Abstract
With the continuing rise of multiresistant pathogens, reliable, cost-effective, and novel diagnostics are urgently required by clinicians and clinical trialists to diagnose conditions such as respiratory tract infections to enable rational antimicrobial choice and enhance clinical outcomes. However, during product development, validation of these in vitro diagnostic devices, a key regulatory hurdle, requires sputum samples in large numbers. The Rapid Point-of-Care test Platform for Infectious Diseases (RAPP-ID) consortium is tasked with producing point of care test (POCT) platforms for rapid diagnosis of lower respiratory tract infections, including tuberculosis and blood stream infections. Validation of diagnostic platforms would ideally use well-characterized samples in a sputum library taken from a range of clinical settings to allow for a wide panel of pathogens to be assessed. These samples would be stored in specific stable conditions (monitored temperature, specific medium) until required for validation. Therefore we reviewed the current literature for details of storage conditions of sputum samples and for previous validation studies of other diagnostic tests using this methodology. However, we conclude that little data exists, and thus the acquisition and successful storage of good quality clinical samples are major roadblocks in the validation of novel POCT platforms, and that while not without limitations, spiked sputum samples appear the best solution until sputum library laboratory techniques allowing careful preservation of pathogens are improved.
Keywords: sputum, storage, rapid diagnostic, respiratory tract infection
Introduction
Rising antimicrobial resistance rates threaten the ability of clinicians to deliver curative therapy to patients with a wide range of infectious diseases and have been highlighted as a key risk that society now faces. Thus, new diagnostic devices for identifying pathogens and their resistance profile are urgently required, not only for clinicians but also for antimicrobial drug developers performing clinical trials.1 However, developers must achieve regulatory approval by following stringent channels, such as the Food and Drug Administration’s 510k approval or the European Medicines Agency’s CE Mark processes, to allow product launch and use in clinical settings and hospitals. A key part of this process is the requirement for validation of any platform with samples of known pathogens. While new platforms are being developed by commercial venture groups, such as the development of a Rapid Point-of-Care test Platform for Infectious Diseases (RAPP-ID) consortium, a consortium of academics, small and medium enterprises, and pharmaceutical companies funded via the Innovative Medicines Initiative.2,3 RAPP-ID has the principle objective of developing point-of-care test (POCT) platforms for rapid detection of clinically important bacteria to proof of concept. These include community, hospital, and ventilator acquired pneumonia (CAP, HAP, and VAP, respectively) as well as tuberculosis (TB). In order to perform validation effectively, clinical specimens (ie, specimens taken from patients) with a wide range of pathogens are needed; therefore, RAPP-ID has identified sample acquisition as the key risk to the project. We therefore performed a focused literature review to define the current roadblocks for sample acquisition for the validation of a POCT platform addressing the diagnosis of lower respiratory tract infections (LRTIs), including TB, which reflected the initial and refined focus of the discovery groups engaged in RAPP-ID.
Pneumonia and TB are commonly occurring infections. Between 1993 and 1994, the hospital infection survey conducted a second national prevalence survey of 37,111 patients across 157 hospitals, demonstrating a prevalence of 2.4% of HAP, and a prevalence of 6.1% of CAP.4 Additionally, it was estimated that around 15% of clinical consultations for ear, cough, sinus, and throat complaints turned out to be respiratory tract infection (RTI)-related.5 HAP and its related condition VAP have mortality rates ranging from 24% to over 70%, depending upon pathogen and comorbidities.6 Added to rising antimicrobial resistance, this demonstrates a significant, unmet medical need for rapid diagnostic tests that can reduce time to enacting the most effective treatment choice. A secondary benefit is an increase in pathogen- and resistance-specific recruitment to anti-infective clinical trials in drug development, where the scale of new drug efficacy is often confounded by subject choice, prolonged time to enrolment, and extended exposure to standard of care while awaiting culture results.
Current diagnostic technologies for RTI have changed slowly in the past two decades, with diagnosis still being confirmed by agar-based bacterial culture, and recently, in most settings, complemented further by nucleic acid testing. Thus, current tests are generally slow to produce meaningful results for clinicians, and their utility is restrained by lower sensitivity and specificity in many real life clinical settings, such as in cases of choosing antibiotic for patients already exposed to antibiotics or with hard to culture pathogens. Furthermore, sputum microscopy and other rapid techniques alone cannot demonstrate bacterial resistance patterns early enough to drive specific treatment choices.
Nevertheless there are a growing number of approved rapid diagnostic tests for use in clinical settings other than RTI. These include GeneXpert® (Cepheid, Sunnyvale, CA, USA), a real-time polymerase chain reaction (PCR) test for multiple organisms and LightCycler® SeptiFast, (Roche, Basel, Switzerland) a real-time PCR bacteremia detection assay. GeneXpert is a self-contained unit that combines on-board preparation of a wide array of primary samples (such as swabs, sputum, gastric aspirates, blood, and stool) with detection functions to provide a fully integrated system for nucleic acid analysis through real-time PCR. The system purifies, concentrates, detects, and identifies specific DNA sequences, thus giving results from unprocessed clinical samples. Due to its modular design, the system has the advantage of having a variety of configurations allowing it to meet a broad range of testing criteria. The speed of this system is also a great advantage, with the majority of test results being available after just 1 hour.7 A critical issue with both these platforms, however, is that they do not detect resistance, with the exception of detected resistance of methicillin (via mecA gene) in Staphylococcus aureus and rifampicin (via rpoB gene mutation) in Mycobacterium tuberculosis, which may be determined using the automated nucleic acid amplification test.8
Although laboratory tests such as urinary antigen testing for Legionella and pneumococcus have high specificity and have become standardly available, their low sensitivity has meant that their impact has been lower than hoped.9 Currently in development are fluorescence in situ hybridization-based assays, which detect specific DNA sequences through the use of fluorescent probes. This too is a relatively slow process, thus further underscoring an unmet need for innovative projects such as RAPP-ID.10
Therefore, it is important that novel in vitro diagnostics accommodate either a wide range of sample types or accept a sample that can be used to potentially detect the majority of target pathogens (eg, sputum). Moreover, they should yield meaningful resistance information with guarantee of accuracy in widespread use, an important commercial concern of diagnostic companies. This is demonstrated by the successful global acceptance of the Xpert® Mycobacterium Tuberculosis/Rifampicin assay. In December 2010, the World Health Organization (WHO) endorsed this product as a major step forward in clinical TB diagnosis despite it being licensed for research use only.11 In a study of 6,648 participants, Xpert testing detected 90.3% of culture-confirmed TB, compared with 67.1% for sputum smear microscopy alone. Test sensitivity was 76.9% in smear-negative, culture-positive patients (where sputum microscopy by definition does not detect) and had 99.0% specificity. This higher sensitivity and specificity gives it a great advantage over microscopy. Moreover, the time taken for TB culture results is a significant barrier to timely treatment, so the provision of prompt results (average time to TB detection with Xpert® system was 0 days compared to 1 day for smear microscopy, 16 days with liquid culture, and 30 days in solid culture) and immediate resistance data (Xpert® rifampicin-resistance sensitivity was 94.4% and specificity was 98.3%) substantially reduces time to effective patient treatment. This rapid technique is revolutionizing TB diagnostics in terms of price, accessibility, and availability of platforms.12
Collection of sputum samples is not the only roadblock to validation. In order to register the POCT with a regulatory body, a validation study is required. This requires real-time collection or, preferably, storage of samples in a library. In the case of VAP, HAP, and CAP there is no such sputum library. The New Diagnostics Working Group has set up a TB specimen bank, which will limit the need for field trials, facilitate the approval process, and allow for head to head comparisons between other diagnostics.13
Storing patients’ samples from clinical trials is potentially appealing because to validate and gain approvals for a new platform, a large number of samples encompassing differing pathogens are required by regulatory agencies. However, this process requires multiple ethical and regulatory approvals, possibly including appropriate prospective consent from patients for sample reuse. Documentation must also exist to support the selected specimens in the study; hence, retrospective use of currently stored samples is unlikely to be practical. Therefore, for many in vitro diagnostics performance studies, archived or discarded clinical specimens may be acceptable in lieu of freshly collected ones, as defined by the guidelines of the Global Harmonization Task Force in 2012 and the International Medical Device Regulators Forum, provided sufficient information on specimen characteristics are available.14 The potential for differential degradation of pathogens within sputum samples over time may lead to bias, with an increase in the error calculating sensitivity and specificity. In addition, using archived or leftover specimens where knowledge of previous use of anti-infective treatment regimens is unknown may bias results. As a result, to reduce the risk of variable treatment regimens obscuring new assay predictive markers, it is preferable to use such specimens when treatments are more uniform among the patient populations, as is often found in clinical trials.
Collecting sufficient samples to produce a validation library may prove to be problematic. However, the literature describes spiking of fresh clinical samples as an alternative acceptable to regulators. For example, the Genomics to combat Resistance against Antibiotics in Community-acquired LRTI in Europe (GRACE) project has developed a spiking protocol as part of its wider objective to study resistant pathogens.15 Serial dilutions of M. tuberculosis-spiked samples were used by Bi et al, producing various concentrations of bacteria in sputum to determine the lower limit of detection for the nucleic acid test under evaluation.16
Furthermore, in the event that storage of sputum is not a viable option, a better option may prove to be a number of artificial sputum media, which can be produced and spiked as required. These artificial media mimic the macroscopic and microscopic structure of sputum and so could potentially be used for validation. Given the known difficulties which arise in attempting to access fresh clinical samples, governing bodies accept this as an alternative method of validation. Of note, the WHO has a protocol in place for an artificial sputum medium to be used in device validation.17
Therefore, we performed a review of the current literature on storage of clinical samples to aid the production of a sputum library for RAPP-ID POCT platform validation. As mentioned, should routine sputum storage not prove viable, we also researched artificial sputum and its storage conditions. We have identified and present in this review major roadblocks, particularly in the acquisition, accessing, and storage of samples.
Methods
This English language literature review was performed using PubMed, Scopus, and Google Scholar search engines (Figure 1). Queries used to find relevant information included searching for sputum storage and artificial sputum in the title, abstract, and keyword list. We also considered methodology for collection of other respiratory tract samples, such as bronchoalveolar lavage, endotracheal aspirates, swabs, and artificial sputum and its storage times in our search strategy.
Figure 1.
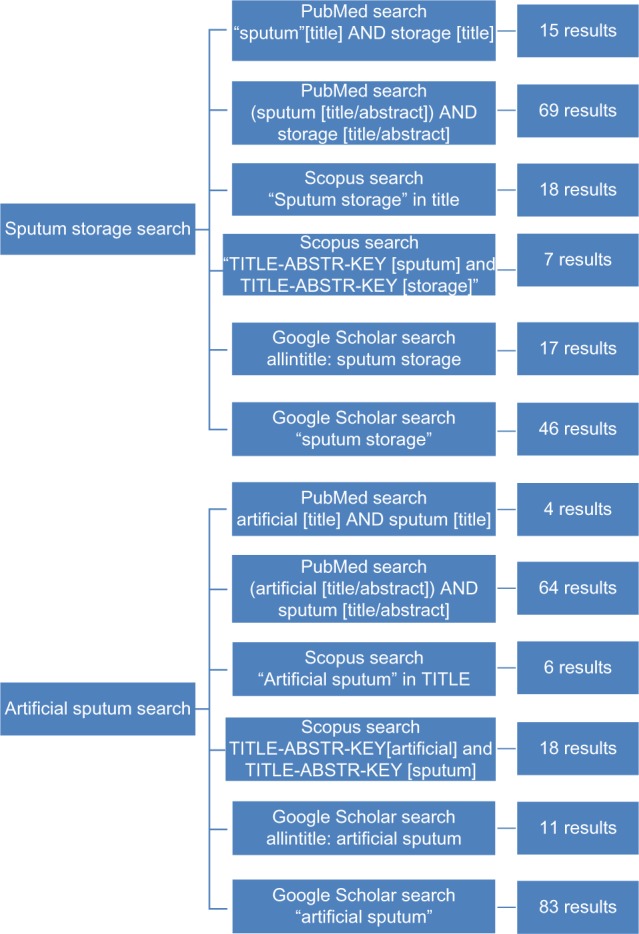
Literature review results summary.
Notes: The availability of literature is limited, as shown by the few results after multiple database searches. Many of the sputum storage articles do not pertain to this review’s selected organisms and disease areas. Furthermore the storage times are significantly shorter than required for a sputum library. Similarly, artificial sputum results often analyzed organisms not detailed in this review, and the severe lack of comparison data further complicated the selection process.
Abbreviations: ABSTR, abstract; KEY, keywords.
Results
Few studies on sputum sample storage time exist
Overall, we found there to be a paucity of data on sputum storage. Only ten studies focused primarily on the length of time for which bacterially-infected sputum samples could be stored at different temperatures, with an average storage time of 48 hours. This is not long enough to provide data for the production of a long-term sputum library, where a number of years of storage may be required.
Searches for studies addressing host cell viability (which might be used in POCT biomarker research or validation) yielded better results, with some studies analyzing sample viability during storage, focusing more on viability of organisms after transport. However, these time periods were no greater than 1 month; therefore, we performed more general searches in article abstracts and keywords (Figure 1). While this did yield a higher number of results, articles which did not include the search terms in the title tended not to focus on storage time, rather having this as a secondary topic, usually in the context of an analysis of how samples deteriorated under various transport conditions. As a result, these studies were deemed not relevant to sample library set-up.
Studies were selected from search results based on their target disease areas and storage times so as to be relevant to sputum storage. Since each bacterial species is likely to react differently to long-term storage conditions, studies focusing on HAP/VAP pathogens and TB were selected as relevant to the aims of RAPP-ID. Studies with storage times longer than 1 week were preferable, and shorter timespans proved useful for the basic theory but did not provide data to inform library set-up. Similar criteria were used to select studies in the artificial sputum search results. Chief among these were studies relating to TB. A number of studies also exist pertaining to artificial medium used for cystic fibrosis (CF); this will be expanded upon later in the review.
Murray et al18 demonstrated that bacterial viability in bronchiectatic sputum stored at 25°C, 4°C, and −20°C was affected by storage for 24 hours and 48 hours before processing. Sputum processed 1 hour after expectoration and stored for 24 hours at 25°C showed an increase in Pseudomonas aeruginosa density. In contrast, at 4°C there was no significant difference in P. aeruginosa density, while at −20°C after 24 hours there was a significant reduction in Pseudomonas density, showing that the organism was less viable at lower storage temperatures. Thus, long-term storage of samples leads to death of this organism under a range of storage temperatures.
A study compared host cell counts (macrophages, neutrophils, and eosinophils) between two different aliquots of induced sputum from the same subject, one aliquot processed 2 hours after expectoration and the other after it was frozen with dimethyl-sulfoxide and stored for up to 10 days at −20°C.19 Thirty-five samples were frozen immediately before cytospin preparation, and ten samples were frozen just after homogenization with sputolysin. Samples were stored for an average of 5 days. Over this period of time, there were no significant differences found in those host cell counts.
Swabs, blood, and urine for use in POCT diagnosis of RTI
Alternatives to sputum for diagnosis of RTI do exist; however, they are less than ideal for the setting of a POCT. For example, the use of nasopharyngeal swabs to diagnose RTI critically depends upon whether the nasopharynx adequately reflects the organisms expectorated or suctioned from deep samples. The GRACE consortium are attempting to reduce antibiotic resistance through genomic research of antibiotic use in LRTI cases; as an additional goal they aim to clarify the use of nasopharyngeal samples as an alternative to sputum to diagnose RTI.20 Traditionally, oropharyngeal, not nasopharyngeal, swabs are taken in clinical practice. Huijskens et al analyzed the diagnostic effectiveness of using blood, sputum, oropharyngeal, and urinary samples to detect various respiratory pathogens.21 The study found that oropharyngeal swabs and sputum were effective at RTI diagnosis but that sputum samples provided the greatest variety of diagnoses of pulmonary diseases. As a result, the authors recommended that for molecular detection of bacterial pathogens, sputum is imperative as a diagnostic tool.
Bacterial density is affected by storage conditions
Pye et al compared bacterial recovery following sputum storage at 20°C (defined as room temperature) and 4°C (refrigerated) for 24 hours. Although the investigators were able to isolate the dominant pathogen in all samples regardless of the storage temperature, samples stored at 4°C had lower numbers of each pathogen. Pathogen recovery rate after storage at 20°C showed no difference to that immediately after collection.22
The importance of storage temperature is further confirmed by two studies performed by Paramasivan et al. Sputum specimens stored at room temperature, with culture performed at 0, 3, 7, 14, 21, and 28 days demonstrated 88%, 83%, 68%, 22%, 13%, and 0% recovery, respectively. In addition, room temperature storage for 3, 5, and 7 days gave culture positivity rates of 83%, 71%, and 63%. So sputum cannot be stored at room temperature for more than 3 days.23
Hence, we can conclude that different pathogens will behave differently during storage, and no relevant publications systematically assessing storage of whole sputum samples for validation of a diagnostic device have been published. This lack of published data shows that to build an infected sample library, a validation study would be required, studying both infected and uninfected sputum storage conditions for important pathogens, under a variety of storage conditions. Furthermore, the lowest storage temperature used in these studies was −20°C, thus further studies should be performed to explore the effects at −80°C on sputum and organisms, with particular consideration to prevention of organism damage, such as ice shearing, at these temperatures.
Sputum samples for TB validation can be stored for subsequent analysis
The maximum length of low-temperature storage found in the literature for M. tuberculosis was 6 months. These stored samples were subsequently used for Ziehl–Nielsen smear and routine culture but not rapid diagnostic (molecular) tests. Tessema et al showed that samples of sputum stored at −20°C for a median of 132 days, without the addition of preservatives, still showed a high rate of recovery when assayed by smear and culture after thawing.24 This suggests a cost effective method of storage of TB samples in contrast to pathogens causing more acute disease. More thorough validation of storage will be required before these data can be used for library sample collection. Alternatively, as mentioned earlier, the Stop TB Partnership has access to the Foundation for Innovative New Diagnostics TB specimen bank, containing sputum samples in 0.5 ml aliquots among other sample types of totally drug resistant TB. Specimens from this library are collected from adults before subjects start any form of TB treatment. Sputum samples are then homogenized with H2O or N-acetyl-L-cysteine/sodium citrate and glass beads. Due to the method of storage used, it is estimated that the samples would remain viable for 100 years, making this a potential method for TB POCT device validation.25
A study using heat-fixed sputum smear samples stored for a maximum of 10 months at ambient temperatures ranging from 25°C–45°C found, in panel testing, only 15 samples had deteriorated enough to cause reading errors, but no speciation or resistance information could be generated using this approach.26
Spiking of sputum samples
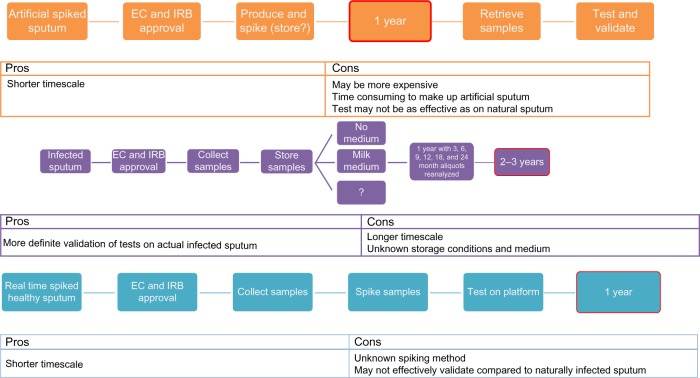
Clinical trials are an efficient way to provide well-characterized sputum specimens. On collection, storage and use of these samples for validation efforts might be possible at a later date. However, our literature review found a paucity of data on long-term storage of such samples, meaning a lengthy validation study of any novel storage procedure would need to be performed. As a result, we conclude that fresh samples should be analyzed in real-time within an ongoing clinical trial, or that known negative samples be stored and spiked later, using a yet to be developed spiking protocol. As shown in Figure 2, this will help to reduce time to validation of the POCT, as samples will be accessible as needed, without prior validation of a storage protocol or a need for informed consent.
Figure 2.
Timelines to proof of concept of rapid diagnostic test.
Notes: Depending on the sample method chosen, timelines can be vastly increased or decreased. Spiked or real time samples could mean that devices could be validated within 1 year, while attempting to validate artificial sputum samples first could delay the project by at least 1 year. Similarly, attempting a validation study on the long-term storage of infected sputum samples, which again may or may not be successful, could cause validation of a rapid POCT to be delayed by anywhere up to 3 years.
Abbreviations: EC, ethics committee; IRB, institutional review board; POCT, point of care test.
Spiking discarded sputum samples with known quantities of specific bacteria for use in validation of POCTs is a potential replacement for infected samples that may only exist in geographically diverse clinical trials sites. Sputum samples are obtained from subjects (or in hospital laboratories) and checked for pathogens using current methodology, such as antibody and PCR assays. Once the samples are confirmed as negative, they would be spiked using a standard operating protocol and used for POCT validation.
Artificial sputum might be suitable for TB diagnostic validation
If we conclude that stored clinical samples are not an option for validation of POCTs and availability of sputum for spiking is time-consuming and expensive, producing artificial sputum and spiking these samples for validation is an attractive option. However, few papers directly comparing artificial and natural sputum storage have been published, both for bacterial pathogens and TB. Limited data are available regarding a polyacrylamide based artificial sputum (PBAS1) stored alone for up to 9 months at both 4°C and −20°C (without additional storage medium). When compared with fresh smears of PBAS1 there were few microscopic or macroscopic differences between the two, showing a good product life.27
While no data are available comparing PBAS1 and sputum, there are data available on the microscopic and macroscopic differences between sputum and a second artificial sputum, PBAS2. The fundamental differences between these two artificial sputa are a new preparation method allowing for more consistency between samples and the presence of a formalin-fixed TB solution which provides oral flora to the samples.28 Yamada et al also state that both macroscopically and microscopically, PBAS2 and sputum were virtually indistinguishable when compared to the fresh TB smears.27 As a result, it is likely that PBAS2 could be used as a sputum replacement, at least in the analysis of TB samples, somewhat helping to decrease the timelines for validation of a diagnostic device if regulatory approval for the technique were gained. It would be advisable to conduct further studies using PBAS2, assessing both longer-term storage at lower temperatures (eg, −80°C) and using a spiking protocol to produce samples in real time. This would help to show the long term viability of samples for use in a library, as well as providing an alternative through spiking of samples should long term storage not be an option.
Furthermore, the WHO has a protocol in place for an artificial sputum media; however, no study could be found comparing this artificial sputum with natural media, meaning that further validation would still be required before this could successfully be used to test a POCT.17
Artificial sputum medium (ASM) mimics the sputum found in CF.29 ASM is an homogenous, nonviscous medium that can be prepared in large quantities. CF biofilms contain cells attached to a solid pulmonary surface allowing P. aeruginosa to grow under microaerophilic to anaerobic conditions in microcolonies mimicked by ASM. Nevertheless, significant differences between CF sputum and patients suffering with RTIs exist, so we feel ASM is not an adequate substitute for infected or spiked sputum. However, polyacrylamide as a sputum substitute, particularly in the form of PBAS2, demonstrates promise, especially in the study of TB. Further data on storage time with PBAS2 is needed.
Conclusion
Due to the lack of robust data found in the review, we conclude that formation of a long-lasting, high-quality sputum sample library is a major roadblock to registration of next-generation diagnostics. While there is still an option to validate storage times for creation of a sputum library, Figure 2 illustrates that since there is no simple solution to storage of sputum samples for production of a HAP/VAP sputum library, this may not be a viable method to pursue; furthermore, were a validation study performed, it may still not provide a guarantee that defrosted samples held under experimental conditions will adequately reflect original microbiology/biomarkers. In order to prevent delay to proof of concept for any novel device, we feel a prototype platform should either be placed in an antibiotic clinical trial site with prior agreement of the investigator and appropriate consents and approvals in place, or spiked samples used, and acknowledge that these do not reflect true patient samples. In contrast, for TB, a sputum library might need less validation to set up based on the information from the Stop TB Partnership and the Foundation for Innovative New Diagnostics bank, and use of artificial sputum may be possible.
Acknowledgments
This work was supported by the Innovative Medicines Initiative, a public-private partnership between the European Union and the European Federation of Pharmaceutical Industries and Associations (RAPP-ID project, grant agreement no 115153). Thanks to Stephan Harbarth for his guidance and advice on this document.
Footnotes
Author contributions
JMD and JAG devised the concept of the review. JMD performed the literature review and the first draft of the paper. Both authors revised the paper critically for important intellectual content.
Disclosure
JAG is employed by and holds stock in GlaxoSmithKline Ltd. JMD is a previous Industrial Placement student at GlaxoSmithKline Ltd. The authors report no other conflicts of interest.
References
- 1.Davies SC, Fowler T, Watson J, Livermore DM, Walker D. Annual Report of the Chief Medical Officer: infection and the rise of antimicrobial resistance. Lancet. 2013;381(9878):1606–1609. doi: 10.1016/S0140-6736(13)60604-2. [DOI] [PubMed] [Google Scholar]
- 2.RAPP-ID [homepage on the Internet] RAPP-ID Consortium; 2011. [Accessed October 25, 2013]. Available from: http://www.ua.ac.be/main.aspx?c=RAPP-ID&n=93905. [Google Scholar]
- 3.Innovative Medicines Initiative [homepage on the Internet] Innovative Medicines Initiative; 2013. [Accessed October 25, 2013]. [Updated November 29, 2013]. Available at: http://www.imi.europa.eu/content/home. [Google Scholar]
- 4.Kelsey MC, Mitchell CA, Griffin M, Spencer RC, Emmerson AM. Prevalence of lower respiratory tract infections in hospitalized patients in the United Kingdom and Eire – results from the Second National Prevalence Survey. J Hosp Infect. 2000;46(1):12–22. doi: 10.1053/jhin.2000.0775. [DOI] [PubMed] [Google Scholar]
- 5.Hak E, Rovers MM, Kuyvenhoven MM, Schellevis FG, Verheij TJ. Incidence of GP-diagnosed respiratory tract infections according to age, gender and high-risk co-morbidity: the Second Dutch National Survey of General Practice. Fam Pract. 2006;23(3):291–294. doi: 10.1093/fampra/cmi121. [DOI] [PubMed] [Google Scholar]
- 6.Laessig KA. End points in hospital-acquired pneumonia and/or ventilator-associated pneumonia clinical trials: food and drug administration perspective. Clin Infect Dis. 2010;51(Suppl 1):S117–S119. doi: 10.1086/653059. [DOI] [PubMed] [Google Scholar]
- 7.Spencer DH, Sellenriek P, Burnham CA. Validation and implementation of the GeneXpert MRSA/SA blood culture assay in a pediatric setting. Am J Clin Pathol. 2011;136(5):690–694. doi: 10.1309/AJCP07UGYOKBVVNC. [DOI] [PubMed] [Google Scholar]
- 8.Al-Darraji HA, Razak HA, Ng KP, Altice FL, Kamarulzaman A. The Diagnostic performance of a single GeneXpert MTB/RIF assay in an intensified tuberculosis case finding survey among HIV-infected prisoners in Malaysia. PLoS One. 2013;8(9):e73717. doi: 10.1371/journal.pone.0073717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcos MA, Jiménez de Anta MT, de la Bellacasa JP, et al. Rapid urinary antigen test for diagnosis of pneumococcal community-acquired pneumonia in adults. Eur Respir J. 2003;21(2):209–214. doi: 10.1183/09031936.03.00058802. [DOI] [PubMed] [Google Scholar]
- 10.Levsky JM, Singer RH. Fluorescence in situ hybridization: past, present and future. J Cell Sci. 2003;116(Pt 14):2833–2838. doi: 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- 11.Rachow A, Zumla A, Heinrich N, et al. Rapid and accurate detection of Mycobacterium tuberculosis in sputum samples by Cepheid Xpert MTB/RIF assay – a clinical validation study. PLoS One. 2011;6(6):e20458. doi: 10.1371/journal.pone.0020458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boehme CC, Nicol MP, Nabeta P, et al. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet. 2011;377(9776):1495–1505. doi: 10.1016/S0140-6736(11)60438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann G, Squire SB, Bissell K, et al. Beyond accuracy: creating a comprehensive evidence base for TB diagnostic tools. Int J Tuberc Lung Dis. 2010;14(12):1518–1524. [PubMed] [Google Scholar]
- 14.Study Group 5 of the Global Harmonization Task Force . Clinical Evidence for ivd Medical Devices – Clinical Performance Studies for In Vitro Diagnostic Medical Devices. Global Harmonization Task Force; 2013. [Accessed August 29, 2013]. Available from: http://www.imdrf.org/docs/ghtf/final/sg5/technical-docs/ghtf-sg5-n8-2012-clinical-performance-studies-ivd-medical-devices-121102.pdf. [Google Scholar]
- 15.Little P, Stuart B, Francis N, et al. GRACE consortium Effects of internet-based training on antibiotic prescribing rates for acute respiratory-tract infections: a multinational, cluster, randomised, factorial, controlled trial. Lancet. 2013;382(9899):1175–1182. doi: 10.1016/S0140-6736(13)60994-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bi A, Nakajima C, Fukushima Y, et al. A rapid loop-mediated isothermal amplification assay targeting hspX for the detection of Mycobacterium tuberculosis complex. Jpn J Infect Dis. 2012;65(3):247–251. doi: 10.7883/yoken.65.247. [DOI] [PubMed] [Google Scholar]
- 17.World Health Organization . Laboratory services in tuberculosis control [webpage on the Internet] Geneva: World Health Organizations; 1999. [Accessed October 25, 2013]. Available from: http://apps.who.int/bookorders/anglais/detart1.jsp?codlan=1&codcol=93&codcch=3167. [Google Scholar]
- 18.Murray MP, Doherty CJ, Govan JR, Hill AT. Do processing time and storage of sputum influence quantitative bacteriology in bronchiectasis? J Med Microbiol. 2010;59(Pt 7):829–833. doi: 10.1099/jmm.0.016683-0. [DOI] [PubMed] [Google Scholar]
- 19.Holz O, Mücke M, Zarza P, Loppow D, Jörres RA, Magnussen H. Freezing of homogenized sputum samples for intermittent storage. Clin Exp Allergy. 2001;31(8):1328–1331. doi: 10.1046/j.1365-2222.2001.01136.x. [DOI] [PubMed] [Google Scholar]
- 20.de Vries M, Deijs M, Canuti M, et al. A sensitive assay for virus discovery in respiratory clinical samples. PLoS One. 2011;6(1):e16118. doi: 10.1371/journal.pone.0016118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huijskens EG, Rossen JW, Kluytmans JA, van der Zanden AG, Koopmans M. Evaluation of yield of currently available diagnostics by sample type to optimize detection of respiratory pathogens in patients with a community-acquired pneumonia. Influenza Other Respir Viruses. 2013 Aug 20; doi: 10.1111/irv.12153. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pye A, Hill SL, Bharadwa P, Stockley RA. Effect of storage and postage on recovery and quantitation of bacteria in sputum samples. J Clin Pathol. 2008;61(3):352–354. doi: 10.1136/jcp.2007.051490. [DOI] [PubMed] [Google Scholar]
- 23.Paramasivan CN, Narayana AS, Prabhakar R, Rajagopal MS, Somasundaram PR, Tripathy SP. Effect of storage of sputum specimens at room temperature on smear and culture results. Tubercle. 1983;64(2):119–124. doi: 10.1016/0041-3879(83)90036-3. [DOI] [PubMed] [Google Scholar]
- 24.Tessema B, Beer J, Emmrich F, Sack U, Rodloff AC. Rate of recovery of Mycobacterium tuberculosis from frozen acid-fast-bacillus smear-positive sputum samples subjected to long-term storage in Northwest Ethiopia. J Clin Microbiol. 2011;49(7):2557–2561. doi: 10.1128/JCM.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vincent V, Rigouts L, Nduwamahoro E, et al. The TDR Tuberculosis Strain Bank: a resource for basic science, tool development and diagnostic services. Int J Tuberc Lung Dis. 2012;16(1):24–31. doi: 10.5588/ijtld.11.0223. [DOI] [PubMed] [Google Scholar]
- 26.Selvakumar N, Sivagamasundari S, Prabhakaran E, et al. Storage of heat-fixed unstained sputum AFB smears for panel testing in a tuberculosis unit in South India. Int J Tuberc Lung Dis. 2005;9(2):223–225. [PubMed] [Google Scholar]
- 27.Yamada H, Matsumoto H, Mitarai S, Fujiki A. [Stability for long-term storage and reproducibility of positivity in the panel test slide prepared with the polyacrylamide-based artificial sputum] Kekkaku. 2008;83(2):65–71. Japanese. [PubMed] [Google Scholar]
- 28.Yamada H, Mitarai S, Wahyunitisari MR, et al. Improved polyacrylamide-based artificial sputum with formalin-fixed tubercle bacilli for training of tuberculosis microscopists. J Clin Microbiol. 2011;49(10):3604–3609. doi: 10.1128/JCM.00370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sriramulu DD. Artificial sputum medium [webpage on the Internet] Nature Publishing Group: Protocol Exchange; 2010. [Accessed October 25, 2013]. Available from: http://www.nature.com/protocolexchange/protocols/1999. [Google Scholar]