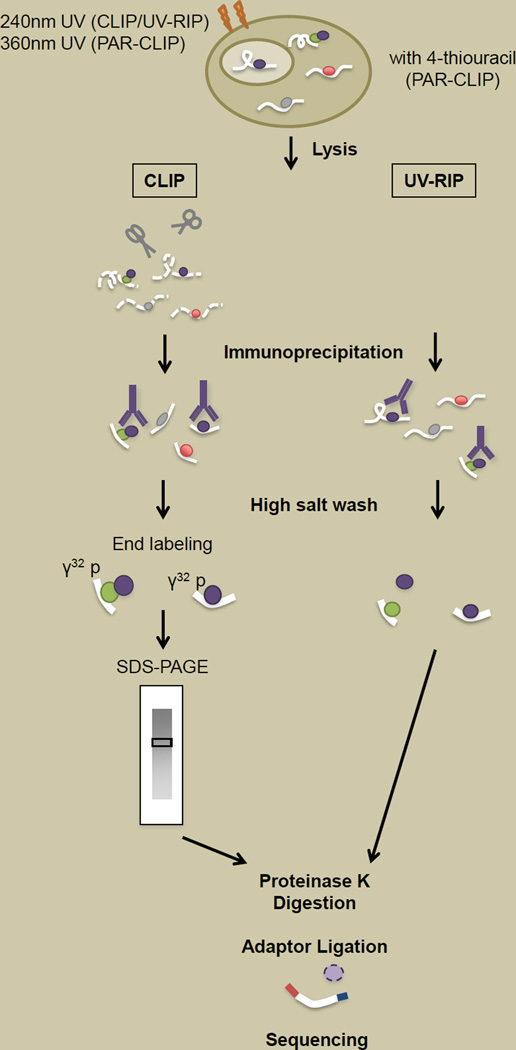
An alternative for lncRNA identification involves enriching for RNA in association with chromatin-bound proteins (e.g. chromatin modifiers) known to be important in gene regulation. Native RIP-seq, for example, has been used to define a transcriptome associated with PRC2, either directly or indirectly [80] (Figure 2). In addition, methodologies such as CLIP (CrossLinking and ImmunoPrecipitation [81, 82]) or its variant PAR-CLIP (PhotoActivable-Ribonucleoside-enhanced CrossLinking and ImmunoPrecipitation [83]) allow for identification of RNA directly bound to protein and potentially providing a short “footprint” suitable for identifying RNA motifs involved in protein binding (Figure 2). CLIP was used to identify an intronic transcript of the H3K4 methyltransferase SMYD3, with antiproliferative effects on cells when overexpressed [84]. Deep sequencing of CLIP products, however, remains a challenging procedure. RNA yields after immunoprecipitation and gel extraction are often very low, making the procedure especially sensitive to RNA contamination at each step of experimental manipulation. Further refinement of the procedure would be critical for establishing CLIP as one of the standard tools in studies of lncRNA-protein interactions.
Figure 2. Techniques for mapping RNA-protein interactions.
In both chromatin crosslinking and immunoprecipitation (CLIP) and UV RNA immunoprecipitation (UV-RIP), cells are irradiated with 254nm UV light, which crosslinks proteins and nucleic acids that are in close proximity. For PAR-CLIP, cells are incubated with 4-thiouracil and crosslinked at 360nm UV. In CLIP, cellular lysate is treated with RNAse after crosslinking to fragment RNA before performing immunoprecipitation. A 3’ adaptor can be added to enriched RNA fragments (not necessary unless for downstream sequencing library preparation). RNA is end labeled with [γ32] p, following which the reaction is subjected to SDS-PAGE. Following transfer to nitrocellulose membranes, the appropriate region of the membrane is excised (dependent on size of protein probed) and treated with proteinase K to release RNA fragments. Denaturing conditions of SDS-PAGE ensures enrichment for RNA fragments in direct interaction with protein of interest. In UV-RIP, cell lysate is subjected to immunoprecipitation to enrich for RNA bound either directly or indirectly to the protein of interest. A high stringency salt wash further enriches for RNA bound both directly and indirectly to the protein of interest by removing uncrosslinked fragments. Proteinase K treatment releases bound RNA. The RNA fraction obtained from both CLIP and UV-RIP can be analyzed by RT-qPCR, array hybridization as well as high throughput sequencing.
Another novel approach for dissecting lncRNA function relies on mapping interaction sites between RNA and genomic DNA. This is especially helpful for understanding the mechanistic details for trans-acting lncRNAs for which target sites are located away from their site of synthesis. Two methods, CHART (capture hybridization analysis of RNA targets) and ChIRP (chromatin isolation by RNA purification) were recently employed to interrogate genome-wide localization sites of lncRNAs such as HOTAIR, the human telomerase RNA TERC, and dosage compensation factors such as Xist and the roX RNAs [48, 49]. These techniques should be broadly applicable to other lncRNAs.
In the RNA world, secondary and tertiary structures are crucial for specificity of interaction with proteins or other nucleic acids [85]. Such RNA structures therefore are expected to regulate the activity and function of lncRNAs, though our ability to map secondary and tertiary structures is presently rudimentary. Analysis of RNA structure of RepA, an internal transcript from the repeat A region of the Xist locus, demonstrated that a 28 nt stem-loop found within one of its repeat sequences bound directly to Ezh2 in vitro, suggesting that the stem-loop is likely to be important for its protein interaction [42, 70]. Repeat A, encompassing approximately 8 repeat units, has itself been proposed to demonstrate higher order structure by forming two larger stem-loops each consisting of 4 repeat units [86]. Importantly, full-length repeat A demonstrated higher affinity for the PRC2 component SUZ12 than truncated versions, suggesting that these larger stemloops are necessary for efficient recruitment of PRC2 to Xist RNA [86]. Interestingly, some promoter-associated transcripts of PRC2-repressed genes also demonstrate similar stem-loop structures [22]. In addition, Uchl1-AS and Uxt-AS, both of which regulate expression of their corresponding genes post-transcriptionally, harbor the SINEB2 repeat element, suggesting that higher order structures of repeat elements could contribute to lncRNA function. These studies highlight the need for more comprehensive mapping of RNA structures to identify lncRNA structural domains that may be relevant for biochemical interactions in the cell. High throughput techniques for lncRNA structural mapping [85] such as fragmentation sequencing (FRAG-seq) [87], SHAPE-seq (which utilizes selective 2’ -hydroxyl acylation chemistry analyzed by primer extension) [88] and parallel analysis of RNA structure (PARS) [89] would be invaluable in this aspect.
lncRNAs in disease: Xist as a model
lncRNAs are implicated in a variety of diseases, especially those involving genomic imprinting and cancer, underscoring their importance in maintaining cellular homeostasis. Transcripts associated with cancer including ANRIL (transcribed from the Ink4b (p15) – ARF (p14) – Ink4a (p16) tumor suppressor loci [39, 90, 91]), PCAT-1 (a pro-proliferation transcript upregulated in prostate cancer samples [79]), HOTAIR of the HOXC locus [92]) and MALAT1 (a prognostic marker of several cancer types [93]). Significantly, Xist RNA has now been directly implicated in human cancers. Since Xist maintains dosage compensation for ~1000 genes on the X chromosome, several of which are putative oncogenes (reviewed in [94]), it is possible that misregulation of Xist contributes to cancer phenotypes through aberrations in expression of X-linked oncogenes.
In line with this idea, cytogenetic studies of human breast, ovarian and cervical cancer samples since the 1950s have noted a high frequency of Barr body (the inactivated X chromosome) loss, particularly in more aggressive breast tumors (reviewed in [95]). Barr body loss was often concomitant with the acquisition of supernumerary active Xs (Xa) and down-regulation of XIST RNA. RNA expression profiling of sporadic basal-like cancers (BLC) which have lost Xi and XIST RNA also demonstrate overexpression of some X-linked genes [96]. These observations suggested that loss of XIST RNA could drive disease progression in these cancer types, possibly through reactivation of Xi.
Direct causality has now emerged from an in vivo study where deletion of Xist in the hematopoietic lineage resulted in the development of leukemia in mice with full penetrance [97]. Gene expression profiling over the course of disease progression revealed significant upregulation of X-linked genes, suggesting the possibility of X reactivation following Xist loss. This sensitivity of hematopoietic cells to Xist misregulation corroborates with previous work in which overexpression of Xist in mice results in lethal anemia due to defective hematopoiesis [98], and overexpression in a lymphoma cell model suppresses tumorigenicity [99]. Further studies are needed to determine whether the tumor suppressive properties of Xist extend to breast and ovarian tissues as well. In any case, these studies support the notion that XIST RNA could be useful as a therapeutic target in female cancers or establish XIST RNA as a diagnostic parameter for stages of tumor progression (Box 1).
Box 1: LncRNAs and therapeutics
While lncRNA targeting for therapeutic purposes is still in early stages of development, RNA-based drugs have been in development for more than two decades. A first such drug, fomivirsen, emerged in 1998 for treatment of cytomegalovirus-mediated eye infections [100]. RNA therapeutics often rely on the use of antisense oligonucleotides (ASO, which hybridize to complementary sequences and induce RNaseH-mediated RNA degradation) or siRNAs (which act through the RNAi machinery) to degrade mRNAs or microRNAs of interest, and feasibility for such methods has already been demonstrated for several drug targets currently in clinical trials. One ASO (Kynamro) was recently FDA-approved for hypercholesterolemia [101]. Many others are still in pre-clinical phase. For example, a proof-of-concept study showed that BDNF-AS, a ~1kb lncRNA transcribed antisense to the BDNF gene (brain-derived neurotrophic factor) could be a useful ASO drug target. Using intracerebroventricular delivery of chemically modified oligonucleotides, the group was able to knockdown BDNF-AS in the mouse brain to relieve BDNF repression and allow for increased neuronal proliferation [102]. This result speaks to the possibility of using antagonizing antisense transcripts for lncRNA depletion. However, given the length of some lncRNAs, it is possible that extensive secondary structures will restrict accessibility of antisense oligos to crucial parts of the transcript. This once again emphasizes the importance of structural mapping for lncRNAs.
Delivery also remains a major obstacle for some RNA-based therapeutics. Recent technical developments such as lipid nanoparticles may facilitate delivery of siRNAs [103], though toxicity has been a concern. Naked delivery of ASOs may also be effective for some tissues and organs, such as liver and kidney, though penetration into other tissues is currently problematic.
An alternative to the oligotherapeutics strategy could involve the use of small molecules to disrupt interactions of lncRNAs with proteins or DNA. In this case, structural motifs of lncRNAs which allow them to recruit chromatin modifiers, or to form triple helixes with DNA, could be targeted by small molecules, thereby rendering the lncRNA ineffective in perturbing gene expression. Chemical library screening, as have been done for microRNAs [104], might be useful in the identification of small molecule inhibitors of lncRNAs. This type of approach is currently in nascent stages.
Concluding remarks
The detection of pervasive transcription and the ensuing discovery of noncoding transcripts have redefined our understanding of how non-genic regions of the genome are involved in the regulation of gene expression profiles within a cell. While the field is still in the early days of assigning biological function to the thousands of transcripts detected, it is clear that lncRNAs add an important layer to the repertoire of regulatory mechanisms used by mammalian cells to modulate gene expression. As more lncRNAs are identified, it will become important to analyze lncRNA sequences and secondary structures to establish structure-function relationships that define mechanisms of lncRNA function. This would move the field forward by both speeding up the identification and discovery of more biologically relevant transcripts, as well as allow for a better understanding of how lncRNA perturbation can be utilized for locus-specific manipulation of gene expression for therapeutic purposes.
Highlights.
Long noncoding RNAs (lncRNAs) provide additional layers of gene regulatory control
Emerging technologies will allow biological function of lncRNAs to be elucidated more efficiently
lncRNAs can be useful diagnostic markers or therapeutic tools for diseases
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Okazaki Y, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science (New York, N.Y.) 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 3.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science (New York, N.Y.) 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 4.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapranov P, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science (New York, N.Y.) 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 6.Kapranov P, et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 7.Rinn JL, et al. The transcriptional activity of human Chromosome 22. Genes & development. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinger ME, et al. NRED: a database of long noncoding RNA expression. Nucleic acids research. 2009;37:D122–D126. doi: 10.1093/nar/gkn617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clamp M, et al. Distinguishing protein-coding and noncoding genes in the human genome. Proceedings of the National Academy of Sciences. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinger ME, et al. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS computational biology. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bánfai B, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome research. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng J, et al. Transcriptional Maps of 10 Human Chromosomes at 5-Nucleotide Resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 16.Venters BJ, Pugh BF. Genomic organization of human transcription initiation complexes. Nature. 2013;502:53–58. doi: 10.1038/nature12535. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Kim T-K, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hah N, et al. Enhancer transcripts mark active estrogen receptor binding sites. Genome research. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taft RJ, et al. Tiny RNAs associated with transcription start sites in animals. Nature genetics. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 22.Kanhere A, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Molecular cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson JM, et al. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends in genetics : TIG. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 24.van Bakel H, et al. Most “Dark Matter” Transcripts Are Associated With Known Genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark MB, et al. The reality of pervasive transcription. PLoS biology. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louro R, et al. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics. 2009;93:291–298. doi: 10.1016/j.ygeno.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nature structural & molecular biology. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 28.Ponjavic J, et al. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome research. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pang KC, et al. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends in genetics : TIG. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Ulitsky I, et al. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nesterova TB, et al. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome research. 2001;11:833–849. doi: 10.1101/gr.174901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Latos PA, et al. Airn Transcriptional Overlap, But Not Its lncRNA Products, Induces Imprinted Igf2r Silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 33.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage512 specific transcriptional silencing through chromatin-level regulation. Molecular cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 34.Kim T-K, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Penny GD, et al. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 36.Nagano T, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 37.Hung T, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature genetics. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotake Y, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pauli A, et al. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12:136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Werven FJ, et al. Transcription of Two Long Noncoding RNAs Mediates Mating-Type Control of Gametogenesis in Budding Yeast. Cell. 2012;150:1170–1181. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao J, et al. Polycomb Proteins Targeted by a Short Repeat RNA to the Mouse X Chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 44.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science (New York, N.Y.) 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 46.Kohlmaier A, et al. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS biology. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon MD, et al. The genomic binding sites of a noncoding RNA. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu C, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Molecular cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dinger ME, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome research. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bertani S, et al. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Molecular cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Sun S, et al. Jpx RNA Activates Xist by Evicting CTCF. Cell. 2013;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmitz K-M, et al. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes & development. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanhere A, Jenner RG. Noncoding RNA localisation mechanisms in chromatin regulation. Silence. 2012;3:2. doi: 10.1186/1758-907X-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwek KY, et al. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nature structural biology. 2002;9:800–805. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- 58.Feng J, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes & development. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bond AM, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nature neuroscience. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ørom UA, et al. Long Noncoding RNAs with Enhancer-like Function in Human Cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang L, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013 doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lai F, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaikkonen MU, et al. Remodeling of the Enhancer Landscape during Macrophage Activation Is Coupled to Enhancer Transcription. Molecular cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martianov I, et al. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 65.Allen Ta, et al. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nature structural & molecular biology. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 66.Yakovchuk P, et al. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beltran M, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes & development. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Setsuie R, Wada K. The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochemistry international. 2007;51:105–111. doi: 10.1016/j.neuint.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 69.Carrieri C, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012 doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 70.Wutz A, et al. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nature genetics. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 71.Kelley D, Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biology. 2012;13:R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amaral PP, et al. lncRNAdb: a reference database for long noncoding RNAs. Nucleic acids research. 2011;39:D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pauli A, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome research. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Willingham aT, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science (New York, N.Y.) 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 77.Sun L, et al. Long noncoding RNAs regulate adipogenesis. Proceedings of the National Academy of Sciences. 2013 doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prensner JR, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nature Biotechnology. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Molecular cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ule J, et al. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods (San Diego, Calif.) 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 82.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guil S, et al. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nature Structural & Molecular Biology. 2012 doi: 10.1038/nsmb.2315. [DOI] [PubMed] [Google Scholar]
- 85.Wan Y, et al. Understanding the transcriptome through RNA structure. Nature reviews. Genetics. 2011;12:641–655. doi: 10.1038/nrg3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maenner S, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS biology. 2010;8:e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Underwood JG, et al. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nature methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lucks JB, et al. Multiplexed RNA structure characterization with selective 2'-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kertesz M, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yap KL, et al. Molecular Interplay of the Noncoding RNA ANRIL and Methylated Histone H3 Lysine 27 by Polycomb CBX7 in Transcriptional Silencing of INK4a. Molecular cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gupta Ra, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ji P, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 94.Spatz A, et al. X-chromosome genetics and human cancer. Nature reviews. Cancer. 2004;4:617–629. doi: 10.1038/nrc1413. [DOI] [PubMed] [Google Scholar]
- 95.Pageau GJ, et al. The disappearing Barr body in breast and ovarian cancers. Nat Rev Cancer. 2007;7:628–633. doi: 10.1038/nrc2172. [DOI] [PubMed] [Google Scholar]
- 96.Richardson AL, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 97.Yildirim E, et al. Xist RNA Is a Potent Suppressor of Hematologic Cancer in Mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Savarese F, et al. Hematopoietic Precursor Cells Transiently Reestablish Permissiveness for X Inactivation. Molecular and cellular biology. 2006;26:7167–7177. doi: 10.1128/MCB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Agrelo R, et al. SATB1 Defines the Developmental Context for Gene Silencing by Xist in Lymphoma and Embryonic Cells. Developmental cell. 2009;16:507–516. doi: 10.1016/j.devcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bonetta L. RNA-Based Therapeutics: Ready for Delivery? Cell. 2009;136:581–584. doi: 10.1016/j.cell.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 101.Raal FJ, et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. The Lancet. 375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 102.Modarresi F, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nature Biotechnology. 2012 doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sheridan C. Proof of concept for next-generation nanoparticle drugs in humans. Nat Biotech. 2012;30:471–473. doi: 10.1038/nbt0612-471. [DOI] [PubMed] [Google Scholar]
- 104.Zhang S, et al. Targeting MicroRNAs With Small Molecules: From Dream to Reality. Clin Pharmacol Ther. 2010;87:754–758. doi: 10.1038/clpt.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]