Abstract
The Xist long noncoding RNA (lncRNA) is essential for X-chromosome inactivation (XCI), the process by which mammals compensate for unequal numbers of sex chromosomes1-3. During XCI, Xist coats the future inactive X (Xi)4 and recruits Polycomb Repressive Complex 2 (PRC2) to the X-inactivation center (Xic)5. How Xist spreads silencing on a 150 Mb scale is unclear. Here we generate high-resolution maps of Xist binding on the X chromosome across a developmental time course using CHART-seq. In female cells undergoing XCI de novo, Xist follows a two-step mechanism, initially targeting gene-rich islands before spreading to intervening gene-poor domains. Xist is depleted from genes that escape XCI but may concentrate near escapee boundaries. Xist binding is linearly proportional to PRC2 density and H3 lysine 27 trimethylation (H3K27me3), suggesting co-migration of Xist and PRC2. Interestingly, when the Xi is acutely stripped off Xist in post-XCI cells, Xist recovers quickly within both gene-rich and -poor domains on a time-scale of hours instead of days, suggesting a previously primed Xi chromatin state. We conclude that Xist spreading takes distinct stage-specific forms: During initial establishment, Xist follows a two-step mechanism, but during maintenance, Xist spreads rapidly to both gene-rich and -poor regions.
Xist RNA is a prototype lncRNA with global epigenetic function1-3,6. The initiation of XCI depends on Xist7 and loading of the Xist-PRC2 complex at a nucleation site within the Xic8. Thereafter, Xist RNA forms a “cloud” over the X-chromosome, signaling the initiation of chromosome-wide silencing4. Concurrently, PRC2 accumulates broadly along the X-chromosome9. Although Xist RNA coats the Xi at cytological resolution, whether and where Xist binds at molecular resolution remains unknown. In one model, Xist targets PRC2 to the Xic, but outward spreading of PRC2 does not involve Xist. Alternatively, both nucleation and spread involve Xist, in which case Xist and PRC2 would co-migrate at a molecular scale.
We mapped genome-wide binding locations of Xist RNA by performing CHART-seq (Capture Hybridization Analysis of RNA Targets with deep sequencing), a technique to localize lncRNAs on chromatin using complementary oligonucleotides to enrich for DNA targets10 (Extended Data Fig. 1a). We designed a cocktail of 11 complementary oligonucleotides for Xist CHART based on conserved or functional Xist domains7,11-13 and RNase H mapping for accessibility (Extended Data Fig. 1b,c; Extended Data Table 1). Allele-specific CHART-seq was performed at four developmental stages (Extended Data Fig. 1d): Before XCI in undifferentiated female mouse embryonic stem (ES) cells (d0; <1% of nuclei XCI positive, showing an Xist cloud or H3K27me3 focus); early-XCI (d3; <10% positive); mid-XCI (d7; 40-50% positive); and post-XCI (mouse embryonic fibroblast (MEF) clone, >95% positive). About 600,000 sequence polymorphisms between the M. musculus (mus) and M. castaneus (cas) X-chromosomes enabled ~35% allele-specific mapping to Xi and Xa (active X), respectively9. Disabling the mus Tsix allele in the female ES cells ensured that the mus X will be Xi14. We validated results by comparing two independent capture oligo submixtures and an alternative 40-oligo cocktail targeting across the length of Xist (Extended Data Fig. 2a-e, Extended Data Table 1). Regions with significant Xist enrichment localized almost exclusively to Xi (>99% X-linked, p<0.001; >90% Xi-skewed, p<0.05, Extended Data Fig. 2f,g,i). On autosomes, binding was minimal and of questionable significance. Enriched segments were not complementary to capture-oligos and showed minimal enrichment on Xa of d0, d3, d7 and MEF cells. Enrichment was not observed using sense control oligos (Extended Data Fig. 2a,c). These experiments excluded artifactual enrichment, validating Xist CHART-seq specificity.
The dominant CHART peak lay in Xist exon 1 and was specific to Xi (Fig. 1a). A developmental time course demonstrated a progression in Xist density, with enriched segments increasing from <0.1% coverage of the X in pre-XCI cells, to ~20% in early- and mid-XCI, and ~54% in post-XCI cells (Fig. 1b,c, Extended Data 2h). Thus, Xist RNA not only forms a cytological cloud but also binds broad swaths of the Xi at molecular resolution. Xist could either spread uniformly along the Xi or target specific regions. Intriguingly, in cells undergoing XCI (d3, d7), Xist preferentially targeted multi-megabase domains (Fig. 1c). In post-XCI MEFs, Xist spread into intervening gene-poor regions throughout the Xi. The d3 and d7 patterns were more similar to each other than to MEF patterns (Fig. 1d, e, Extended Data Fig. 3a). Furthermore, comparative analysis identified MEF-specific domains not found during XCI (Fig. 1e). Despite heterogeneity in the onset of XCI in the ex vivo ES differentiation system, the highly similar d3 and d7 distributions show that Xist targets gene-rich domains first. Extension of ES differentiation to d10 showed statistically significant filling in of gene-poor domains (Extended Data Fig. 3b,c), though not to the extent observed in somatic cells (MEFs). We infer that full spreading across Xi may only be achieved later in development, once differentiation into somatic lineages occurs. Thus, during de novo XCI in the embryo, Xist likely follows a two-step pattern of spreading, first targeting gene-rich clusters (hereafter, “early” domains) and eventually spreading to intervening gene-poor regions (“late” domains). Throughout the process, gene bodies of escapees15,16 were depleted of Xist, but occasionally demonstrated Xist enrichment in flanking regions (Fig. 1f, Extended Data Fig. 4), suggesting boundaries that sequester Xist and prevent spreading into neighboring privileged escapee loci.
Figure 1. CHART-seq reveals a two-step mechanism of Xist spreading during de novo XCI.
a, Xist RNA is enriched on Xi. Normalized read densities displayed in mus, cas, and composite (comp) tracks. b, Coverage of enriched segments on chrX and autosomes. c, Xist coverage at indicated timepoints relative to gene silencing. Enriched segments shown beneath in gray. Brackets, y-axis scale. Xist peaks at d0 have less amplitude and density, but reflect d3 and d7 patterns, and are Xi-enriched (Extended Data Fig. 2f), consistent with initial Xist spreading to local regions, suggesting initial differentiation in a subfraction of cells. RNAseq of d7 and MEF shown below. Skewed allelic expression consistent with Xi-silencing (value −0.5 = 3-fold expression difference between Xi and Xa). d-e, Xist CHART signals (40 kb bins) from d7 correlate with d3 (d) and MEF (e)(see Extended Data Fig. 3). Regions showing >10-fold differences after normalization are colored purple and displayed on the X (lower panels). f, Depletion of Xist at a representative escapee. g, Xist preferentially targets genes in active chromatin (H3K4me3-marked on d7). Xist densities shown for gene bodies of active (n=532), inactive (n=475), and escapee genes (n=10). Medians are indicated. Individual data points overlaid on boxplot; error bars, 1.5-fold interquartile range. *p<0.05, **p<1×10−8, ***p<2.2×10−16 , Mann-Whitney U tests. h, Xist RNA distribution from d7 cells relative to chromatin features.
We investigated what might target Xist to early domains by comparisons with various chromatin features (see Methods)9,17-22. Interestingly, Xist is more likely to target genes in regions of active chromatin in ES cells. Allele-specific RNA-seq analysis demonstrated Xist's preference for genes that are active (e.g., on the Xa and in d0 and d7 cells) and showed skewed expression in d7 ES cells and in MEFs (Fig. 1c, g, Extended Data Fig. 5a; p<2.2×10−16; Pearson's r=0.46). Furthermore, there were positive correlations with DNase I hypersensitive sites (Pearson's r=0.43), SINEs (r=0.55), and early replicating regions (r=0.45; Fig. 1h, Extended Data Fig. 5b). Strong anti-correlations were observed with LTRs (r=−0.39), LINE1 (r=−0.54), and lamin-associated domains (LADs, r=−0.48)23. Xist partitioning did not correlate with cytogenetic banding on the X-chromosome (Fig. 1h)19,24. LINE1s have been proposed as spreading elements25, but repetitive reads from Xist CHART-seq aligning to LINE1 were not enriched over input (Extended Data Fig. 5c). Xist's localization showed modest positive correlation with Xic looping contacts inferred from HiC (high-throughput chromosome conformation capture)18 via an anchor within the Xist locus (Fig. 1h, Extended Data Fig. 5b). Together, these data support a role for open chromatin in guiding Xist, with Xist coming into contact with gene-rich regions (early domains) first, and spreading secondarily to more distal gene-poor inter-regions (late domains).
Given co-nucleation of Xist and PRC2 at the Xic8, we asked whether Xist continues to associate with PRC2 during spreading. Comparison of Xist, EZH2, and H3K27me3 enrichment revealed strikingly similar chromosome profiles across time (Fig. 2a, Extended Data Fig. 6a-c). By contrast, PRC2 and H3K27me3 densities on Xa did not correlate with Xist, nor did those on Chr13, a representative autosome (Extended Data Fig. 6a). Consistent with the idea that Xist directs PRC2 localization onto Xi5, Xist densities demonstrated an extensive linear relationship with EZH2 and its product H3K27me3 across the X in mid-XCI but not pre-XCI cells (Fig. 2b). Correlation with the H3K4me3 control (active mark) was poor. In MEFs, densities of H3K27me3 and Xist remained highly correlated, while reduced densities of PRC2 were observed during maintenance. Interestingly, Xist densities were not necessarily greater at previously defined “PRC2 strong sites”9 (Extended Data Fig. 6d,e); instead, Xist densities showed a general correlation with Xi-specific PRC2 enrichment (Extended Data Figs. 6d,h; 5b). This supports the idea that strong sites are Xist-independent (as indeed they are present in d0 cells9) and indicate that Xist and PRC2 co-migrate to new regions within the early domains on the Xi.
Figure 2. Co-spreading of Xist RNA and PRC2.
a, Normalized read densities of Xist, EZH2, and H3K27me3 on chrX in d7 cells. b, Xist densities (200 kb bins) correlated with EZH2, H3K27me3 and H3K4me3 signals at different stages of XCI. Pearson's r displayed. EZH2/H3K27me3 R2 values: 0.3/0.37 for d0, 0.77/0.88 for d7, and 0.23/0.66 for MEFs, respectively. H3K4me3 R2 values: < 0.15 across all samples.
We then asked if localization mechanisms were inherent to Xist RNA or chromatin context. In perturbation experiments, we stripped away Xist RNA and observed recovery on the Xi of MEFs at 1h, 3h, and 8h. Locked nucleic acids (LNA) directed against Xist RNA's Repeat C prevented nucleation and therefore spreading13. RNA fluorescence in situ hybridization (FISH) showed that LNA-4978 did not overtly perturb Xist at 1h, but led to full Xist displacement by 3h, with Xist reassociation at 8h (Fig. 3a). As reassociation requires newly synthesized Xist rather than relocalization of displaced Xist13, reassociation must depend on outward spreading of new RNA from the Xic, just as during XCI establishment.
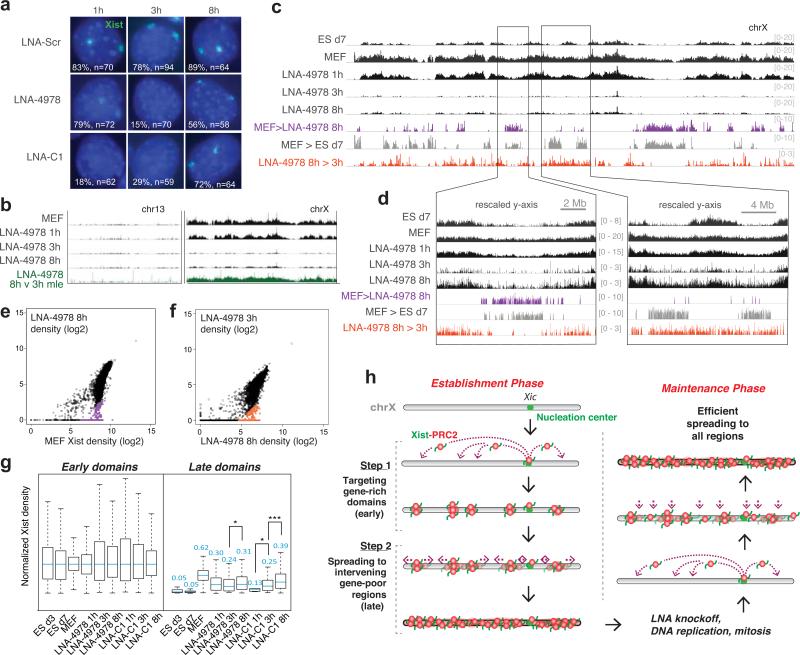
Figure 3. Xist knockoff uncovers a distinct spreading method during the maintenance phase.
a, RNA FISH shows depletion and recovery of Xist RNA (green) in MEF cells after Xist knockoff. %nuclei with Xist clouds and sample size (n) shown. Scr, scrambled LNA. b, Chromosome-wide recovery of Xist after LNA-4978 knockoff on chrX and chr13. Regions of recovery comparing 8h over 3h LNA-4978 were determined using a maximum likelihood enrichment estimate29. c,d, Xist knockoff and recovery across chrX. Colored regions show >10-fold median-normalized differences between samples. (c) Entire chrX (d) zoom of one region with more late domain recovery (right) than the other (left). e,f, Xist CHART signals (40 kb bins) from LNA-4978 8h correlated with MEF (e); LNA-4978 3h correlated against LNA-4978 8h (f). Regions showing >10-fold differences after normalization are colored as shown in panel d. g, Xist recovery in indicated samples, with 40-kb binned Xist densities normalized to median levels of early domains of each sample, to determine how early and late domains recover from knockoff compared to during de novo XCI. Normalized median values for each sample indicated above box. *, p<0.05; ***, p<10−8, Wilcoxon test, as in Extended Data Figs. 7c, 3c. h, Model: distinct methods of Xist spreading during establishment and maintenance.
Interestingly, however, CHART-seq revealed a pattern not evident cytologically by RNA FISH. At 1h, when Xist was still visualized on Xi (Fig. 3a), CHART-seq demonstrated a relative loss in late domains (Fig. 3b-d), suggesting that Xist binds more weakly to gene-poor than gene-rich regions and consistent with Xist's banded pattern on the metaphase Xi observed cytologically24. At 3h, Xist was strongly depleted from both regions. At 8h, partial recovery was evident in both regions. However, unlike spreading during de novo XCI (d3, d7), spreading of Xist during the somatic maintenance phase (MEF) did not follow a two-step process, as Xist reassociation in early and late domains occurred simultaneously (Fig. 3b-d). Therefore, spreading during de novo XCI was restricted to early domains and occurred on a time-scale of days in the ex vivo system; in contrast, re-covery and respreading in post-XCI cells occurred more generally in both domains and on a time-scale of hours. This quantitative difference is significant, with accumulation in late domains appearing on the same time-scale as early domains during the recovery period after Xist knockoff (Fig. 3e-g; Extended Data Figs. 7, 8). Similar results were observed using an independent LNA, LNA-C1, targeted to a different sequence in the Repeat C region and in multiple replicates (Fig 3a, Extended Data Figs. 2d, 3a, 7, 8). Despite LNA-C1 being faster-acting13 (Fig. 3a), LNA-C1 and LNA-4978 treatment resulted in remarkably similar Xist knockoff and recovery on Xi.
Taken together, these data argue for distinct mechanisms of Xist spreading during establishment (de novo XCI) in early embryonic cells, when spreading occurs in a two-step fashion (early to late domains), and during maintenance in somatic cells, when Xist spreads more generally into both early and late domains (Fig. 3h). The Xi may retain epigenetic memory of Xist26, enabling more efficient spreading during maintenance. As Xist mostly dissociates from the Xi during mitosis4, epigenetic memory could facilitate Xist's resynthesis and re-spreading in G1, and duplication of Xist patterns after DNA replication. Indeed, Xist's continued action is essential for maintenance of XCI27. In summary, we have illuminated the mechanism by which Xist spreads on a 150 Mb scale. Comparing localization dynamics of Xist relative to other lncRNAs (Hasiculeyman and Rinn, personal communication) and 3D conformations28 may prove highly informative for understanding general mechanisms of RNA-directed chromatin change.
METHODS SUMMARY
Cells were grown as previously described9. RNA was isolated and sequenced using a modified adaptor ligation protocol, and Xist localization determined by CHART10 with several modifications as detailed in the extended Methods. LNAs that target Xist were previously described13.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Overton, I. Tikhonova, and A. Lopez of Yale Center for Genome Analysis, and the MGH Bioinformatics Core. We also acknowledge funding from the Deutsche Forschungsgemeinschaft to S.F.P. and V.K.M, the MGH Fund for Medical Discovery to S.F.P., and the NIH [F32-GM090765 to K.S., RO1-GM043901 to R.E.K., and RO1-GM090278 to J.T.L.]. J.T.L. is an Investigator of the HHMI.
Footnotes
AUTHOR CONTRIBUTIONS
R.E.K., J.T.L., M.D.S. and S.F.P. designed the experiments; S.F.P., K.S. and R.F. performed cell culture; R.F. and M.D.S. established CHART conditions and performed CHART; S.K.B. prepared CHART libraries; K.S. performed LNA knockoffs; S.F.P. and V.K.M. performed RNA-seq; S.F.P. performed bioinformatic analyses with M.D.S., M.R.S., R.F. and B.A.K.; M.D.S., S.F.P., R.F., J.T.L., and R.E.K. interpreted the results; J.T.L, M.D.S, R.E.K, S.F.P. and R.F. wrote the manuscript.
DATA DEPOSITION
CHART-seq and RNA-seq datasets have been deposited in GEO (accession number GSE48649).
The authors declare no competing financial interests.
REFERENCES
- 1.Disteche CM. Dosage compensation of the sex chromosomes. Annu Rev Genet. 2012;46:537–560. doi: 10.1146/annurev-genet-110711-155454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12(8):542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 3.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 4.Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132(3):259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pontier DB, Gribnau J. Xist regulation and function explored. Hum Genet. 2011;130(2):223–236. doi: 10.1007/s00439-011-1008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 8.Jeon Y, Lee JT. YY1 Tethers Xist RNA to the Inactive X Nucleation Center. Cell. 2011;146(1):119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinter SF, Sadreyev RI, Yildirim E, Jeon Y, Ohsumi TK, Borowsky M, Lee JT. Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res. 2012 doi: 10.1101/gr.133751.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 2011;108(51):20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71(3):515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 12.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30(2):167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 13.Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci U S A. 2010;107:22196–22201. doi: 10.1073/pnas.1009785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogawa Y, Sun BK, Lee JT. Intersection of the RNA Interference and XInactivation Pathways. Science. 2008;320(5881):1336–1341. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berletch JB, Yang F, Xu J, Carrel L, Disteche CM. Genes that escape from X inactivation. Hum Genet. 2011;130(2):237–245. doi: 10.1007/s00439-011-1011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in Xlinked gene expression in females. Nature. 2005;434(7031):400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 17.Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y, Kaaij LJ, van Ijcken W, Gribnau J, Heard E, de Laat W. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25(13):1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chadwick BP, Willard HF. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc Natl Acad Sci U S A. 2004;101(50):17450–17455. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chadwick BP, Willard HF. Chromatin of the Barr body: histone and non-histone proteins associated with or excluded from the inactive X chromosome. Hum Mol Genet. 2003;12(17):2167–2178. doi: 10.1093/hmg/ddg229. [DOI] [PubMed] [Google Scholar]
- 21.Marks H, Chow JC, Denissov S, Francoijs KJ, Brockdorff N, Heard E, Stunnenberg HG. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009;19(8):1361–1373. doi: 10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calabrese JM, Sun W, Song L, Mugford JW, Williams L, Yee D, Starmer J, Mieczkowski P, Crawford GE, Magnuson T. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell. 2012;151(5):951–963. doi: 10.1016/j.cell.2012.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152(6):1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Duthie SM, Nesterova TB, Formstone EJ, Keohane AM, Turner BM, Zakian SM, Brockdorff N. Xist RNA exhibits a banded localization on the inactive X chromosome and is excluded from autosomal material in cis. Hum Mol Genet. 1999;8(2):195–204. doi: 10.1093/hmg/8.2.195. [DOI] [PubMed] [Google Scholar]
- 25.Lyon MF. The Lyon and the LINE hypothesis. Semin Cell Dev Biol. 2003;14(6):313–318. doi: 10.1016/j.semcdb.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2(7):E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev R, Scadden DT, Lee JT. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26(12):1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wernersson R, Nielsen HB. OligoWiz 2.0--integrating sequence feature annotation into the design of microarray probes. Nucleic Acids Res. 2005;33:W611–615. doi: 10.1093/nar/gki399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flickinger JL, Gebeyehu G, Buchman G, Haces A, Rashtchian A. Differential incorporation of biotinylated nucleotides by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 1992;20:2382. doi: 10.1093/nar/20.9.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yildirim E, Sadreyev RI, Pinter SF, Lee JT. X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat. Struct. Mol. Biol. 2012;19:56–61. doi: 10.1038/nsmb.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowman SK, et al. Multiplexed Illumina sequencing libraries from picogram quantities of DNA. BMC Genomics. 2013;14:466. doi: 10.1186/1471-2164-14-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. doi:10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang W, et al. Efficiently identifying genome-wide changes with next-generation sequencing data. Nucleic Acids Research. 2011;39:e130. doi: 10.1093/nar/gkr592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji X, Li W, Song J, Wei L, Liu XS. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 2006;34:W551–554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. doi:10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hiratani I, et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peric-Hupkes D, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuhn RM, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.