Abstract
In the US alone, around 60,000 lives/year are lost due to colon cancer. Diet and environment have been implicated in the development of sporadic colon tumors. The objective of this study was to determine how dietary fat potentiates the development of colon tumors through altered B(a)P biotransformation, using the Adenomatous polyposis coli with Multiple intestinal neoplasia mouse model. Benzo(a)pyrene was administered to mice through tricaprylin, and unsaturated (USF; peanut oil) and saturated (SF; coconut oil) fats at doses of 50 and 100 μg/kg via oral gavage over a 60-day period. Blood, colon, and liver were collected at the end of exposure period. The expression of B(a)P biotransformation enzymes [cytochrome P450 (CYP)1A1, CYP1B1 and glutathione-S-transferase] in liver and colon were assayed at the level of protein, mRNA and activities. Plasma and tissue samples were analyzed by reverse phase high-performance liquid chromatography for B(a)P metabolites. Additionally, DNA isolated from colon and liver tissues was analyzed for B(a)P-induced DNA adducts by the 32P-postlabeling method using a thin-layer chromatography system. Benzo(a)pyrene exposure through dietary fat altered its metabolic fate in a dose-dependent manner, with 100 μg/kg dose group registering an elevated expression of B(a)P biotransformation enzymes, and greater concentration of B(a)P metabolites, compared to the 50 μg/kg dose group (P<.05). This effect was more pronounced for SF group compared to USF group (P<.05). These findings establish that SF causes sustained induction of B(a)P biotransformation enzymes and extensive metabolism of this toxicant. As a consequence, B(a)P metabolites were generated to a greater extent in colon and liver, whose concentrations also registered a dose-dependent increase. These metabolites were found to bind with DNA and form B(a)P-DNA adducts, which may have contributed to colon tumors in a subchronic exposure regimen.
Keywords: Benzo(a)pyrene, Polycyclic aromatic hydrocarbons, ApcMin mouse, Colon cancer, Biotransformation, Metabolites, B(a)P-DNA adducts
1. Introduction
Colorectal cancer (CRC) is one of the most common cancers in the Western world. In the United States alone, nearly 143,460 new cases of CRC have been diagnosed recently and 51,690 will die of this cancer [1]. People get colon cancer through hereditary (familial) and sporadic means. Sporadic gene mutation seems to play an important role in the development of tumors in 90% of the colon cancer cases in which there is no familial history of colon cancer [2]. Dietary factors and environmental agents have been suspected of causing sporadic gene mutations and therefore involved in the induction of sporadic colon carcinomas [3]. Global cancer statistics reveal that this cancer is a major cause of cancer deaths in the western world [4].
Specific components of western diet including consumption of meat (particularly red and/or well-done meat) and dietary fat (particularly polyunsaturated and saturated fatty acids) have been proposed as risk factors that influence susceptibility to colorectal cancer [5,6]. An overwhelming epidemiological evidence indicates that red meat intake and excessive adiposity increase susceptibility to colorectal neoplasia [7–9].
Of the several environmental chemicals reported to contribute to toxicity of the gastrointestinal system, the polycyclic aromatic hydrocarbons (PAHs) have garnered a lot of interest as they are formed in barbecued meat [10–12]. In addition to their formation during cooking, PAHs are also emanated from environmental [13,14] and occupational [15,16] sources, thus contributing significantly to dietary contamination, intake and development of CRC in humans [17,18]. The concentrations of PAHs found in products of plant and animal origin have extensively been reviewed and the intake ranged from 0.02 to 3.6 μg per person per day [10]. Grilled and barbecued meats were reported to contain high levels of benzo(a)pyrene [B(a)P; a prototypical PAH compound] compared to pan-fried and boiled foods [19] and contributes to 21% of mean daily intake of B(a)P [20].
Epidemiological studies provide evidence for a consistent interaction between PAH-associated fatty diet/red meat intake and CRC development. Findings from a clinic-based case–control study bolster the hypothesis that dietary intake of PAHs is associated with CRC risk [21]. Using the same study design and a sample size of about 4000 adenoma cases, this research group [22,23] also showed that consumption of well-done red meat was associated with increased risks for colon adenomas. Similar to the above-mentioned studies, another sigmoidoscopy- based study (275 CRC cases) reported an association among high intake of barbecued red meat, B(a)P, and colorectal adenomas [24]. In a colonoscopy study that involved more than 2500 subjects, a statistically significant dose–response relationship between adenoma incidence in colon and dietary exposure to B(a)P was revealed [6]. In another study involving 370 cases of CRC, high intake of B(a)P was associated with an increased risk of CRC among individuals carrying the CT genotype for UGT1A (UDP-glucuronosyltransferase1A), a phase II enzyme involved in the detoxification of B(a)P. Additionally, correlation between total mutagenic activity and adenomas was found to be high for B(a)P [25]. A recent study that comprised of 1008 subjects revealed an elevated risk of rectal adenoma (early neoplasia) in association with B(a)P intake through meat [26].
The aforementioned epidemiological studies have already established an association between PAH intake and incidence of CRC in human populations. However, studies in animal models are warranted to replicate phenotypic manifestation of the disease most similar to that of humans and to identify the mechanisms of environmental toxicant-induced colon cancers. Of the rodent models, Adenomatous polyposis coli with Multiple intestinal neoplasia (ApcMin) has been widely used to study the onset and progression of gastrointestinal cancers. In this mouse, the loss of a single copy of the tumor suppressor gene APC predisposes the mice to the development of a large number of polyps in the intestine [27], which grow into full-blown when challenged with chemical carcinogens. Studies from our laboratory have already shown the incidence of colon tumors caused by B(a)P in ApcMin mouse with more tumors occurring in mice that received B(a)P through saturated fat (SF) compared to unsaturated fat (USF) [28].
Biotransformation plays a pivotal role in the conversion of chemical carcinogens into reactive species that damage cellular macromolecules, interfere with signaling pathways and cause cancer [29–31]. Hence, it is conceivable that colorectal cancers are promoted by the increased intake of PAHs through dietary fat that in turn influences the biotransformation and metabolic processing of toxic chemicals. Therefore, the objective of this study was to examine whether dietary lipid type and administered dose levels of B(a)P will alter the biotransformation of this toxicant in this mouse model. In this study we report that the biotransformation enzyme [cytochrome P450 (CYP)1A1, 1B1 and glutathione-S-transferase (GST)] activities and expression was pronounced when B(a)P was administered through saturated fat, relative to its administration through unsatu-rated fat or tricaprylin. Additionally, the biotransformation profiles, products of biotransformation [B(a)P metabolites], and B(a)P-DNA adducts (markers of premutagenesis) were found to be B(a)P dose-dependent. Our studies indicate a relationship between biotransformation and colonic tumors in this mouse model.
2. Materials and methods
2.1. Chemicals
Benzo(a)pyrene (CAS No. 50-32-8; 98% pure), peanut oil, coconut oil, sodium dodecyl sulfate (SDS), Ponceau S solution and tricaprylin were purchased from Sigma-Aldrich Chemical Company (St. Louis, MO, USA). Lithium chloride, urea, sodium phosphate (monobasic and dibasic), methanol, chloroform, ethanol and 10% formalin, isopropyl alcohol were purchased from Fisher Scientific Company (Kennesaw, GA). Sucrose, EDTA and Tris–HCl were purchased from Curtin Matheson Scientific (Houston, TX, USA). The CYP1A1, CYP1B1, actin, and rabbit anti-goat IgG–horseradish peroxodase (HRP) antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). The GST-P antibody was purchased from Assay Designs (Ann Harbor, MI, USA). The Quick Start Bradford Protein Assay Kit, ethidium bromide, Precision Plus Protein blue standards, tetramethyl-ethylenediamine, ammonium persulfate, 30% acrylamide and bis-acrylamide solution, Laemmli sample buffer, agarose, EZ load 100 bp molecular ruler, Immun-Star HRP substrate and 2-mercaptoethanol (βME) were purchased from Bio-Rad Laboratories (Richmond, CA, USA). The HyGLO ECL Spray Chemiluminescent HRP antibody detection reagent was purchased from Denville Scientific (Metuchen, NJ, USA). Easy-DNA kit, RNase/DNase-free water and Trizol reagent were purchased from Invitrogen (Carlsbad, CA, USA). The magnesium chloride, Go Taq Flexi DNA Polymerase, random primers, reverse transcription buffer, deoxynucleotide triphosphates (dNTPs), RNase inhibitor, avian myeloblastosis virus reverse transcriptase, Tris/Borate/EDTA buffer, and Tris-acetate-EDTA buffer were purchased from Promega (Madison, WI, USA). The CYP1A1 and 1B1 enzyme assay kits were purchased from Promega (Madison, WI, USA). The GST assay kit was purchased from Biovision (Mountain View, CA, USA). HeLa whole cell lysate (positive control for CYP1A1) and mouse kidney extract (positive control for CYP1B1) were obtained from Santa Cruz Biotechnology. Mouse kidney extract (positive control for GST) was also purchased from Assay Designs.
2.2. Animal exposure to benzo(a)pyrene
Six-week-old male ApcMin mice (Jackson Labs, Bar Harbor, ME, USA) weighing approximately 24 g were used in this study. The mice were housed in groups of 2–3 per cage, maintained on a 12/12 hour light/dark cycle (lights on at 0600 hour) and allowed free access to rodent chow (2016 Teklad Global 16% protein rodent diet [3.5% fat]; Harlan Laboratories, Indianapolis, IN, USA) and water. All animals were allowed a seven-day acclimation period prior to being randomly assigned to a control (n=7) or treatment group (n=7). Mice were housed in polycarbonate cages (Lab Products, Seaford, DE, USA) with Soft Cellulose bedding (laboratory grade 7089 Teklad; Harlan Laboratories, Indianapolis, IN, USA) in Meharry animal care facility. The animal care facility is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International and is under the oversight of Institutional Animal Care and Use Committee (IACUC). The Meharry IACUC ensures that animal-related experiments adhere to the National Institutes of Health (NIH) guidelines for the humane care and use of laboratory animals [32].
Two doses (50 and 100 μg/kg) of B(a)P (97% pure, Sigma Chemical, St. Louis, MO, USA) dissolved in peanut and coconut oils (representatives of unsaturated and saturated fat respectively; research grade, Sigma) were administered to mice. Mice from the control groups received only the oils without B(a)P. Mice, which received no oils and B(a)P were grouped as negative controls, whereas mice that received no oils, but administered B(a)P through tricaprylin [vehicle for B(a)P] were grouped as positive controls. Benzo(a)pyrene was administered to mice daily via a single oral gavage (200 μl/animal) for 60 days. The rationale for choosing B(a)P doses and dosing vehicles were mentioned elsewhere [28].
Since B(a)P and its metabolites are fall under the category of human carcinogens Group I, designated by the International Agency for Research on Cancer [33], they were handled according to US Environmental Protection Agency health effects testing guidelines (40CFR 798) and NIH [34] guidelines. In addition to wearing the appropriate protective equipment which includes a laboratory coat, gloves and mask, all handling was conducted in an exhaust hood and in subdued light to prevent photodegradation of B(a)P. Any unused B(a)P was placed in a hazardous waste container for proper disposal.
2.3. Tissue sample collection
After the above mentioned period of exposure, the mice were sacrificed post induction of anesthesia. The chemicals used for anesthesia were pharmaceutical grade ketamine (80 mg/kg) and xylazine (16 mg/kg) administered via intraperitoneal injection. Blood, colon and liver tissues were collected, transferred into cryovials and stored frozen in liquid nitrogen until analyzed.
2.4. RNA Isolation
Tissues (colon or liver) weighing 100 mg were homogenized in 1 ml of Trizol reagent using a Polytron PT 3100 homogenizer (Capitol Scientific, Austin, TX, USA). The homogenate was centrifuged at 12,000×g for 10 min using a Sorvall Legend Micro 17 R rotor (DJB Labcore Ltd, Buckinghamshire, England). Two hundred microliters of chloroform was added to the supernatant, mixed well and incubated at room temperature for 3 min. Samples were then centrifuged at 12500×g for 15 min at 4°C to promote separation of the sample into three phases. The upper aqueous phase containing the RNA was transferred to a fresh mircofuge tube and the RNA was precipitated by mixing with 500 μl of isopropyl alcohol. Samples were then incubated at room temperature for 10 min and centrifuged at 12500×g for 10 min at 4°C to precipitate the RNA. Then RNA pellet was then washed once by vortexing in 1 ml of 75% ethanol and centrifuged at 12500×g for 20 min at 4°C. The ethanol was decanted, and the RNA pellet was allowed to dry to remove any residual ethanol. After the pellet was dried, the RNA was redissolved in RNase-DNase free water and heated for 10 min in a 55 °C water bath. The RNA concentration and integrity was determined using a Nanodrop Spectrophotometer (Thermo Scientific, Wilmington, DE, USA), and RNA was stored at –20°C.
2.5. Reverse transcriptase polymerase chain reaction (RT-PCR)
Total RNA was isolated using Trizol reagent as previously described. The cDNA and production of PCR amplification from 1 μg of RNA were synthesized using QIAGEN One-Step RT-PCR kit (QIAGEN Inc, Valencia, CA, USA). The final concentrations of reagents in the PCR system were as follows: 1X QIAGEN One-Step RT-PCR buffer, 400 μM of each dNTP, 20 pmol forward and reverse primer mix, 1× Q-solution (containing MgCl2), an enzyme mix containing Omniscript and Sensiscript Reverse transcriptase for reverse transcription and HotStarTaq polymerase for PCR, and RNase–DNase-free water to yield a final reaction master mix volume of 24 μl. The reaction tube was gently mixed by tapping and then briefly spun down. Twenty-four microliters of reaction master mix was added to 1 μl (1 μg/μl) of each sample and mixed by tapping and briefly spun. The primers used for amplification of CYP1A1, CYP1B1, GSTP1 and 18 s were listed in Table 1.
Table 1.
Primers used for amplification of CYP1A1, CYP1B1, GST and 18 sRNA
| Gene | Primer | Length (BP) | NCBI Reference Sequence |
|---|---|---|---|
| CYP1A1 | 5'-TATBACCATGATGACCAAGAGC−3'forward 5'-TAACGGAGGACAGGAATGAAGT−3' reverse | 107 | NM_09992.3 |
| CYP1B1 | 5'-CCCCATAGGAAACTGCAGTAAG−3' forward 5'-AAGCAAGCTGTCTCTTGGTAGG−3' reverse | 115 | NM_009994.1 |
| GSTP1 | 5'-GGCAAATATGTCACCCTCATCT−3' forward 5'-AGCAGGTCCAGCAAGTTGTAAT−3' reverse | 176 | NM_013541.1 |
| 18s | 5'-GAATGGTGCTACCGGTCATT−3' forward 5'-ACCTCTCTTACCCGCTCTCC−3' reverse | 193 | NR_003278 |
Cycling conditions for the RT-PCR were done in one step. Reverse transcription and initial PCR activation were 1 cycle (30 min at 50° and 15 min at 95°). The PCR cycle for CYP1A1, 1B1 and GSTP1 was as follows: 40 cycles (each 1 min at 95°, 1 min at 55° and 1 min at 72°) and 1 cycle of 10 min at 72°. For 18 s, the PCR cycle was as follows: 30 cycles (each 1 min at 95°, 1 min at 55° and 1 min at 72° as an internal reference to control for variability in all steps leading up to PCR amplification. PCR products were visualized in 2% agarose gels stained with ethidium bromide and visualized and captured in IVP Bio-Imaging System using LabWorks Image Acquisitions Analysis Software (UVP Laboratory products, Upland, CA, USA). Densitometric scores for CYP1A1, 1B1 and GSTP1 were calculated and normalized against values for 18sRNA as an internal standard.
2.6. Protein isolation and assay
Colon or liver tissues were washed in PBS and weighed to obtain 100 mg for analysis. RIPA Buffer 9× vol (900 μl) containing a protease inhibitor was added to samples. Tissues were homogenized on ice for 1 minute until the tissue has been disrupted. Samples were centrifuged at 12×1000×g at 4°C for 10 min. The supernatant containing protein was transferred to fresh microcentrifuge tube and stored at –20°C. Protein concentration was determined by using the Pierce bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Rockford, IL, USA).
2.7. Preparation of protein and Western blots
Protein samples were mixed 1:1 with Laemmli Sample Buffer (BioRad, Hercules, CA, USA) and boiled for 10 min for denaturation. Equivalent amounts of protein (15 μg) were loaded into each lane of Ready Gel 4-20% precast Gel (BioRad, Hercules, CA, USA) and separated by SDS polyacrylamide gel electrophoresis at 100 V for 1 h. The protein bands were transferred onto a Sequi-Blot polyvinyldiene difluoride (PVDF) membrane (BioRad, Hercules, CA, USA) at 80 V for 1 h. The protein bands were visualized by Ponceau stain to assess the accuracy of protein loading. The PVDF membranes were blocked at room temperature for 1 hour in 5% non-fat milk in TBST (100 mM Tris–HCl, 1.5 M NaCl, 0.5% Tween-20) with subsequent incubation with anti-CYP1A1 (200 μg/ml), CYP1B1, or GSTP1 overnight. After extensive washing, blots were incubated with the corresponding HRP-coupled secondary antibody (1:10,000) for 1 h followed by additional washing in TBST and detected using ECL western blotting detection system (Denville Scientific, Metuchen, NJ, USA). Immunoreactive bands were quantified using UN-SCAN IT analysis software (Silk Scientific, Orem, UT, USA). Subsequently, blots were probed with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies as a loading control.
2.8. Drug metabolizing enzyme assays
The GST assay was performed using the BioVision (Mountain View, CA, USA) GST assay kit. Microsomes were isolated using the Endoplasmic Reticulum Isolation kit (Sigma-Aldrich, St. Louis, MO, USA). The P450-Glo assay (Promega, Madison, WI, USA) was used to assay for CYP1A1 and 1B1 enzyme activities in microsomes of liver and colon.
2.9. B(a)P metabolite disposition, extraction and analysis from plasma, colon and liver tissues
Blood (plasma), colon and liver samples harvested from control, B(a)P-exposed (administered through tricaprylin, and dietary fat) were processed for analysis of B(a)P metabolites by liquid–liquid extraction and reverse-phase high-performance liquid chromatography coupled with fluorescence detection methods as described previously in Ramesh et al. [35].
2.10. DNA isolation
DNA was isolated from liver and colon tissues of ApcMin mice exposed to B(a)P through USF and SF. DNA isolation was done by using Invitrogen DNA-easy kit. Briefly, 100 mg of tissue was homogenized in 350 μl of lysis buffer and incubated at 65°C for 10 min. Each homogenate was then placed on ice and 150 μl of precipitation reagent, and 500 μl of chloroform was added to homogenate. Samples were vortexed for about 1 min and centrifuged at 13×1000×g at 4°C for 20 min. The supernatant was transferred to a new centrifuge tube, and 1 ml of 100% ethanol (stored at –20 °C) was added to it and stored on ice for 30 min. After 30 min, the supernatant was subjected to centrifugation at 13×1000×g for 15 min at 4 °C. The ethanol was removed and 500 μl of 80% ethanol (stored at –20°C) was added; the samples were mixed by inverting the tube 5 times and subjected to centrifugation at 13×1000×g for 5 min at 4°C. Following centrifugation, the ethanol was again removed, and samples were centrifuged briefly to collect any residual ethanol. Residual ethanol was removed by pipetting and samples were air dried for 8 min. Each DNA pellet was resuspended in 100 μl TE buffer. The concentration of DNA in the tissues was determined by using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The DNA yield in tissues was ≥3 μg/μl.
2.11. 32P-Postlabeling of DNA and Identification of B(a)P–DNA adducts
The methods of Gupta and Randerath [36] were adopted for analysis of DNA adducts. The B(a)P–DNA adducts were identified as detailed in Walker et al. [37].
2.12. Statistical treatment of data
To evaluate the contribution of the two experimental variables [B(a)P doses and dietary fat types) on drug metabolizing enzymes for mRNA transcription, protein, enzyme activities, B(a)P metabolite and B(a)P-DNA adduct concentrations; the data were analyzed using a 2-way analysis of variance with repeated measures, and differences among means were tested with the Newman-Keuls post-test. This approach was used to discern if there are any data interactions associated with the experimental approach.
3. Results
3.1. Benzo(a)pyrene treatment-related effects on drug metabolizing enzyme transcription, expression and activity in liver
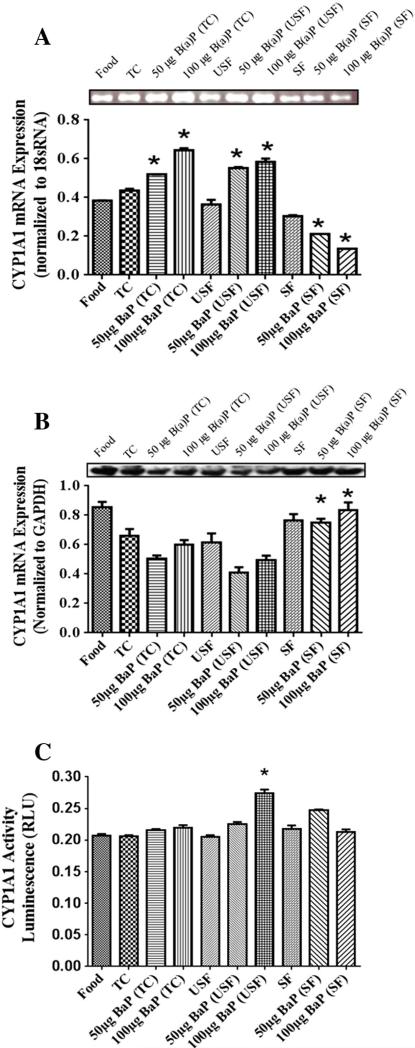
To determine RNA levels of the phase I (CYP1A1 and 1B1) and phase II (GST) enzymes, RNA was isolated from liver tissue and subjected to RT-PCR analysis. The mRNA for CYP1A1 showed that B(a) P increased CYP1A1mRNA in the B(a)P treatment groups in comparison to their respective vehicles except in the SF group (Fig. 1A). There was a higher CYP1A1 mRNA in B(a)P+tricaprylin (TC) groups compared to B(a)P+USF and B(a)P+SF. In addition, CYP1A1 expression was high in B(a)P+USF group in comparison to B(a)P+SF group.
Fig. 1.
(A) RT-PCR analysis of liver CYP1A1 mRNA expression in ApcMin-exposed mice. Mice were exposed to either vehicles (TC, USF or SF) or B(a)P (50 μg TC, 100 μg TC, 50 μg USF, 100 μg USF, 50 μg SF and 100 μg SF) for 60 days via oral gavage. The relative expression of CYP1A1 was quantified by densitometric quantitation of CYP1A1 bands and normalized to level of 18 sRNA. The bars represent mean±S.D. for three independent experiments (n = 7 animals for this and all other assays unless otherwise mentioned). *P<.005, when B(a)P+TC is compared to B(a)P+USF and B(a)P+SF; and when B(a)P+USF is compared to B(a)P+SF; 100 μg B(a)P+TC and 50 μg B(a)P+SF are compared to 50 μg B(a)P+TC and 100 μg B(a)P+SF, respectively; and when SF control is compared to 50 μg B(a)P+SF and 100 μg B(a)P+SF. (B) Western blot analysis of liver CYP1A1 protein expression in ApcMin-exposed mice. The relative expression of CYP1A1 was quantified by densitometric quantitation of CYP1A1 bands and GAPDH was used as the loading control. The bars represent mean±S.D. for three independent experiments. *P<.005, when B(a)P+SF is compared to other B(a)P treatment groups. (C) Enzyme activity of liver CYP1A1 in ApcMin-exposed mice. The activity of CYP1A1 was determined using luminescence detection. The bars represent mean±S.D. for three independent experiments.
The mRNA expression was greater in the 100 μg B(a)P+TC and 50 μg B(a)P+SF groups compared to 50 μg B(a)P+TC and 100 μg B(a) P+SF groups, respectively. The SF control group had higher transcription compared to 50 μg B(a)P+SF and 100 μg B(a)P+SF groups individually. Western blot analysis showed (Fig. 1B) highest expression of CYP1A1 in the B(a)P+SF group compared to the other B(a)P treatment groups (P<.05; dose×treatment interaction). The CYP1A1 enzyme activities (Fig. 1C) were evident in all treatment groups. There was an increase in activity in the B(a)P+USF treatment groups compared to their USF control and a dose-dependent increase in the 100 μg B(a)P+USF group compared to 50 μg B(a)P+USF group. On the other hand, the 50 μg B(a)P+SF group had higher activity compared to 100 μg B(a)P+SF group.
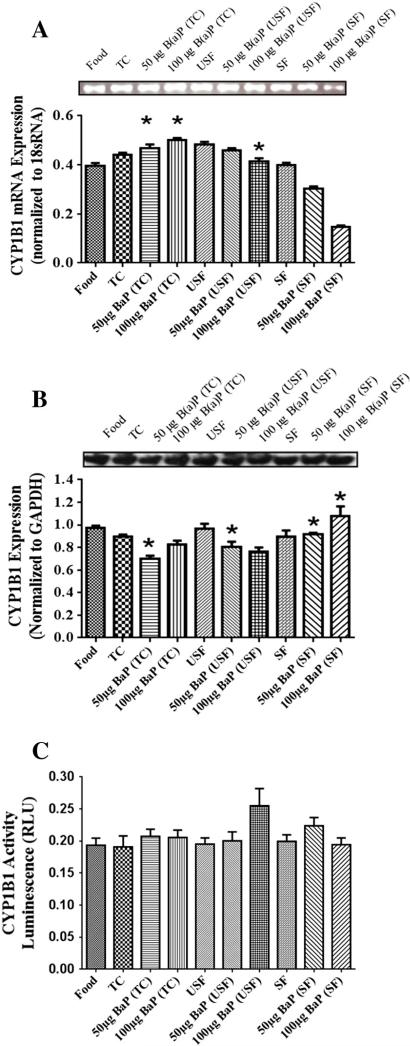
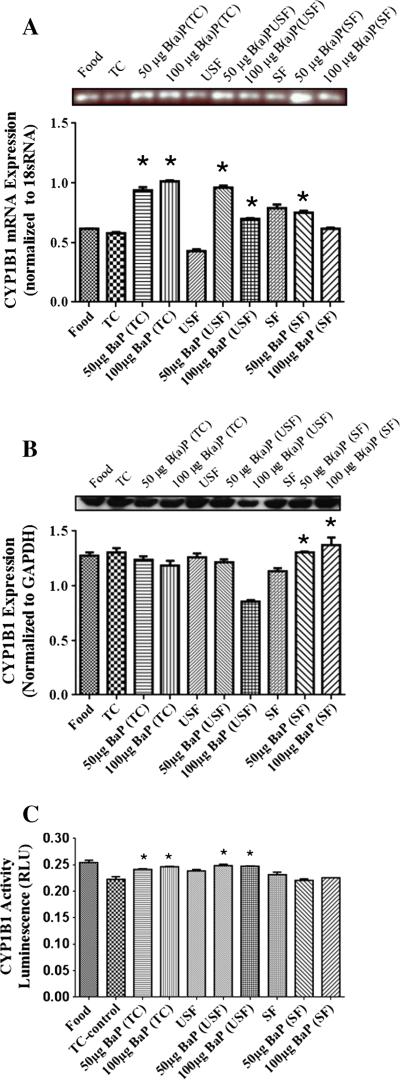
The mRNA for CYP1B1 revealed that 50 μg B(a)P+TC, 50 μg B(a)P+USF and 50 μg B(a)P+SF groups had a higher increase in CYP1B1 mRNA (Fig. 2A) compared to 100 μg B(a)P+TC, 100 μg B(a)P+USF and 100 μg B(a)P+SF groups, respectively (P<.05; dose×treatment interaction). In addition, there was a B(a)P-induced increase in mRNA transcript in the B(a)P+TC treatment groups compared to TC alone.
Fig. 2.
(A) RT-PCR analysis of liver CYP1B1 mRNA expression in ApcMin-exposed mice. The relative expression of CYP1B1 was quantified by densitometric quantitation of CYP1B1 bands and normalized to level of 18 sRNA. The bars represent mean±S.D. for three independent experiments. *P<.005, when 50 μg B(a)P+TC, 50 μg B(a)P+USF and 50 μg B(a)P+SF compared to 100 μg B(a)P+TC, 100 μg B(a)P+USF and 100 μg B(a)P+SF, respectively; and when B(a)P+TC is compared to TC. (B) Western blot analysis of liver CYP1B1 protein expression in ApcMin-exposed mice. Mice were exposed to either vehicles (TC, USF or SF) or B(a)P (50 μg-TC, 100 μg-TC, 50 μg USF, 100 μg USF, 50 μg SF and 100 μg SF) for 60 days via oral gavage. The relative expression of CYP1B1 was quantified by densitometric quantitation of CYP1B1 bands and GAPDH was used as the loading control. The bars represent mean±S.D. for three independent experiments.*P<.005, when 50 μg B(a)P+SF and 50 μg B(a)P+USF are compared to 50 μg B(a)P+TC and 50 μg B(a)P+SF, respectively; and when 100 μg B(a)P+SF group is compared to both 100 μg B(a)P+USF and 100 μg B(a)P+TC groups. (C) Enzyme activity of liver CYP1B1 in ApcMin-exposed mice. The activity of CYP1B1 was determined using luminescence detection. The bars represent mean±S.D. for three independent experiments.
Western blot analysis also showed CYP1B1 expression (Fig. 2B) in all treatment groups. However, there was a significant B(a)P treatment- related increase in CYP1B1 expression in the 50 μg B(a)P+SF and 50 μg B(a)P+USF groups compared to 50 μg B(a)P+TC and 50 μg B(a)P+SF groups, respectively. In addition there was an increase in expression in 100 μg B(a)P+SF group compared to both 100 μg B(a)P+USF and 100 μg B(a)P+TC groups. The CYP1B1 activity (Fig. 2C) was noted in all treatment groups. Though the activity was apparently high in the 100 μg B(a)P+USF group, it was not statistically significant compared to any of the treatment groups.
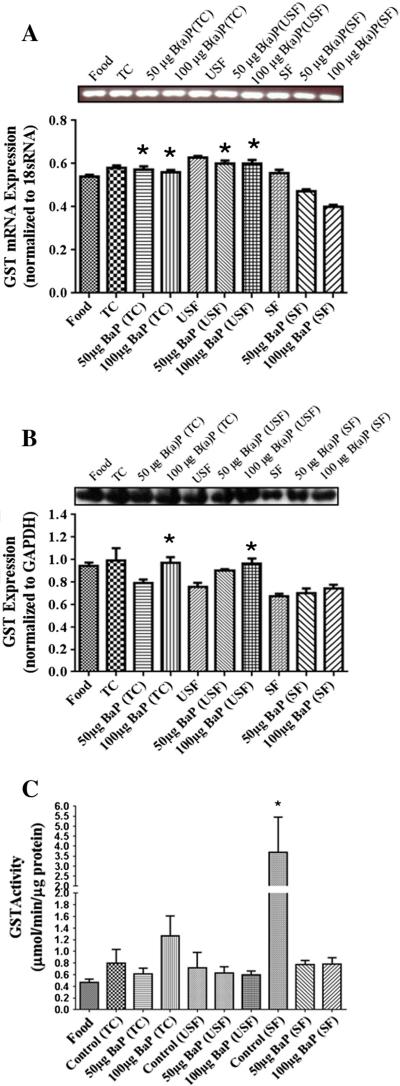
GST mRNA transcription occurred in all the treatment groups (Fig. 3A). The 50 μg B(a)P+TC GST transcription was similar to its high dose counterpart (100 μg B(a)P+TC group). In addition, the transcription for 50 μg B(a)P+TC group was also higher than 50 μg B(a)P+USF and 50 μg B(a)P+SF groups. Similar trend was noticed in the 100 μg B(a)P+TC group as well. The 50 and 100 μg B(a)P+USF groups, had significantly higher transcription compared to their 50 and 100 μg B(a)P+SF counterparts (P<.05; dose×treatment interaction). Though B(a)P caused an induction of GST protein compared to the USF and SF controls, the expression was not statistically significant (Fig. 3B). However, there was a significant GST induction in the 100 μg B(a)P+USF group compared to its USF control. All treatment groups exhibited GST activity (Fig. 3C). The SF control had significantly high GST activity than both 50 μg B(a)P+SF and 100 μg B(a)P+SF groups.
Fig. 3.
(A) RT-PCR analysis of liver GST mRNA expression in ApcMin-exposed mice. The relative expression of GST was quantified by densitometric quantitation of GST bands and normalized to level of 18 sRNA. The bars represent mean±S.D. for three independent experiments. *P<.005 when 50 μg B(a)P+TC is compared to 50 μg B(a) P+USF and 50 μg B(a)P+SF groups; 50 μg B(a)P+TC is compared to 100 μg B(a)P+TC, 50 and 100 μg B(a)P+USF groups is compared to 50 and 100 μg B(a)P+SF counterparts. (B) Western blot analysis of liver GST protein expression in ApcMin-exposed mice. The relative expression of GST was quantified by densitometric quantitation of GST bands and GAPDH was used as the loading control. The bars represent mean±S.D. for three independent experiments. *P<.005 when 100 μg B(a)P+TC is compared to 50 μg B(a)P+TC; 100 μg B(a)P+USF compared to 50 μg B(a)P+USF, and 50 and 100 μg B(a)P+SF respectively. (C) Enzyme activity of liver GST in ApcMin-exposed mice. The activity of GST was determined using a spectrophotometer. The bars represent mean±S.D. for three independent experiments. *P<.005, when SF is compared to both 50 μg B(a)P+SF and 100 μg B(a)P+SF.
3.2. Benzo(a)pyrene treatment-related effects on drug metabolizing enzyme transcription, expression and activity in colon
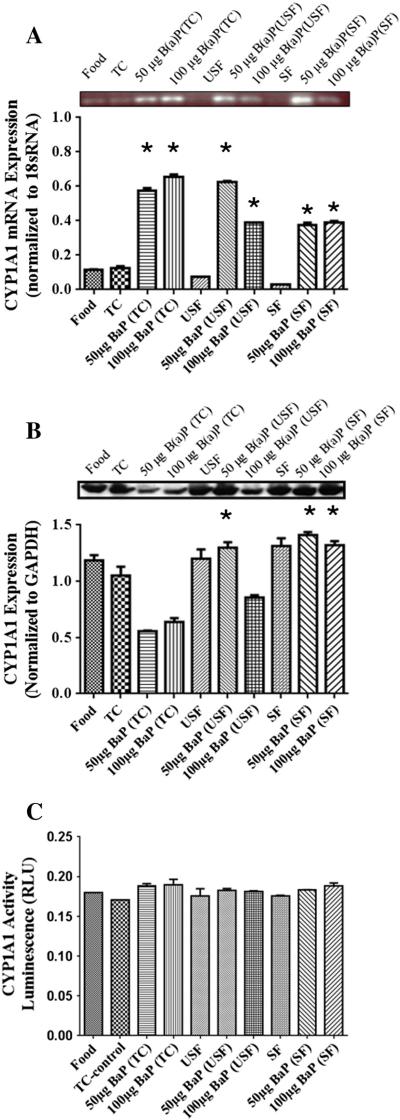
Benzo(a)pyrene caused an induction of CYP1A1 mRNA levels in the B(a)P treatment groups in comparison to their respective vehicles controls (Fig. 4A). The mRNA levels were significantly high in the 50 μg B(a)P+USF group compared to 100 μg B(a)P+USF, whereas no difference was noted between 50 and 100 μg B(a)P+SF groups. However, there was a dose-dependent increase in the 100 μg B(a)P+TC group compared to 50 μg B(a)P+TC group. The 50 μg B(a)P+USF treatment group had higher 1A1 transcription than the 50 μg B(a)P+TC group. In addition, the 100 μg B(a)P+TC group had higher transcription than both the 100 μg B(a)P+USF and 100 μg B(a)P+SF groups. Western blot analysis revealed that CYP1A1 expression was highest in TC control group compared to B(a)P+TC group (Fig. 4B). The 50 μg B(a)P+USF group had higher 1A1 protein levels than 50 μg B(a)P+TC group. However, the 50 μg B(a)P+SF group had higher protein expression than both 50 μg B(a)P+USF and 50 μg B(a)P+TC groups. In addition, the 100 μg B(a)P+SF group had higher expression than both the 100 μg B(a)P+USF and 100 μg B(a)P+TC groups (P<.05; dose×treatment interaction). CYP1A1 enzyme activity (Fig. 4C) in the colon showed no statistically significant difference among any of the treatment groups examined.
Fig. 4.
(A) RT-PCR analysis of colon CYP1A1 mRNA expression in ApcMin-exposed mice. The relative expression of CYP1A1 was quantified by densitometric quantitation of CYP1A1 bands and normalized to level of 18 sRNA. The bars represent mean±S.D. for three independent experiments. *P<.005, when 100 μg B(a)P+TC is compared to 50 μg B(a)P+TC; 50 μg B(a)P+USF is compared to 50 μg B(a)P+TC and 50 μg B(a)P+SF; and when 100 μg B(a)P+USF is compared to 100 μg B(a)P+SF groups. (B) Western blot analysis of colon CYP1A1 protein expression in ApcMin-exposed mice. The relative expression of CYP1A1 was quantified by densitometric quantitation of CYP1A1 bands and GAPDH was used as the loading control. The bars represent mean±S.D. for three independent experiments. *P<.005, when 50 μg B(a)P+USF is compared to 50 μg B(a)P+TC; 50 μg B(a)P+SF is compared to 50 μg B(a)P+USF and 50 μg B(a)P+TC; and when 100 μg B(a)P+SF is compared to 100 μg B(a)P+USF and 100 μg B(a)P+TC. (C) Enzyme activity of colon CYP1A1 in ApcMin-exposed mice. The activity of CYP1A1 was determined using luminescence detection. The bars represent mean±S.D. for three independent experiments.
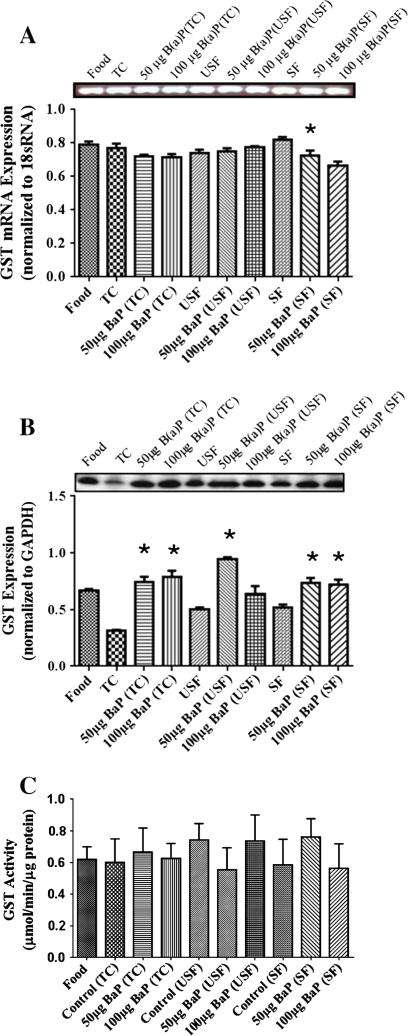
The mRNA for CYP1B1 (Fig. 5A) shows that B(a)P caused an induction of CYP1B1 mRNA in the B(a)P+TC and B(a)P+USF groups compared to their respective controls but did not cause a significant increase in the B(a)P+SF group compared to its respective SF control. The 100 μg B(a)P+TC group caused a dose-dependent increase in CYP1B1 mRNA compared to 50 μg B(a)P+TC group. However, the 50 μg B(a)P+USF and 50 μg B(a)P+SF groups had higher CYP1B1 mRNA than 100 μg B(a)P+USF and 100 μg B(a)P+SF groups, respectively. Western blot analysis of colon CYP1B1 expression (Fig. 5B) revealed that there were no significant differences between B(a)P+TC or B(a)P+USF compared to TC and SF control groups, respectively. There was a significant reduction of CYP1B1 protein levels in both the 50 μg B(a)P+USF and 100 μg B(a)P+USF compared to 50 μg B(a)P+TC and 100 μg B(a)P+TC groups, respectively. A significant increase in the CYP1B1 enzyme activity (Fig. 5C) in the B(a)P+TC groups compared to their TC control was noted. In addition, 50 μg B(a)P+TC and 100 μg B(a)P+TC had had higher CYP1B1 activity than 50 μg B(a)P+SF and 100 μg B(a)P+SF, respectively. Both doses (50 and 100 μg) of B(a)P+USF groups had higher CYP1B1 activity than B(a)P+SF groups (P<.05; dose×treatment interaction).
Fig. 5.
(A) RT-PCR of colon CYP1B1 mRNA expression in ApcMin-exposed mice. The relative expression of CYP1B1 was quantified by densitometric quantitation of GST bands and normalized to level of 18 sRNA. The bars represent mean±S.D. for three independent experiments. *P<.005, when the 100 μg B(a)P+TC is compared to 50 μg B(a)P+TC; and when 50 μg B(a)P+USF and 50 μg B(a)P+SF are compared to 100 μg B(a)P+USF and 100 μg B(a)P+SF, respectively. (B) Western blot of colon CYP1B1 protein expression in ApcMin-exposed mice. The relative expression of CYP1B1 was quantified by densitometric quantitation of CYP1B1 bands and GAPDH was used as the loading control. The bars represent mean±S.D. for three independent experiments. *P<.005, when 50 μg B(a)P+USF and 100 μg B(a)P+USF are compared to 50 μg B(a)P+TC and 100 μg B(a)P+TC, respectively. (C) Enzyme activity of colon CYP1B1 in ApcMin-exposed mice. The activity of CYP1B1 was determined using luminescence detection. The bars represent mean±S.D. for three independent experiments. *P<.005, when B(a)P+TC compared to TC control; 50 μg B(a)P+TC and 100 μg B(a)P+TC are compared to 50 μg B(a)P+SF and 100 μg B(a)P+SF, respectively; and when 50 and 100 μg B(a)P+USF are compared to their respective B(a)P+SF.
The GST mRNA was detected in all treatment groups and controls (Fig. 6A). There was a reduction in GST transcription in the 50 μg B(a) P+SF group compared to SF group. Western blot analysis showed GST expression in all treatment groups (Fig. 6B). The B(a)P+TC and B(a)P+SF group caused an induction in GST expression compared to their respective control groups (P<.005) for both the doses administered. Additionally, the 50 μg B(a)P+USF group had higher GST induction than 100 μg B(a)P+USF, 50 μg B(a)P+SF and 100 μg B(a)P+SF groups (P<.05; dose×treatment interaction). The GST activity was evident in all treatment groups but there was no significant difference in activity within or between groups (Fig. 6C).
Fig. 6.
(A) RT-PCR analysis of colon GST mRNA expression in ApcMin-exposed mice. The relative expression of GST was quantified by densitometric quantitation of GST bands and normalized to level of 18 sRNA. The bars represent mean±S.D. for three independent experiments. *P<.005 when 50 μg B(a)P+SF is compared to SF, and 100 μg B(a)P+SF. (B) Western blot analysis of colon GST protein expression in ApcMin-exposed mice. The relative expression of GST was quantified by densitometric quantitation of GST bands and GAPDH was used as the loading control. The bars represent mean±S.D. for three independent experiments. *P<.005, when B(a)P+TC and B(a)P+SF are compared to their respective control groups for both the doses; and when 50 μg B(a)P+USF are compared to 100 μg B(a)P+USF, 50 μg B(a)P+SF and 100 μg B(a)P+SF. (C) Enzyme activity of colon GST in ApcMin-exposed mice. The activity of GST was determined using a spectrophotometer. The bars represent mean± S.D. for three independent experiments.
3.3. Benzo(a)pyrene treatment-related effects on disposition of B(a)P metabolites in plasma, colon and liver
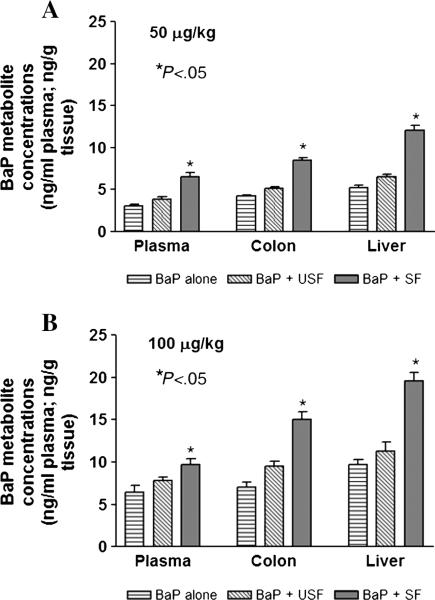
Since metabolism of ingested PAHs is the first step prior to initiating toxicity or carcinogenesis of the gastrointestinal (GI) tract, the colon tissues and plasma were analyzed for B(a)P and its metabolites. Measuring B(a)P metabolite concentrations will help us address the question of whether tissue burden of B(a)P reactive metabolites (total metabolite load and specific metabolite types) is greater when B(a)P is administered through saturated versus unsaturated fat. The concentrations of metabolites in plasma and colon tissues following subchronic B(a)P administration in unsatu-rated and saturated fat is shown in Fig. 7A and B.
Fig. 7.
Effect of the type of dietary fat on total B(a)P metabolite concentrations in plasma, colon and liver of ApcMin mice that received 50 μg B(a)P/kg of bw (A) and 100 μg B(a)P/kg of bw via oral gavage (B). The bars represent mean±S.D. for three independent experiments. *P<.05 in metabolite concentrations between saturated fat category containing B(a)P, compared to B(a)P alone and unsaturated fat.
Disposition studies revealed no traces of parent B(a)P compound either in plasma or in target tissues. Mice that received B(a)P through saturated fat registered greater amounts of B(a)P metabolites both in liver and colon compared to mice that ingested B(a)P through unsaturated fat. The metabolite concentrations were found to be B(a) P dose-dependent (Fig. 7A and B; P<.05; dose×treatment interaction).
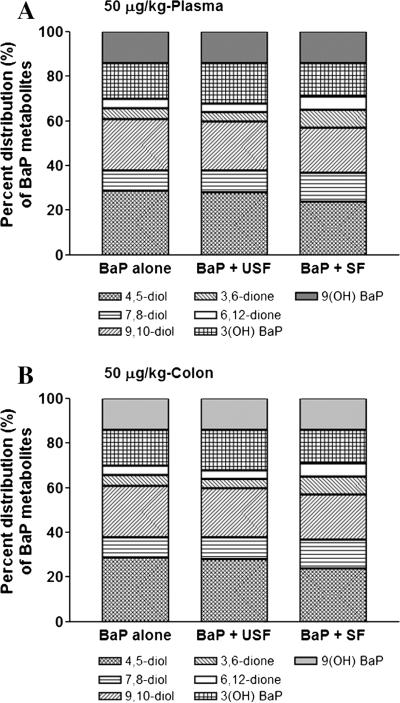
Since the B(a)P metabolite types and percentages will reveal the balance between metabolic activation and detoxification processes as a result of B(a)P administration, our studies were focused on knowing to what extent does dietary fat alter the B(a)P metabolite composition. The metabolites identified were: B(a)P 4,5-diol; B(a)P 7,8-diol; B(a)P 9,10-diol; B(a)P 3,6-dione, B(a)P 6,12-dione; 3(OH) B(a)P and 9(OH) B(a)P. The first five are toxicologically relevant metabolites and the last two are detoxification metabolites. Among the B(a)P metabolite concentrations, the proportion of its reactive metabolites such as 7,8-diol-9,10-epoxide; B(a)P-3,6-dione, and B(a)P-6,12-dione were high (Fig. 8 A and B). Additionally, there was no major difference between 50 and 100 μg/kg body weight (bw) groups in the proportion of different B(a)P metabolites generated.
Fig. 8.
Effect of the type of dietary fat on B(a)P metabolite types in plasma (A) and colon (B) of ApcMin mice that received 50 μg B(a)P/kg bw via oral gavage. Inasmuch as there were no differences between 50 and 100 μg/kg B(a)P/kg bw in percentage composition of B(a)P metabolites, the 50 μg/kg B(a)P/kg bw was chosen as a representative of both 50 and 100 μg/kg B(a)P/kg bw. Similarly, there being no differences between colon and liver in the types of metabolites generated, only colon sample data are shown here as representative of colon and liver.
3.4. Benzo(a)pyrene treatment-related effects on disposition of B(a)P-DNA adducts in colon and liver tissues
As B(a)P-DNA adduct formation is one of the important steps en route to carcinogenesis, the B(a)P-DNA adducts levels in these tissues were measured as these adducts serve as a markers of B(a)P-induced damage to colonocytes following oral B(a)P ingestion.
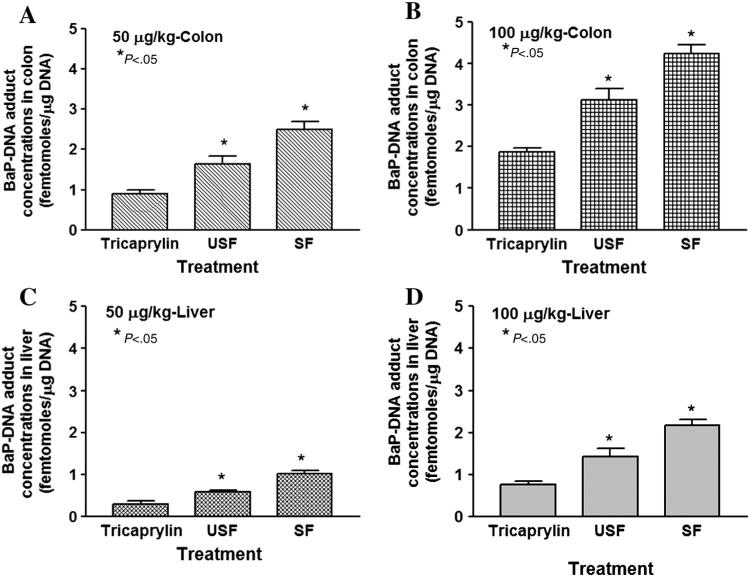
No B(a)P-DNA adducts were detected either in colon or liver tissues of control mice. On the other hand, B(a)P-DNA adducts were detected in colon, and liver tissues of mice treated with B(a)P in saturated and unsaturated fat. The adduct concentrations in colon and liver for both the doses employed are shown in Fig. 9A–D. Benzo(a) pyrene ingested through saturated fat showed the greatest concentration of DNA adducts levels compared to its unsaturated and B(a)P alone counterparts and a dose–response relationship as well (P<.05; dose×treatment interaction).
Fig. 9.
Benzo(a)pyrene-DNA adduct total concentrations in colon (A and B) and liver (C and D) of ApcMin mice exposed to 50 μg/kg and 100 μg/kg through TC, USF and SF. The bars represent mean±S.D. for three independent experiments. *P<.05 in adduct concentrations for the respective B(a)P+USF or B(a)P+SF treatment group compared to B(a)P+TC treatment group.
The major adducts observed in each tissue were derived from the 7,8,9,10-tetrahydrobenzo(a) pyrene (BPDE), which are the deoxyguanosine (BPDE-N2-dG-3′P) adducts. On the other hand, the proportion of deoxyadenosine (dA) adducts formed from BPDE (BPDE-N2-dA-3′P) were minor in both colon and liver tissues. The 3-OH-B(a)P; 9-OH–B(a)P; B(a)P-3,6-dione, and B(a)P-6,12-dione metabolites were not found to contribute to adduct formation in both the tissues studied.
The distribution of BPDE adduct types in both colon and liver for the three B(a)P treatment groups [B(a)P+TC, B(a)P+USF, B(a)P+SF] is shown in Table 2. The adducts identified were dA and deoxyguanosine (dG) ones. A predominance of dG adducts was seen in both colon and liver tissues at both the doses employed for all the treatment groups mentioned above. Inasmuch as there is no difference in dA/dG ratio of adducts between the two doses, the distribution of adduct types at 50 μg/kg is shown as a representative example in Table 2.
Table 2.
Effect of dietary fat on major B(a)P-DNA adduct types in colon and liver tissues of ApcMin mice
| Treatment | Colon |
Liver |
||
|---|---|---|---|---|
| dA | dG | dA | dG | |
| B(a)P+SF | 15 | 85 | 10 | 90 |
| B(a)P+USF | 20 | 80 | 10 | 90 |
| B(a)P | 25 | 75 | 20 | 80 |
4. Discussion
It is well known that biotransformation drives the toxicity or carcinogenesis caused by some environmental chemicals [29,38]. The regulation of drug metabolizing enzyme (DME) gene expression at the transcriptional and post translational levels impact the elimination of toxicants, and affect disease pathogenesis as well [39]. The susceptibility of rodent models to chemical carcinogen exposure is correlated with expression of DME in target tissues [40]. This characteristic warrants quantification of magnitude of induction of DME to demonstrate susceptibility of colon to B(a)P exposure. Though the association of dietary PAHs with GI tract tumors in epidemiological and animal model studies has already been established, information on underlying mechanisms is lacking. This study attempts to gain an insight into the mechanism of colon tumorigenesis as a result of B(a)P ingestion through two different types of dietary fat.
The mRNA expression of CYP1A1 and CYP1B1 was induced in livers of B(a)P-treated mice with B(a)P administered through USF registering high levels of expression compared to its SF counterparts and this pattern was found to be dose-dependent. The B(a)P-induced changes in enzyme activities are due to classic increases in CYP expression at the transcriptional level, as we have observed increases in mRNA levels that correspond directly to the increases in activity levels of the DME, suggestive of enzyme induction. However, the protein expressions for both the isozymes was found to be high in B(a)P+SF treatment group, compared to B(a)P+USF and B(a)P+TC treatment groups. The differences between mRNA and protein expression patterns for both CYP1A1 and 1B1 suggests that the extent of CYP induction upon exposure to B(a)P may be regulated differently depending on the dose and type of dietary fat. Regardless of the matrix (SF, USF and TC) used, B(a)P exposure resulted in greater CYP1A1 transcription in liver compared to CYP1B1. Findings of our study are in agreement with those of Harrigan et al. [41] who showed several fold greater induction of CYP1A1 in liver compared to CYP1B1 following exposure to B(a)P. Additionally, our findings in the ApcMin mouse model are in support of our earlier observations in F-344 rat model where dietary exposure to B(a)P resulted in higher induction of aryl hydrocarbon hydroxylase (CYP1A1 and 1B1) activities in liver [42].
The liver GST mRNA expression was slightly elevated by B(a)P exposure only through USF, but not the other treatment groups. Also, the absence of a dose-dependent GST mRNA expression for B(a)P administered through saturated fat suggests that B(a)P detoxification through glutathione conjugation is limited. These findings clearly indicate that the administered doses of B(a)P were not able to induce the expression of detoxification enzymes in liver to a considerable extent than bioactivation enzymes.
The expression of CYP1A1 and CYP1B1 in colon mirrored the trend seen in the case of liver. Contrary to that seen in the case of liver, the B(a)P+SF group showed a dose-dependent increase for GST mRNA expression in colon, but not for other groups. Published reports reveal an association between the biotransformation enzymes and the type and content of dietary lipids through which PAHs were administered (reviewed in [10]). The contrasting trend observed in CYP transcription in the case of SF versus USF suggest that some DME are elevated in activity following B(a)P exposure due to protein stabilization or via an yet unknown mechanism.
The lack of concordance between increase in mRNA, protein expression and DME activity could be attributed to potential saturation of DME activity as well as signal for protein expression. Additionally, variations in phospholipid composition of endoplasmic reticulum affecting the turnover number of CYPs, and/or limiting CYP450 reductase levels [43] cannot be excluded. For example, several factors, including the rate of transcription initiation, mRNA stability, as well as protein stability and modification, may account for discrepancies between gene expression at mRNA, protein levels and enzyme activity [44]. Also, modification of DME activity via post-translational modifications, including phosphorylation of enzymes and ROS-evoked structural modifications cannot be ruled out. In this context it needs to be mentioned that post translational regulation of CYP1A1 has been reported, including phosphorylation of individual CYPs via diverse kinases, which led to enzyme-specific alterations in catalytic activity [45]. The differential effects of B(a)P on CYP gene expression and enzyme activity were also noticed in other rodent models by Floreani et al. [46].
The CYP1A1, CYP1B1 and GST activities in the colon showed no statistically significant variation among the various B(a)P treatment groups. Studies conducted earlier by Sattar et al. [47] also revealed no change in GST levels and activities in adenomas and normal tissues of the small intestine of ApcMin mouse treated with 100 mg/kg B(a)P through corn oil.
Our findings indicate that CYP1A1 may play an important role in the metabolic processing of B(a)P by liver. Studies conducted by Uno et al. [48,49] and Shi et al. [50,51] have ascribed the role of CYP1A1 to detoxication than metabolic activation. When CYP1A1 (–/–) global knockout mice were exposed to 1.25 mg/kg per day of B(a)P, they survived for 4–5 months, while the CYP1A1 (+/+) wild-type mice survived for a year [50]. On the other hand, when these groups of mice received 125 mg/kg per day of B(a)P, the knockout mice lived for 18 days, while their counterparts survived for 1 year [51]. Additionally, the studies of Uno et al. [48] and Shi et al. [51] demonstrated that small intestinal CYP1A1 processes much of the toxicant load, before it reaches the liver for the CYP1A1 in liver tissues to be induced. Immunohistochemistry studies conducted by Uno et al. [52] also were in support of this supposition. These studies revealed that CYP1A1 is localized throughout the mice small intestine, and this isozyme was located close to lumen than CYP1B1, thus being able to process B(a)P during its residence in intestine. A similar localization of CYP1B1 can be expected in the large intestine. Taken together these studies argued that CYP1A1 is required for detoxication, while CYP1B1 is needed for metabolic activation.
Even though the aforementioned studies advocate the predominant role played by CYP1A1 in biotransformation, expression of CYP1B1 cannot be underestimated as it is an important extrahepatic enzyme that converts B(a)P to dihydrodiol epoxides [53] and, hence, contribute significantly to the global metabolite burden. Its role in metabolic activation, especially the conversion of B(a)P-7,8-diol to the ultimate carcinogen BPDE [41] appears to be vital in the neoplastic transformation of adenomas in the ApcMin mouse colon. Halberg et al. [54] have demonstrated an elevated expression of CYP1B1 in crypts and stroma of the intestine adjacent to tumors in the ApcMin mouse. Also relevant in this context was the reported observation that CYP1B1 deficient Min (multiple intestinal neoplasia) mice developed a two fold increase in tumors compared to controls [54], underscoring the role played by CYP1B1 in tumor progression. It has been reported that B(a)P exposure leads to an increase in CYP1B1 induction, resultant generation of metabolites and cytotoxicity in mice tissues [55,56].
In addition to the B(a)P metabolites transported from liver, colon cells are also capable of metabolizing PAHs by the cytochrome P450 [57,58]. In other words, B(a)P metabolites such as dihydrodiols formed in the liver could enter colon via vascular transport and take up residence in colon crypts where they could be further metabolized into dihydrodiol epoxides. In this regard, the studies of Herbst et al. [59] are worth mentioning. When human colon epithelial cells subsequent to incubation with dihydrodiol epoxides of phenanthrene (a PAH compound) were injected into SCID mice, these mice developed tumors. Present study also corroborated our previous findings that microsomes (one of the subcellular fractions) of tumorous and non-tumorous colon tissues of ApcMin mouse were capable of metabolizing B(a)P and fluoranthene (FLA) [60] into dihydrodiols and diones. Increased generation of dimethylbenz(a) anthracene metabolites by CYP1B1 has been implicated in a concomitant increase in intestinal tumorigenesis in the ApcMin mouse model [54]. Additionally, an elevated expression of CYP1B1 and other CYP isozymes have been reported in human colon cancer tissues [61] as well. Taken together, these studies furnish evidence to support the contention that PAH metabolites are capable of malignant transformation of colon epithelial cells.
The dietary fat type and B(a)P doses upon long term exposure may lead to a differential induction of CYP450 both in liver and colon. As a consequence, continuous production of metabolites is expected, the extent of which may depend on B(a)P dose, and fat type.
Benzo(a)pyrene assimilation through saturated dietary lipid is more likely to contribute to an increased residence time at high doses. The lipid-soluble nature of B(a)P may facilitate its uptake when ingested through lipid-rich diet. The hydrophobic domain of the dietary fat to which PAHs are bound [62] remains intact during lipolysis [63]. This property may enable the delivery of BaP through lipid influx into the intestinal epithelium subsequent to digestion [64]. Using the Caco-2 cell model, Vasiluk et al. [65] have demonstrated that uptake of B(a)P released into the gut lumen occurs through passive diffusion and its transfer into the bloodstream is aided by the fugacity gradient generated by the lipids. Additionally, works of Laurent et al. [66,67] and Cavret et al. [68] have shown a robust relationship between B(a)P and phenanthrene absorption and fat absorption. These studies reinforce the likelihood that prolonged exposure to B(a)P through dietary fat contributes to increased bioavailability of B(a)P and/or its metabolites in the GI tract and cause localized damage to the colon epithelium.
The greater concentrations of B(a)P metabolites measured in plasma, colon and liver of mice that received the B(a)P through SF relative to other treatment groups suggest that lipid type influences the disposition kinetics of ingested B(a)P. Additionally, our findings on dietary lipid-influenced increase in B(a)P metabolite burden in colon tissues is in agreement with our earlier findings that showed a saturated fat-induced increase in FLA (a PAH compound) metabolite concentrations in various tissues of F-344 rats [69].
Our findings support the notion that dietary fat modulates the biotransformation of B(a)P leading to development of adenomas and carcinomas. Our observations are consistent with previous reports from our laboratory [28] and others [70], which demonstrated a relationship between B(a)P exposure and tumor development in mice. Therefore, it is reasonable to speculate that B(a)P metabolites influence the steps involved in the adenoma-carcinoma sequence, which encompasses transition from normal mucosa to adenoma and proceed to invasive carcinoma as well [71].
Overall, the B(a)P metabolite types identified in our study are in agreement with published literature [55,72–75] which documented that orally administered B(a)P undergoes extensive biotransformation producing an array of metabolites and the concentrations of these metabolites were B(a)P dose-dependent. The relatively high concentrations of B(a)P diols and quinones in plasma and colon of mice that ingested B(a)P through saturated fat compared to unsaturated fat suggest that B(a)P incorporated through saturated fat is likely to generate more toxic metabolites. This assumption also derives support from the low concentrations of 3-, and 9-OH B(a)P metabolites measured in plasma and tissues of mice that ingested saturated compared to unsaturated fat. The data generated thus far suggest that repeated exposure to B(a)P leads to an increase in residual body burden of B(a)P/metabolites at high doses.
Binding of PAHs with cellular macromolecules such as proteins, and nucleic acids constitute a step in the mechanism by which PAHs cause toxicity and/or cancer [76]. The induction of CYP1A1, CYP1B1 mRNA and protein following exposure of ApcMin mice to B(a)P in this study also resulted in an increase in generation of B(a)P metabolites and a concomitant increase in B(a)P-DNA adduct formation in colon. The adduct concentrations revealed a matrix (dietary fat or tricaprylin)-specific and dose- specific disposition. It is of interest to note that the B(a)P-DNA adduct concentrations in colon mirrored the B(a)P metabolite concentrations in this target tissue. The concordance between metabolite and adduct levels strongly implicate that B(a)P reactive metabolite accumulation in colon plays an important role in determining the likelihood of causing damage to cellular macromolecules.
The low B(a)P-DNA adduct concentrations in colon and liver tissues of mice in the unsaturated fat treatment group reflect a decreased biotransformation of B(a)P in target tissues that received B(a)P through unsaturated fat relative to saturated fat. The higher affinity of PAHs and their metabolites for high-density lipoproteins (HDL; [77]), may enable the HDL-sequestered B(a)P and its metabolites undergo cellular internalization [78]. This process prevents the reactive metabolites from undergoing hydrolysis in mice that received B(a)P through saturated fat. In support of this supposition was the finding that B(a)P-DNA binding was greater in tissues of rats that received diets containing lard and cod liver oil compared to their counterparts that were fed fat-free diet [79]. On similar lines, Curfs et al. [80] have demonstrated that high-fat cholesterol diet accelerated DNA damage due to B(a)P exposure in a hyperlipidemic mouse model. The increased concentration of B(a)PDNA adducts in colon and liver from saturated fat group in the present study suggest that these organ systems are susceptible to damage from intake of B(a)P. Also of worth mentioning in this regard was our earlier studies in a fluoranthene (a PAH compound)-administered F-344 rat model, wherein we observed association of a SF-elevated DNA damage compared to USF [69]. Not only animal studies but studies in human subjects lend credence to our argument. Diet-derived B(a)P has been reported to contribute to B(a)P-DNA adduct formation in human colon mucosa [81]. Additionally, calorie intake, which is inextricably linked to lipid-rich diet showed a strong association with B(a)P-DNA adduct levels in white blood cells isolated from human subjects [82]. Taken together, all the above-mentioned studies reiterate that the genotoxic effects of ingested B(a)P are governed by the intake of lipid type.
An interesting observation that stemmed from our studies is the strong association of B(a)P-DNA adduct levels with adenomas in colon reported by us earlier [28]. We have observed an increased incidence of adenomas in mice that received B(a)P through saturated fat compared to unsaturated fat and controls. Also, the adenoma counts displayed a dose–response relationship with the 100 μg/kg dose group registering more adenomas compared to its 50 μg/kg counterpart for B(a)P, B(a)P+USF and B(a)P+SF treatment groups. A similar linkage between B(a)P-DNA adducts concentrations and adenomas have been observed for A/J mouse lung [83] and ApcMin mouse small intestine [47]. Additionally, in a clinic-based study of colorectal adenomas, a positive association between leucocyte PAH-DNA adducts concentrations and adenoma prevalence was seen [84]. The coincidence of B(a)P-DNA adducts concentrations and adenomas strongly suggest that adenoma development is a function of robust bioactivation of B(a)P leading to accumulation of mutations and subsequent progression to tumor development.
The preponderance of deoxyguanosine (dG) adducts relative to that of dA adducts are consistent with the results of studies reported from our laboratory [72] and those of others [83,85]. Since the carcinogenicity of B(a)P results from this toxicant's propensity to form dG adducts [86], the greater incidence of these adducts at 100 μg/kg, compared to 50 μg/kg, seen in the present study, indicate the vulnerability of tissues to damage resulting from long term exposure to high doses of B(a)P through dietary fat.
Taken together, the findings of our study suggest that (a) the extent of B(a)P biotransformation depend on dietary lipid administered through oral gavage; (b) dietary lipid modulates the disposition of B(a)P metabolites; (c) more accumulation of B(a)P metabolites in plasma, colon and liver tissue of ApcMin mice administered with saturated fat relative to unsaturated fat; (d) the distribution of B(a)P metabolite types in the body is governed by the lipid type, with more accumulation of metabolites in mice that received saturated fat, compared to unsaturated fat; (e) the distribution trend of B(a)P-DNA adduct concentrations in colon and liver tissues mirroring that of the metabolites; and (f) a strong association between adduct levels and incidence of adenomas previously reported in this animal model.
Future studies will focus on interaction of B(a)P and dietary fat on oxidative injury to colon and liver tissues using F2-isoprostanes as biomarkers. These studies are expected to furnish new information on the role of oxidative stress in inflammation-associated chronic diseases such as sporadic CRC caused by exposure to dietary carcinogens.
Acknowledgments
Research reported in this publication was supported by the NIH grants 5R01CA142845-02 (AR), 1F31ES017391-01 (DLD), 1RO3CA130112-01 (AR), 5T32HL007735-12 and 18 (DLD, LDB; PI: Adunyah), 2R25GM059994-13 (LDB; PI: Lima), 1S11ES014156-01A1 (DBH, AR), and 1R56ES017448-01A1(DBH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Authors’ Contributions: Drs. DLD and AR designed the study and applied for Institutional Animal Care and Use Committee approval. Drs. DLD, JNM, MSN, AR and Ms. LDB performed the experiments and collected the data. Drs. DLD and AR analyzed the data and prepared draft figures and tables. Drs. DLD and AR prepared the manuscript draft with intellectual input from Drs. LJR and DBH. All authors approved the final manuscript.
Conflicts of interest: The authors declare that they have no conflicts of interest.
References
- 1.Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Chen HS, Feuer EJ, Cronin KA, editors. SEER Cancer Statistics Review, 1975–2009 (Vintage 2009 Populations) National Cancer Institute; Bethesda, MD: 2012. http://seer.cancer.gov/csr/1975_2009_pops09/, based on November 2011 SEER data submission, posted to the SEER web site. [Google Scholar]
- 2.Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22:191–7. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Cancer Fund and the American Institute for Cancer Research . Food, nutrition and the prevention of cancer: a global perspective. American Institute for Cancer Research; Washington: 1997. [DOI] [PubMed] [Google Scholar]
- 4.Parkin DM, Whelan SL, Ferlay J, Raymond L, Young J, editors. Cancer incidence in five continents. VII. IARC Scienlific Publications; Lyon: 1997. p. 143. IARC. [Google Scholar]
- 5.Burnett-Hartman AN, Newcomb PA, Mandelson MT, Adams SV, Wernli KJ, Shadman M, Wurscher MA, Makar KW. Colorectal endoscopy, advanced adenomas, and sessile serrated polyps: implications for proximal colon cancer. Nutr Cancer. 2011;63:583–92. [Google Scholar]
- 6.Fu Z, Shrubsole MJ, Smalley WE, Wu H, Chen Z, Shyr Y, Ness RM, Zheng W. Lifestyle factors and their combined impact on the risk of colorectal polyps. Am J Epidemiol. 2012;176:766–76. doi: 10.1093/aje/kws157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross AJ, Ferrucci LM, Risch A, Graubard BI, Ward MH, Park Y, Hollenbeck AR, Schatzkin A, Sinha R. A large prospective study of meat consumption and colorectal cancer risk: an investigation of potential mechanisms underlying this association. Cancer Res. 2010;70:2406–14. doi: 10.1158/0008-5472.CAN-09-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J, Kwak S, Myung S, Jee S. Obesity and colorectal adenomatous polyps: A cross-sectional study in Korean adults. Obesity. 2013 doi: 10.1002/oby.20390. (in press) [DOI] [PubMed] [Google Scholar]
- 9.Vongsuvanh R, George J, Qiao L, van der Poorten D. Visceral adiposity in gastrointestinal and hepatic carcinogenesis. Cancer Lett. 2013;330:1–10. doi: 10.1016/j.canlet.2012.11.038. [DOI] [PubMed] [Google Scholar]
- 10.Ramesh A, Walker SA, Hood DB, Guillen MD, Schneider H, Weyand EH. Bioavailability and risk assessment of orally ingested polycyclic aromatic hydrocarbons. Int J Toxicol. 2004;23:301–33. doi: 10.1080/10915810490517063. [DOI] [PubMed] [Google Scholar]
- 11.Diggs DL, Huderson AC, Harris KL, Myers JN, Banks LD, Rekhadevi PV, Niaz MS, Ramesh A. Polycyclic aromatic hydrocarbons and digestive tract cancers: a perspective. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2011;29:324–57. doi: 10.1080/10590501.2011.629974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamin EL, Riu A, Douki T, Debrauwer L, Cravedi JP, Zalko D, Audebert M. Combined genotoxic effects of a polycyclic aromatic hydrocarbon [B(a)P] and an heterocyclic amine (PhIP) in relation to colorectal carcinogenesis. PLoS One. 2013;8:e58591. doi: 10.1371/journal.pone.0058591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramesh A, Archibong AE, Hood DB, Guo Z, Loganathan BG. Global Environmental distribution and human health effects of polycyclic aromatic hydrocarbons. In: Loganathan BG, Lam PK-S, editors. CRC Press; Boca Raton, Florida: 2011. pp. 95–124. [Google Scholar]
- 14.Ramesh A, Archibong AE, Huderson AC, Diggs DL, Myers JN, Hood DB, Rekhadevi PV, Niaz MS. Polycyclic aromatic hydrocarbons. In: Gupta RC, editor. Veterinary toxicology. 2nd ed. Academic Press; London, UK: 2012. [Google Scholar]
- 15.Svecova V, Topinka J, Solansky I, Rossner Jr P, Sram RJ. Personal exposure to carcinogenic polycyclic aromatic hydrocarbons in the Czech Republic. J Expo Sci Environ Epidemiol. 2013;23:350–5. doi: 10.1038/jes.2012.110. [DOI] [PubMed] [Google Scholar]
- 16.Scarselli A, Di Marzio D, Marinaccio A, Iavicoli S. Assessment of work-related exposure to polycyclic aromatic hydrocarbons in Italy. Am J Ind Med. 2013 doi: 10.1002/ajim.22172. (in press) [DOI] [PubMed] [Google Scholar]
- 17.Cross AJ, Sinha R. Meat-related mutagens/carcinogens in the etiology of colorectal cancer. Environ Mol Mutagen. 2004;44:44–55. doi: 10.1002/em.20030. [DOI] [PubMed] [Google Scholar]
- 18.Veyrand B, Sirot V, Durand S, Pollono C, Marchand P, Dervilly-Pinel G, Tard A, Leblanc JC, Le Bizec B. Human dietary exposure to polycyclic aromatic hydrocarbons: results of the second French Total Diet Study. Environ Int. 2013;54:11–7. doi: 10.1016/j.envint.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Miller PE, Lazarus P, Lesko SM, Cross AJ, Sinha R, Laio J, Zhu J, Harper G, Muscat JE, Hartman TJ. Meat-related compounds and colorectal cancer risk by anatomical subsite. Nutr Cancer. 2013;65:202–26. doi: 10.1080/01635581.2013.756534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazerouni N, Sinha R, Hsu CH, Greenberg A, Rothman N. Analysis of 200 food items for benzo(a)pyrene and estimation of its intake in an epidemiologic study. Fd Chem Toxic. 2001;39:423–36. doi: 10.1016/s0278-6915(00)00158-7. [DOI] [PubMed] [Google Scholar]
- 21.Sinha R, Kulldorff M, Gunter MJ, Strickland P, Rothman N. Dietary benzo(a)pyrene intake and risk of colorectal adenoma. Cancer Epidemiol Biomarkers Prev. 2005;14:2030–4. doi: 10.1158/1055-9965.EPI-04-0854. [DOI] [PubMed] [Google Scholar]
- 22.Sinha R, Peters U, Cross AJ, Kulldorff M, Weissfeld JL, Pinsky PF, Rothman N, Hayes RB. Prostate, Lung, Colorectal and Ovarian Cancer Project Team. Meat, meat cooking methods and preservation and risk for colorectal adenoma. Cancer Res. 2005;65:8034–41. doi: 10.1158/0008-5472.CAN-04-3429. [DOI] [PubMed] [Google Scholar]
- 23.Sinha R, Cross A, Curtin J, Zimmerman T, McNutt S, Risch A, Holden J. Development of a food frequency questionnaire module and databases for compounds in cooked and processed meats. Mol Nutr Food Res. 2005;49:648–55. doi: 10.1002/mnfr.200500018. [DOI] [PubMed] [Google Scholar]
- 24.Gunter MJ, Probst-Hensch NM, Cortessis VK, Kulldorff M, Haile RW, Sinha R. Meat intake, cooking-related mutagens and risk of colorectal adenoma in a sigmoidoscopy-based case–control study. Carcinogenesis. 2005;26:637–42. doi: 10.1093/carcin/bgh350. [DOI] [PubMed] [Google Scholar]
- 25.Gilsing A, Berndt S, Ruder E, Graubard B, Ferrucci L, Burdette L, Weissfeld J, Cross A, Sinha R. Meat-related mutagen exposure, xenobiotic metabolizing gene polymorphisms and the risk of advanced colorectal adenoma and cancer. Carcinogenesis. 2012;33:1332–9. doi: 10.1093/carcin/bgs158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrucci LM, Sinha R, Huang WY, Berndt SI, Katki HA, Schoen RE, Hayes RB, Cross AJ. Meat consumption and the risk of incident distal colon and rectal adenoma. Br J Cancer. 2012;106:608–16. doi: 10.1038/bjc.2011.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamprecht SA, Lipkin M. Mouse models of gastrointestinal tumorigenesis for dietary cancer prevention studies. Nutr Rev. 2003;61:255–8. doi: 10.1301/nr.2003.jul.255-258. [DOI] [PubMed] [Google Scholar]
- 28.Harris DL, Washington MK, Hood DB, Roberts LJ, 2nd, Ramesh A. Dietary fat-influenced development of colon neoplasia in ApcMin mice exposed to benzo(a) pyrene. Toxicol Pathol. 2009;37:938–46. doi: 10.1177/0192623309351722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guengerich FP. Forging the links between metabolism and carcinogenesis. Mutat Res. 2001;488:195–209. doi: 10.1016/s1383-5742(01)00059-x. [DOI] [PubMed] [Google Scholar]
- 30.Baird WM, Hooven LA, Mahadevan B. Carcinogenic polycyclic aromatic hydrocarbon-DNA adducts and mechanism of action. Environ Mol Mutagen. 2005;45:106–14. doi: 10.1002/em.20095. [DOI] [PubMed] [Google Scholar]
- 31.Marchand LL. The role of chemical carcinogens and their biotransformation in colorectal cancer. In: Potter JD, Lindor NM, editors. Genetics of colorectal cancer. Springer; Berlin: 2009. pp. 261–76. [Google Scholar]
- 32.National Research Council [NRC] Guide for the care and use of laboratory animals. National Academy Press; Washington, DC: 1996. [Google Scholar]
- 33.IARC . Some non-heterocyclic polycyclic aromatic hydrocarbons and some related exposures. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Volume 92. World Health Organization & International Agency for Research on Cancer; 2010. p. 868. [PMC free article] [PubMed] [Google Scholar]
- 34.NIH guidelines for the laboratory use of chemical carcinogens . NIH Publication No. 81–2385. U.S. Government Printing Office; Washington, DC: 1981. [Google Scholar]
- 35.Ramesh A, Inyang F, Hood DB, Archibong AE, Knuckles ME, Nyanda AM. Metabolism, bioavailability, and toxicokinetics of benzo[a]pyrene [B(a)P] in F-344 rats following oral administration. Exp Toxic Pathol. 2001;53:253–70. doi: 10.1078/0940-2993-00192. [DOI] [PubMed] [Google Scholar]
- 36.Gupta RC, Randerath K. Analysis of DNA adducts by 32P labeling and thin layer chromatography. In: Friedberg EC, Hanawalt PC, editors. DNA Repair. Vol. 3. Marcel Dekker, Inc.; 1988. pp. 399–418. [Google Scholar]
- 37.Walker MP, Jahnke GD, Snedeker SM, Gladen BC, Lucier GW, DiAugustine RP. 32P-postlabeling analysis of the formation and persistence of DNA adducts in mammary glands of parous and nulliparous mice treated with benzo[a]pyrene. Carcinogenesis. 1992;11:2009–15. doi: 10.1093/carcin/13.11.2009. [DOI] [PubMed] [Google Scholar]
- 38.Buhler DR, Williams DE. The role of biotransformation in the toxicity of chemicals. Aquat Toxicol. 1988;11:19–28. [Google Scholar]
- 39.Aitken AE, Richardson TA, Morgan ET. Regulation of drug-metabolizing enzymes and transporters in inflammation. Annu Rev Pharmacol Toxicol. 2006;46:123–49. doi: 10.1146/annurev.pharmtox.46.120604.141059. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu Y, Nakatsuru Y, Ichinose M, Takahashi Y, Kume H, Mimura J, Fujii-Kuriyama Y, Ishikawa T. Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. Proc Natl Acad Sci. 2000;97:779–82. doi: 10.1073/pnas.97.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrigan JA, McGarrigle BP, Sutter TR, Olson JR. Tissue specific induction of cytochrome P450 (CYP) 1A1 and 1B1 in rat liver and lung following in vitro (tissue slice) and in vivo exposure to benzo(a)pyrene. Toxicol In Vitro. 2006;20:426–38. doi: 10.1016/j.tiv.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 42.Ramesh A, Inyang F, Hood DB, Knuckles ME. Aryl hydrocarbon hydroxylase activity in F-344 rats subchronically exposed to benzo(a)pyrene and fluoranthene through diet. J Biochem Mol Toxicol. 2000;14:155–61. doi: 10.1002/(sici)1099-0461(2000)14:3<155::aid-jbt5>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi K, Urashima K, Shimada N, Chiba K. Substrate specificity for rat cytochrome P450 (CYP) isoforms: screening with cDNA-expressed systems of the rat. Biochem Pharmacol. 2002;63:889–96. doi: 10.1016/s0006-2952(01)00843-7. [DOI] [PubMed] [Google Scholar]
- 44.Glanemann C, Loos A, Gorret N, Willis LB, O'Brien XM, Lessard PA, Sinskey AJ. Disparity between changes in mRNA abundance and enzyme activity in Corynebacterium glutamicum: Implications for DNA microarray analysis. Appl Microbiol Biotechnol. 2003;61:61–8. doi: 10.1007/s00253-002-1191-5. [DOI] [PubMed] [Google Scholar]
- 45.Marchand A, Barouki R, Garlatti M. Regulation of NAD(P)H: quinone oxidoreductase 1 gene expression by CYP1A1 activity. Mol Pharmacol. 2004;65:1029–37. doi: 10.1124/mol.65.4.1029. [DOI] [PubMed] [Google Scholar]
- 46.Floreani M, Gabbia D, Barbierato M, DeMartin S, Palatini P. Differential inducing effect of benzo(a)pyrene on gene expression and enzyme activity of cytochromes P4501A1 and 1A2 in Sprague Dawley and Wistar rats. Drug Metab Pharmacokinet. 2012;27:640–52. doi: 10.2133/dmpk.dmpk-12-rg-035. [DOI] [PubMed] [Google Scholar]
- 47.Sattar A, Hewer A, Phillips DH, Campbell FC. Metabolic proficiency and benzo(a) pyrene DNA adduct formation in ApcMin mouse adenomas and uninvolved mucosa. Carcinogenesis. 1999;20:1097–101. doi: 10.1093/carcin/20.6.1097. [DOI] [PubMed] [Google Scholar]
- 48.Uno S, Dalton TP, Derkenne S, Curran CP, Miller ML, Shertzer HG, Nebert DW. Oral exposure to benzo[a]pyrene in the mouse: detoxication by inducible cytochrome P450 is more important than metabolic activation. Mol Pharmacol. 2004;65:1225–37. doi: 10.1124/mol.65.5.1225. [DOI] [PubMed] [Google Scholar]
- 49.Uno S, Dalton TP, Dragin N, Curran CP, Derkenne S, Miller ML, Shertzer HG, Gonzalez FJ, Nebert DW. Oral benzo[a]pyrene in Cyp1 knockout mouse lines: CYP1A1 important in detoxication, CYP1B1 metabolism required for immune damage independent of total-body burden and clearance rate. Mol Pharmacol. 2006;69:1103–14. doi: 10.1124/mol.105.021501. [DOI] [PubMed] [Google Scholar]
- 50.Shi Z, Dragin N, Galvez-Peralta M, Jorge-Nebert LF, Miller ML, Wang B, Nebert DW. Organ-specific roles of CYP1A1 during detoxication of dietary benzo[a]pyrene. Mol Pharmacol. 2010;78:46–57. doi: 10.1124/mol.110.063438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi Z, Dragin N, Miller ML, Stringer KF, Johansson E, Chen J, Uno, Gonzalez FJ, Rubio C, Nebert DW. Oral benzo[a]pyrene-induced cancer: two distinct types in different target organs dependent on the mouse Cyp1 genotype. Int J Cancer. 2010;10:2334–50. doi: 10.1002/ijc.25222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uno S, Dragin N, Miller ML, Dalton TP, Gonzalez FJ, Nebert DW. Basal and inducible Cyp1 mRNA quantitation and protein localization throughout the mouse gastrointestinal tract. Free Radic Biol Med. 2008;44:570–83. doi: 10.1016/j.freeradbiomed.2007.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimada T, Murayama N, Okada K, Funae Y, Yamazaki H, Guengerich FP. Different mechanisms for inhibition of human cytochromes P450 1A1, 1A2, and 1B1 by polycyclic aromatic inhibitors. Chem Res Toxicol. 2007;20:489–96. doi: 10.1021/tx600299p. [DOI] [PubMed] [Google Scholar]
- 54.Halberg RB, Larsen MC, Elmergreen TL, Ko AY, Irving AA, Clipson L, Jefcoate CR. Cyp1b1 exerts opposing effects on intestinal tumorigenesis via exogenous and endogenous substrates. Cancer Res. 2008;68:7394–402. doi: 10.1158/0008-5472.CAN-07-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moorthy B, Miller KP, Jiang W, Williams ES, Kondraganti SR, Ramos KS. Role of cytochrome P4501B1 in benzo[a]pyrene bioactivation to DNA-binding metabolites in mouse vascular smooth muscle cells: evidence from 32P-postlabeling for formation of 3-hydroxybenzo[a]pyrene and benzo[a]pyrene-3,6-quinone as major proximate genotoxic intermediates. J Pharmacol Exp Ther. 2003;305:394–401. doi: 10.1124/jpet.102.044271. [DOI] [PubMed] [Google Scholar]
- 56.Galvan N, Teske DE, Zhou G, Moorthy B, MacWilliams PS, Czuprynski CJ, Jefcoate CR. Induction of CYP1A1 and CYP1B1 in liver and lung by benao(a)pyrene and 7,12-dimethylbenz(a)anthracene do not affect distribution of polycyclic hydrocarbons to target tissue: role of AhR and CYP1B1 in bone marrow cytotoxicity. Toxicol Appl Pharmacol. 2005;202:244–57. doi: 10.1016/j.taap.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 57.McKay JA, Murray GI, Weaver RJ, Ewen SW, Melvin WT, Burke MD. Xenobiotic metabolising enzyme expression in colonic neoplasia. Gut. 1993;34:1234–9. doi: 10.1136/gut.34.9.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding X, Kaminsky LS. Human extrahepatic cytochrome P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol. 2003;43:149–73. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- 59.Herbst U, Fuchs JI, Teubner W, Steinberg P. Malignant transformation of human colon epithelial cells by benzo[c]phenanthrene dihydrodiolepoxides as well as 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Toxicol Appl Pharmacol. 2006;212:136–45. doi: 10.1016/j.taap.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 60.Diggs DL, Harris KL, Rekhadevi PV, Ramesh A. Tumor microsomal metabolism of the food toxicant, benzo(a)pyrene, in ApcMin mouse model of colon cancer. Tumour Biol. 2012;33:1255–60. doi: 10.1007/s13277-012-0375-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kulasingham M, Rooney PH, Dundas SR, Telfer C, Melvin WT, Curran S, Murray GI. Cytochrome p450 profile of colorectal cancer: identification of markers of prognosis. Clin Cancer Res. 2005;11:3758–65. doi: 10.1158/1078-0432.CCR-04-1848. [DOI] [PubMed] [Google Scholar]
- 62.Landrum PF, Fisher SW. Influence of lipids on the bioaccumulation and trophic transfer of organic contaminants in aquatic organisms. In: Wainman BC, Arts MT, editors. Lipids in freshwater ecosystems. Springer-Verlag; New York, USA: 1999. pp. 203–34. [Google Scholar]
- 63.Verkade HJ, Tso P. Biophysics of intestinal luminal lipids. In: Mansbach CM II, Tso P, Kuksis A, editors. Intestinal lipid metabolism. Kluwer Academic/Plenum Publishers; New York: 2001. pp. 1–19. [Google Scholar]
- 64.Schlummer M, Moser GA, McLachlan MS. Digestive tract absorption of PCDD/Fs, PCBs, and HCB in humans: mass balances and mechanistic considerations. Toxicol Appl Pharmacol. 1998;152:128–37. doi: 10.1006/taap.1998.8487. [DOI] [PubMed] [Google Scholar]
- 65.Vasiluk L, Pinto LJ, Tsang WS, Gobas FA, Eickhoff C, Moore MM. The uptake and metabolism of benzo[a]pyrene from a sample food substrate in an in vitro model of digestion. Food Chem Toxicol. 2008;46:610–8. doi: 10.1016/j.fct.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 66.Laurent C, Feidt C, Lichtfouse E, Grova N, Laurent F, Rychen G. Milk-blood transfer of 14C-tagged polycyclic aromatic hydrocarbons (PAHs) in pigs. J Agric Food Chem. 2001;49:2493–6. doi: 10.1021/jf0014011. [DOI] [PubMed] [Google Scholar]
- 67.Laurent C, Feidt C, Grova N, Mpassi D, Lichtfouse E, Laurent F, Rychen G. Portal absorption of 14C after ingestion of spiked milk with 14C-phenanthrene, 14C-benzo(a)pyrene or 14C-TCDD in growing pigs. Chemosphere. 2002;48:843–8. doi: 10.1016/s0045-6535(02)00145-5. [DOI] [PubMed] [Google Scholar]
- 68.Cavret S, Laurent C, Feidt C, Laurent F, Rychen G. Intestinal absorption of 14C from 14C-phenanthrene, 14C-benzo(a)pyrene and 14Ctetrachlorodibenzo-para-dioxin: approaches with the Caco-2 cell line and portal absorption measurements in growing pigs. Reprod Nutr Dev. 2003;43:145–54. doi: 10.1051/rnd:2003014. [DOI] [PubMed] [Google Scholar]
- 69.Walker SA, Addai AB, Mathis M, Ramesh A. Effect of dietary fat on metabolism and DNA adduct formation after acute oral exposure of F-344 rats to fluoranthene. J Nutr Biochem. 2007;18:236–49. doi: 10.1016/j.jnutbio.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 70.Hakura A, Seki Y, Sonoda J, Hosokawa S, Aoki T, Suganuma A, Kerns WD, Tsukidate K. Rapid induction of colonic adenocarcinoma in mice exposed to benzo[a]pyrene and dextran sulfate sodium. Food Chem Toxicol. 2011;49:2997–3001. doi: 10.1016/j.fct.2011.07.057. [DOI] [PubMed] [Google Scholar]
- 71.Hou L, Chatterjee N, Huang WY, Baccarelli A, Yadavalli S, Yeager M, Bresalier RS, Chanock SJ, Caporaso NE, Ji BT, Weissfeld JL, Hayes RB. CYP1A1 Val462 and NQO1 Ser187 polymorphisms, cigarette use, and risk for colorectal adenoma. Carcino-genesis. 2005;26:1122–8. doi: 10.1093/carcin/bgi054. [DOI] [PubMed] [Google Scholar]
- 72.Ramesh A, Knuckles ME. Dose-dependent benzo(a)pyrene [B(a)P]-DNA adduct levels and persistence in F-344 rats following subchronic dietary exposure to B(a) P. Cancer Letters. 2006;240:268–78. doi: 10.1016/j.canlet.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 73.Brown LA, Khoshbouei H, Goodwin SJ, Irvin-Wilson C, Ramesh A, Sheng L, McCallister M, Jiang GT, Aschner M, Hood DB. Downregulation of early ionotrophic glutamate receptor subunit developmental expression as a mechanism for observed plasticity deficits following gestational exposure to benzo(a) pyrene. Neurotoxicology. 2007;28:965–78. doi: 10.1016/j.neuro.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McCallister NM, Maguire M, Ramesh A, Aimin Q, Liu S, Khoshbouei H, Aschner M, Ebner FF, Hood DB. Prenatal exposure to benzo(a)pyrene impairs later-life cortical neuronal function. Neurotoxicology. 2008;29:846–54. doi: 10.1016/j.neuro.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jules GE, Pratap S, Ramesh A, Hood DB. In utero exposure to benzo(a)pyrene predisposes offspring to cardiovascular dysfunction in later-life. Toxicology. 2012;295:56–67. doi: 10.1016/j.tox.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luch A. The carcinogenic effects of polycyclic aromatic hydrocarbons. Imperial College Press; London: 2005. p. 516. [Google Scholar]
- 77.Polyakov LM, Chasovskikh MI, Panin LE. Binding and transport of benzo[a]pyrene by blood plasma lipoproteins: the possible role of apolipoprotein B in this process. Bioconjug Chem. 1996;4:396–400. doi: 10.1021/bc960005e. [DOI] [PubMed] [Google Scholar]
- 78.Busbee DL, Norman JO, Ziprin RL. Comparative uptake, vascular transport, and cellular internalization of aflatoxin B1 and benzo(a)pyrene. Arch Toxicol. 1990;64:285–90. doi: 10.1007/BF01972988. [DOI] [PubMed] [Google Scholar]
- 79.Willis ED. Effects of dietary lipids on the metabolism of polycyclic hydrocarbons and the binding of their metabolites to DNA. Biochem Soc Trans. 1983;11:258–61. doi: 10.1042/bst0110258. [DOI] [PubMed] [Google Scholar]
- 80.Curfs DMJ, Beckers L, Godschalk RWL, Gijbels MJJ, van Schooten FJ. Modulation of plasma lipid levels affects benzo(a)pyrene-induced DNA damage in tissues of two hyperlipidemic mouse models. Environ Mol Mutagen. 2003;42:243–9. doi: 10.1002/em.10200. [DOI] [PubMed] [Google Scholar]
- 81.Alexandrov K, Rojas M, Kadlubar FF, Lang NP, Bartsch H. Evidence of antibenzo[a] pyrene diolepoxide-DNA adduct formation in human colon mucosa. Carcinogenesis. 1996;17:2081–3. doi: 10.1093/carcin/17.9.2081. [DOI] [PubMed] [Google Scholar]
- 82.Rundle A, Madsen A, Orjuela M, Mooney L, Tang D, Kim M, Perera F. The association between benzo[a]pyrene-DNA adducts and body mass index, calorie intake and physical activity. Biomarkers. 2007;12:123–32. doi: 10.1080/13547500601010418. [DOI] [PubMed] [Google Scholar]
- 83.Ross JA, Nelson GB, Wilson KH, Rabinowitz JR, Galati A, Stoner GD, Nesnow S, Mass MJ. Adenomas induced by polycyclic aromatic hydrocarbons in strain A/J mouse lung correlate with time-integrated DNA adduct levels. Cancer Res. 1995;55:1039–44. [PubMed] [Google Scholar]
- 84.Gunter MJ, Divi RL, Kulldorff M, Vermeulen R, Haverkos KJ, Kuo MM, Strickland P, Poirier MC, Rothman N, Sinha R. Leukocyte polycyclic aromatic hydrocarbon-DNA adduct formation and colorectal adenoma. Carcinogenesis. 2007;28:1426–9. doi: 10.1093/carcin/bgm022. [DOI] [PubMed] [Google Scholar]
- 85.Dreij K, Seidel A, Jernstrom B. Differential removal of DNA adducts derived from anti-diol epoxides of dibenzo[a, l]pyrene and benzo(a)pyrene in human cells. Chem Res Toxicol. 2005;18:655–64. doi: 10.1021/tx0497090. [DOI] [PubMed] [Google Scholar]
- 86.Kramata P, Zajc B, Sayer JM, Jerina C, Wei CS- J. A single site-specific trans-opened 7,8,9,10-tetrahydrobenzo(a)pyrene 7,8-diol 9,10-epoxide N2-deoxyguanosine adduct induces mutations at multiple sites in DNA. J Biol Chem. 2003;278:14940–8. doi: 10.1074/jbc.M211557200. [DOI] [PubMed] [Google Scholar]