Abstract
Glycosylphosphatidylinositols (GPIs) from several protozoan parasites are thought to elicit a detrimental stimulation of the host innate immune system aside their main function to anchor surface proteins. Here we analyzed the GPI biosynthesis of an avirulent Toxoplasma gondii type 2 strain (PTG) by metabolic radioactive labeling. We determined the biological function of individual GPI species in the PTG strain in comparison with previously characterized GPI-anchors of a virulent strain (RH). The GPI intermediates of both strains were structurally similar, however the abundance of two of six GPI intermediates was significantly reduced in the PTG strain. The side-by-side comparison of GPI-anchor content revealed that the PTG strain had only ∼34% of the protein-free GPIs as well as ∼70% of the GPI-anchored proteins with significantly lower rates of protein N-glycosylation compared to the RH strain. All mature GPIs from both strains induced comparable secretion levels of TNF-α and IL-12p40, and initiated TLR4/MyD88-dependent NF-κBp65 activation in macrophages. Taken together, these results demonstrate that PTG and RH strains differ in their GPI biosynthesis and possess significantly different GPI-anchor content, while individual GPI species of both strains induce similar biological functions in macrophages.
Introduction
The obligate intracellular protozoan Toxoplasma gondii ranks among the most common parasites of animals and humans, with an estimated worldwide incidence of 30% of chronically infected individuals [1]. The canonical lineages, designated types 1, 2, 3 (formerly I, II, and III) and the haplogroup 12, comprise the genus Toxoplasma in Europe and North America [2], [3], [4]. These haplogroups differ genetically by 1% or less but diverge in a number of phenotypes such as growth, migration potential, and most notably acute virulence in laboratory mice [5], [6]. Type 1 strains are highly virulent and uniformly lethal with LD100 of <10 parasites, whereas type 2 strains are avirulent in most, but not all, inbred mouse lines with a median lethal dose (LD50) of ∼103 parasites. Type 3 strains are typically avirulent with LD50≥105 parasites [7], [8].
Toxoplasma polymorphic effector molecules subvert key host defense mechanisms and account for much of the strain-specific difference in virulence in mice [9], [10]. These are introduced during invasion into the host cytosol from secretory compartments, namely rhoptries and dense granules, and traffic to the host nucleus and to the nascent parasitophorous vacuole membrane (PVM). The high virulence of type 1 strains is thus largely due to the rhoptry protein (ROP)18 kinase, the closely related ROP5 pseudokinase, and the ROP16 kinase [6], [11], [12], [13], [14]. The former kinases target host immunity by disrupting the function of immunity-related GTPase (IRG)-mediated parasite clearance in macrophages, while this process is not affected by type 2 and 3 strains, where IRGs cause disruption of the PVM and subsequent parasite destruction [15], [16], [17]. The latter alters host gene transcription through sustained activation of signal transducer and activator of transcription (STAT)3/STAT6 and promotes alternative macrophage activation (M2), thereby limiting protective Th1 responses, which in turn favor parasite growth, causing possibly a secondary response that leads to cytokine-mediated pathology [18], [19], [20], [21]. This detrimental overstimulation of host immunity is likely triggered by parasite components, such as profilin, cyclophilin-18, and glycosylphosphatidylinositol (GPI) anchors [22], [23], [24].
GPI anchors are common glycolipids in all eukaryotes that mainly function to covalently anchor surface proteins to membranes, while synthesized at much higher rates in parasitic protozoa than in other eukaryotes [25]. Over the last two decades many biological and immunological properties have been found associated with GPI anchors and GPI-related molecules of protozoan parasites [26], [27], [28]. Both protein-free GPIs and/or several GPI-anchored proteins, including SAG1-related sequence proteins (SRS) and SAG-unrelated surface antigens (SUSA) have been reported to decorate the surface of all T. gondii life cycle stages, however the molecular function and biological importance of most surface proteins remain poorly defined [29], [30], [31], [32], [33]. The majority of known SRS and SUSA proteins have dual functions, either as adhesins mediating initial host cell attachment, and/or immune decoys eliciting or modulating inflammatory responses during acute infection [29], [30], [31], [33]. The GPI biosynthesis has been shown to be essential for T. gondii viability but it remains unknown whether the observed lethality is due to the lack of protein-free GPIs, GPI-anchored proteins or both [34]. The protein-free GPI intermediates from the virulent T. gondii RH strain have been structurally analyzed and demonstrated to induce a TLR2/TLR4/MyD88-mediated TNF-α production in macrophages that is dependent on the expression of galectin-3 [23], [35], [36], [37]. In contrast, little was known about the structure and the biological functions of GPIs from avirulent T. gondii strains.
In this report, we characterized the GPI content and assessed the biological function of individual GPI species of an avirulent type 2 strain (PTG). We demonstrated that this strain possesses significantly less GPI content, lower levels of GPI-anchored proteins, and lower rates of GPI-anchored protein N-glycosylation than the virulent RH strain, while protein-free GPIs of both strains activate identical TLR signaling pathways and induce comparable levels of inflammatory cytokines in macrophages.
Materials and Methods
Analysis, extraction, and purification of individual GPIs
GPIs of the strains RH and PTG of T. gondii were metabolically labeled by incubating parasites grown in Mycoplasma-free Vero (American Type Culture Collection, ATCC) cells, in 20 ml glucose-free DMEM containing 20 mM sodium pyruvate and supplemented with 0.5 mCi D-[6-3H]-glucosamine hydrochloride (Hartmann Analytic GmbH, Braunschweig) for 6 h at 37°C in 5% CO2 atmosphere. Remaining intracellular parasites were released from host cells with the help of glass beads in a Mixer Mill homogenizer (Retsch, Haan) and subsequently purified from host cell debris by glass wool filtration [38]. Labeled and unlabeled GPIs were purified from a determined number of parasites using the method previously described to obtain both protein-free and GPI-anchored proteins [39]. Briefly, glycolipids were extracted with chloroform-methanol-water 10∶10∶3 (v/v), partitioned between water and water-saturated butan-1-ol, and precipitated gently under a nitrogen stream. Finally, GPIs recovered from the butan-1-ol phase were separated by thin-layer chromatography (TLC) on Merck Si 60 HPTLC plates using chloroform-methanol-hexane-water-acetic acid 10∶10∶3∶2∶1 (v/v) as solvent and D-[6-3H]-glucosamine labeled GPIs used as tracers. Chromatograms were scanned for radioactivity using a Berthold LB 2842 linear analyzer and areas corresponding to individual GPIs [35] were scraped off, re-extracted with chloroform-methanol-water 10∶10∶3 (v/v), and recovered in the butan-1-ol phase after water-saturated butan-1-ol/water partition to obtain individual purified GPIs. The absence of endotoxin in purified GPIs was validated with the Limulus Amebocyte Lysate kit QCL-100 (Bio-Whittaker, Walkersville, MD). GPIs were stored at −20°C in butan-1-ol until use.
Structural analysis of GPI core glycans
Extracted GPIs were dephosphorylated, deaminated, and reduced as described elsewhere [40]. Briefly, GPIs were dephosphorylated with 48% aqueous hydrofluoric acid for 60 h at 0°C and the reaction was stopped by blowing off hydrofluoric acid under a nitrogen stream. Molecules were suspended in 400 µl of freshly prepared 0.1 M sodium acetate (pH 3.5) containing 0.25 M NaNO2 and incubated at room temperature (RT) for 4 h. The reaction was then terminated by addition of 300 µl of 0.4 M boric acid and 130 µl of 1 M NaOH. The material was reduced overnight at 4°C using 2 M NaBH4 in 0.3 M NaOH and the reaction was stopped with 5% acetic acid in methanol that was removed by two evaporations with 50 µl toluene. The resulting material was desalted on a Chelex 100 (Na+) - AG50X12 (H+) - AG3X4 (OH−) - QAE Sephadex A25 (OH−) ion-exchange column and filtered through a 0.2 µm filter. D-[6-3H]-glucosamine-labeled neutral glycans were digested by 2 units of Jack bean α-mannosidase or N-acetylhexosaminidase in 100 µl of 50 mM sodium acetate (pH 4.5) containing 0.2 mM ZnCl2 and 0.02% sodium azide for 24 h at 37°C [41]. Analysis was performed by HPAEC using a Bio-LC Dionex module (Dionex Corporation, Sunnyvale, CA) before and after digestion. Samples were mixed with 0.6 mg of partially hydrolyzed dextran [42] prior to injection into a Carbopac® PA1 anion-exchange column equilibrated with 100 mM NaOH. Elution was accomplished with 100% buffer A (100 mM NaOH) for 6 min, followed by a linear increase of buffer B (100 mM NaOH, 0.25 M NaOAc) from 0 to 30% in 30 min at a flow rate of 1 ml/min. Positions of glucose monosaccharide and oligomer standards were obtained using pulsed amperometric detection, while eluted radiolabeled glycans were detected by scintillation counting.
GPI quantification and carbohydrate composition analysis
The method is based upon quantification of the GlcN residues of the GPIs being converted to AHM as described elsewhere [43] with slight modifications. An internal standard of scyllo-inositol was added to each triplicate sample containing either protein-free GPIs or GPI-anchored proteins. Controls with various GlcN and myo-inositol concentrations were prepared and processed in parallel. Samples were dephosphorylated, deaminated, reduced, and transferred to glass capillaries in 100 µl methanol-water 1∶1 (v/v) and dried in a vacuum concentrator. Methanolysis was conducted by adding 50 µl of dry methanol containing 0.5 M HCl, flame sealing the capillaries under vacuum and incubating them at 95°C for 4 h. The glass capillaries were opened, pyridine (10 µl) was added to neutralize the HCl and re-N-acetylation was performed with acetic anhydride (10 µl) for 30 min at RT. Samples were dried twice from dry methanol (20 µl) in a vacuum concentrator. Derivatization was performed with 10 µl of TMS (trimethylchlorosilane-hexamethyldisilazane-dry pyridine, 1∶3∶10, v/v) for 20 min prior to analysis of 2 µl of the products by GC-MS on a MS detector-5973 (GC-6890N, Agilent Technologies, Santa Clara, CA) with a HP-5 column (30 m×25 mm) at 80°C for 2 min followed by a gradient up to 140°C at 30°C/min and a second gradient up to 265°C at 5°C/min and held at 265°C for a further 10 min. Ion monitoring of m/z 273 was selected to detect AHM and m/z 318 to detect both scyllo- and myo-inositol. The peak areas of the corresponding standards were used to calculate the molar relative response, allowing quantification of both AHM and myo-inositol in the samples. The same method was used to determine monosaccharide content. Analyses were conducted in triplicate on 0.5 nM of myo-inositol containing glyconjugate/glycoprotein, with 1.0 nM scyllo-inositol as an internal standard. A mixture of sugar standards (each at 0.5 nM) was run in parallel to determine the molar relative response factor.
Structural analysis of PIs
Extracted GPIs were subjected to deamination, followed by organic extraction prior to analysis by ES-MS on a Quattro Ultima triple quadrapole instrument. MassLynx was used to record and process the data [44].
Immunofluorescence microscopy
HFF (ATCC) cells were grown to confluence on glass coverslips and infected either with T. gondii RH or PTG strains. Cells were fixed (48 h post infection, h p.i.) with 4% paraformaldehyde and subsequently permeabilized with 0.02% Triton-X 100, 10% bovine serum albumin (BSA) for 30 min, washed three times with PBS, and incubated for 1 h in 0.02% Triton-X 100, 10% BSA with mAb T54 E10 [45] that recognizes the EtN-PO4 and GlcGalNAc epitopes of protein-free GPIs [35]. Cells were washed again for three times with PBS and incubated for 1 h with secondary FITC-conjugated anti-mouse immunoglobulin antibody (DakoCytomation, Glostrup) in 0.02% Triton-X 100, 10% BSA. Finally, cells were washed three times with PBS and mounted in Fluoprep (BioMérieux, Marcy-l'Etoile).
Cytokine measurements
RAW 264.7 (ATCC) and J774 (ATCC) mouse macrophages were used to measure TNF-α or IL-12p40 levels, respectively. Macrophages were incubated with protein-free GPIs purified by TLC from the RH or PTG strain. For that purpose, the GPI material needed was dried under a stream of nitrogen to volatilize the butan-1-ol. The GPIs were suspended by sonication in PANSERIN 401 serum-free medium. Measurements of TNF-α and IL-12p40 in cell culture supernatants were determined using a specific sandwich ELISA (BD Pharmingen), following the manufacturer's directions. GPIa, a chemically synthesized GPI molecule of T. gondii [46] kindly provided by Prof. Richard R. Schmidt (Department of Chemistry, University of Konstanz, Germany), was tested on J774 cells.
NF-κB p65 transcription factor assay
RAW 264.7 macrophages were stimulated for 15, 30, and 60 min with or without protein-free GPIs purified from 4×108 parasites of the PTG strain or with medium alone. Macrophage proteins were extracted and activation of NF-κB was measured using the TransAM™ NF-κBp65 transcription factor assay kit according to the manufacturer's instructions (Active Motif, Rixensart).
FACS analysis
The CHO reporter cell lines (CHO/CD14, expressing endogenous functional TLR4; 7.19/CD14/TLR2, expressing stably transfected human TLR2; and the 7.19 clone, expressing neither TLR2 nor functional TLR4) contain a cd25 gene reporter under the control of the human E-selectin promoter, which contains an NF-κB-binding site [47], [48]. The CHO reporter cell lines were generated as described elsewhere [48]. Cells were plated at 1×105 per well in 24-well tissue-culture dishes. After 18 h stimulation with medium alone or with protein-free GPIs purified from 1×109 parasites of the PTG strain, cells were stained with PE-labeled anti-CD25 (mouse mAb to human CD25, PE conjugate; Caltag Laboratories, Burlingame, CA), examined by FACS analysis and evaluated with CellQuest software (BD Biosciences, San Jose, CA).
Statistical analysis
An unpaired Student t-test was used for statistical evaluation and P≤0.05 was considered statistically significant.
Results
PTG and RH strains synthesize different sets of protein-free GPIs
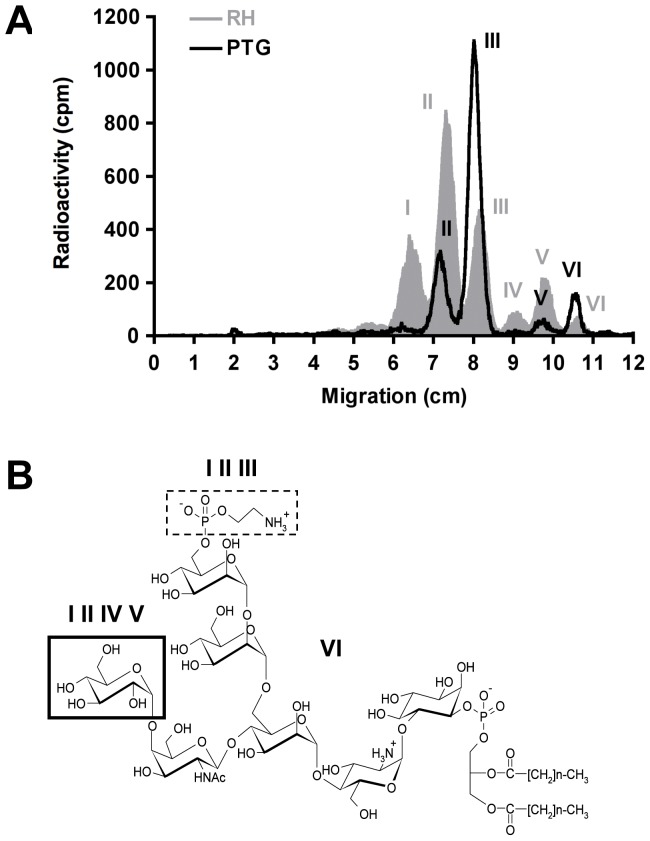
We first sought to examine whether the virulent RH strain and the avirulent PTG strain synthesize identical GPI structures. To this end, we metabolically labeled these two strains with D-[6-3H]-glucosamine. Figure 1A shows the separation of [3H]-labeled protein-free GPIs of both strains by thin layer chromatography (TLC). The RH strain synthesized six different GPI structures with previously described structures [35]. In contrast, the PTG strain synthesized only four major GPIs that seem to correspond to GPIs II, III, V and VI of the RH strain. The Glc-GalNAc-substituted GPIs I and IV were strongly reduced to near background levels in the PTG strain (Figure 1A). The peaks of both strains migrated with nearly identical Rf (rate of flow) values, suggesting only minor differences in their structures (Figure 1A).
Figure 1. RH and PTG strains have different GPI profiles.
(A) Metabolically labeled ([3H]-glucosamine) parasite glycolipids were extracted and separated by TLC as described in Materials and Methods. TLC chromatograms were scanned for radioactivity using a Berthold LB 2842 linear analyzer. Six different peaks representing individual GPIs are detected in the RH strain [35], whereas only four major peaks are present in the PTG strain. (B) Structures of T. gondii RH GPIs. GPI VI (Man-Man-[GalNAc]Man-GlcN-PI) is neither substituted with an EtN-PO4 nor with the Glc linked to the GalNAc residue. GPIs I, II and III contain an EtN-PO4 residue (dashed frame line). GPIs I, II, IV and V are substituted with a Glc that is linked to the GalNAc side-branch in α1–4 linkage (thickened frame line). GPIs I, II and IV, V, respectively, have identical carbohydrate moieties and differ in their fatty acid composition.
GPIs of PTG and RH strains have similar core glycan structures
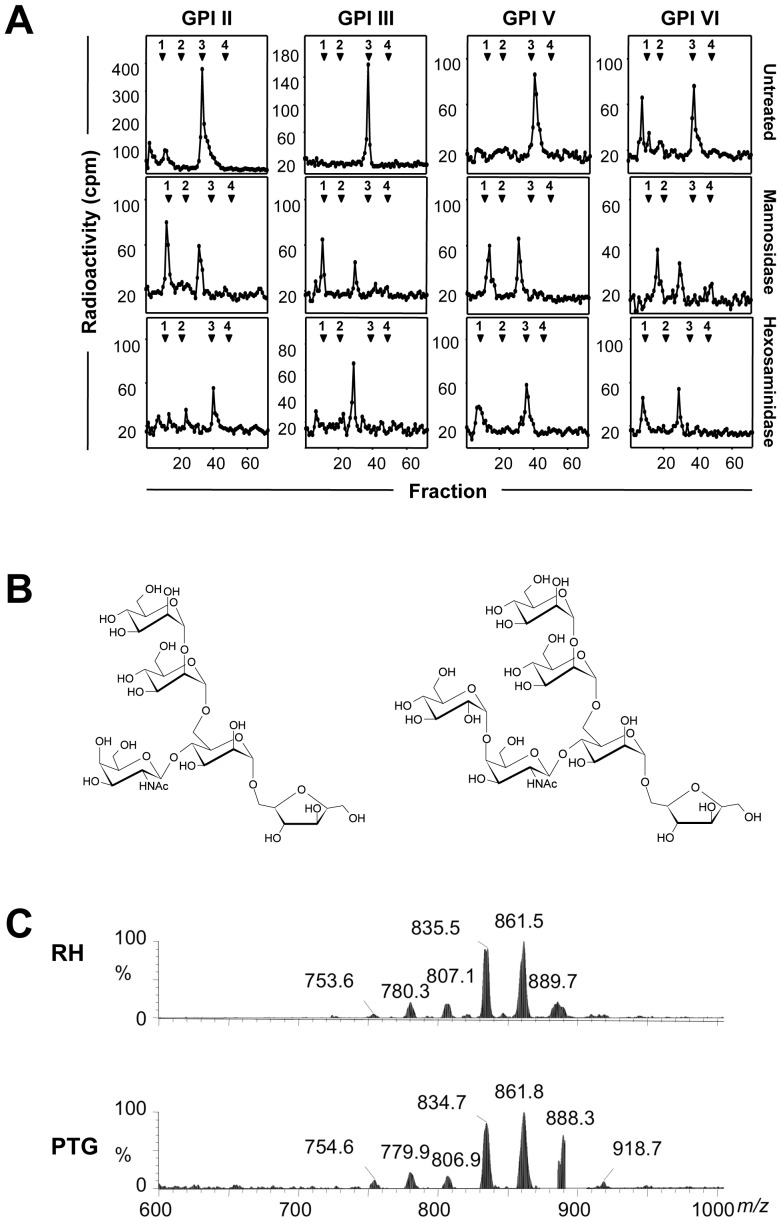
To investigate whether the protein-free GPIs synthesized by the PTG strain are structurally similar to those of the RH strain, we first analyzed their core glycan structures. The GPIs II, III, V and VI of the PTG strain were individually subjected to dephosphorylation, deamination and reduction to generate Man3-anhydromannitol (AHM) core glycans. They were then analyzed by high-performance anion-exchange chromatography (HPAEC) together with an internal dextran hydrolysate standard (dextran unit, DU). All four core glycans co-eluted with the 3 DU standard (Figure 2A, upper panel), similar to those of the RH strain [35]. To detect the presence of α-D-mannosyl residues, core glycans were individually digested with α-mannosidase. Core glycans individually treated with α-mannosidase eluted at 2.4 DU instead of 0.9 DU, the elution position of AHM (Figure 2A, middle panel). This incomplete digestion suggests the presence of a non-mannose substituent linked to the glycan cores, which may represent N-acetylgalactosamine (GalNAc) bound to GPI core glycan structures of the RH strain [35]. The presence of GalNAc residues on PTG strain core glycans was assessed by N-acetylhexosaminidase digestion. The core glycans derived from GPIs II and V were resistant to digestion with N-acetylhexosaminidase (elution at 3 DU, Figure 2A, lower panel), whereas core glycans derived from GPIs III and VI were sensitive to this treatment (elution of Man3-AHM at 2.5 DU and of GalNAc at 1 DU, Figure 2A, lower panel). This suggests the presence of an extra residue that is linked to the GalNAc, thus hindering its enzymatic removal by N-acetylhexosaminidase, which is similar to GPIs II and V of the RH strain that have an additional Glc residue [35]. We propose that the GPIs of the PTG and RH strains have similar GPI core glycan structures, namely Man-Man-(Glc-GalNAc)Man-anhydromannitol for GPIs II and V, and Man-Man-(GalNAc)Man-anhydromannitol for GPIs III and VI (Figure 2B).
Figure 2. GPI core glycans and PIs of RH and PTG strains have similar structures.
(A) HPAEC analysis of the core glycans generated from T. gondii PTG strain protein-free GPIs II, III, V and VI. TLC-purified D-[3H]-glucosamine labeled GPIs were dephosphorylated, deaminated and reduced. The resulting neutral glycans were analysed before (untreated) and after Jack bean α-mannosidase or hexosaminidase treatments. The elution positions of the co-injected glucose oligomer standards are indicated at the top of each profile (bold digits) and given as DU. (B) Predicted Man-Man-(Glc-GalNAc)Man-anhydromannitol core glycan structure derived from protein-free GPIs II and V (right panel), and predicted Man-Man-(GalNAc)Man-anhydromannitol core glycan structure derived from protein-free GPIs III and VI (left panel) of the PTG strain. (C) ES-MS spectra of the PI moieties released by deamination from purified protein-free GPIs from RH (upper panel) and PTG (lower panel) strains.
GPIs of RH and PTG strains have similar PI structures
We next analyzed the structures of the phosphatidylinositols (PI) moieties of the protein-free GPIs from both RH and PTG strains by negative ion electrospray mass spectrometry (ES-MS). The ES-MS analysis revealed that the PI moieties from both strains were very similar, with the vast majority of the heterogeneous PI species containing a sn-2 C18∶1 acyl chain (Figure 2C, and Table S1). The two most abundant PI species found in both RH and PTG strain GPIs were C34∶1 (C16∶0, C18∶1; 835 m/z) and C36∶2 (C18∶1, C18∶1; 861 m/z). The only significant difference was the increased abundance of the PI species at 889 m/z, C38∶2 (C20∶1, C18∶1) in GPIs of the PTG strain (Figure 2C). This difference in heterogeneity was also observed in PI moieties obtained from the protein-anchoring GPIs of both strains (data not shown).
Glc-GalNAc-substituted GPIs of RH and PTG strains are exclusively present on extracellular parasites
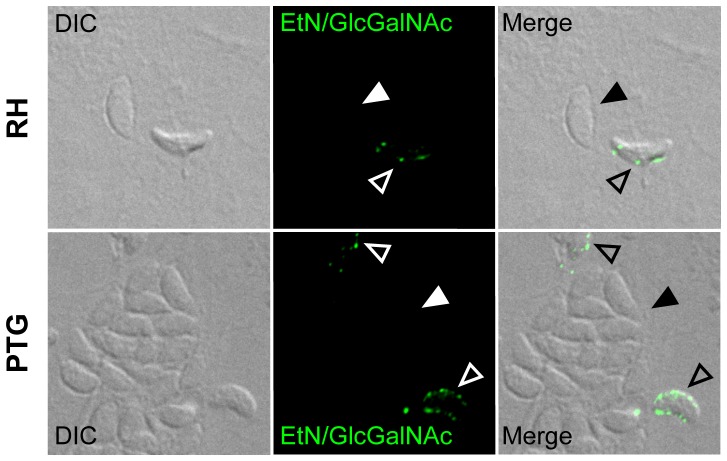
We have previously shown that the Glc-GalNAc-substituted GPIs remain protein-free and clustered in detergent resistant membrane domains (DRMs) that are enriched in cholesterol and sphingolipids, whereas the GalNAc-substituted GPIs are used to anchor surface proteins in RH strain parasites [32]. To investigate the distribution pattern of protein-free Glc-GalNAc-substituted GPIs in the PTG strain, we performed parasite labeling with mAb specific for the EtN-PO4 and the Glc-GalNAc side-branch of GPIs. Figure 3 illustrates that protein-free Glc-GalNAc-substituted GPIs are present on parasites of the PTG strain with similar distribution pattern and to the level found on the surface of RH strain parasites. Interestingly, we could not detect any labeling specific for protein-free Glc-GalNAc-substituted GPIs on intracellular parasites, whatever the strain type. This could be due to an insufficient permeabilization procedure before labeling. To exclude this possibility, the labeling procedure was repeated with both intracellular parasites mechanically released from their host cells and extracellular parasites harvested from cell culture supernatants. This approach confirmed that the Glc-GalNAc-containing GPIs are exclusively found on extracellular parasites (Figure S1). The labeling of the main surface protein SAG1 confirmed the accessibility and integrity of parasite surface molecules (Figure S1).
Figure 3. Protein-free Glc-GalNAc-substituted GPIs are clustered on extracellular parasites.
Staining after permeabilization of HFF cells infected (72 h p.i.) with RH (upper panel) and PTG (lower panel) strains was performed using mAb T54 E10, recognizing both the EtN-PO4 and the Glc-GalNAc side-branch epitopes of protein-free GPIs. Filled arrowheads point to intracellular parasites residing inside parasitophorous vacuoles. Unfilled arrowheads point to extracellular parasites. DIC, differential interference contrast.
RH strain has more protein-free GPI and protein-anchored GPI content than the PTG strain
To evaluate quantitative difference between GPIs of both strains, we analyzed both protein-free GPIs and protein-anchored GPIs of RH and PTG strains by gas chromatography-mass spectrometry (GC-MS). The method is based upon quantification of the GlcN residues of the GPIs being converted to AHM [43]. The close correlation between the AHM and the myo-inositol content of the GPIs indicated a high quality of the extraction and purification methods employed (Table 1). This analysis revealed that the PTG strain had about 70% of the total amount of GPI-anchored proteins of the RH strain, while the total protein-free GPI content was reduced to 34%, most probably due to the missing GPI I and IV. This quantitative reduction of GPI-anchor content in the PTG strain may lead to a reduction of SRS and/or SUSA proteins. Non-quantitative peptide-mass fingerprinting analysis of the GPI-anchored proteins identified identical SRS proteins, namely SAG1, SAG2, and SRS3 in both strains (Table S2) despite different SDS/PAGE profiles of GPI-anchored protein fractions of both strains (Figure S2). Together, the quantitative comparison of AHM revealed that the RH strain has significantly higher protein-free GPI, as well as protein-anchored GPI content than the PTG strain.
Table 1. Comparison of AHM and inositol quantifications of protein-free GPIs and GPI-anchored proteins of RH and PTG strains.
| Sample | AHM quantificationa | Inositol quantificationa |
| RH protein-free GPIs | 4.25±0.21×106 | 4.54±0.16×106 |
| PTG protein-free GPIs | 1.46±0.13×106 (34%) | 1.84±0.22×106 (41%) |
| RH GPI-anchored proteins | 1.12±0.11×106 | 1.42±0.26×106 |
| PTG GPI-anchored proteins | 0.78±0.04×106 (70%) | 1.02±0.07×106 (72%) |
Copies per cell. In parentheses are percentages of PTG values compared to RH values. Values are means ± SD of three replicates.
GPI-anchored proteins of the RH strain have a higher degree of N-glycosylation than GPI-anchored proteins of the PTG strain
To determine whether the glycosylation rates of protein-free GPIs and GPI-anchored proteins correlate with the GPI content, we compared protein-free GPIs and GPI-anchored proteins of both strains for altered glycosylation levels by GC-MS. The carbohydrate composition analysis of protein-free GPIs of the RH and PTG strains revealed a molar ratio of Ino∶Man∶GlcNAc∶GalNAc of 1∶3∶2∶1 and 1∶3∶1∶1, respectively (Table 2). Significant higher molar ratios of glucose were detected in protein-free GPIs of the RH strain (Table 2). This is consistent with the TLC-analysis of metabolically labeled PTG strain GPIs that demonstrated that two of four Glc-GalNAc-substituted GPIs were dramatically reduced in this strain (Figure 1A). We also found unexpected presence of galactose, fucose, and xylose in protein-free GPIs with higher molar ratios for the RH strain. The latter may be evidence of co-purification of an as yet unidentified glycoconjugate or a minor contamination by a GPI-anchored protein, which is N- and/or O-glycosylated. The analysis of RH and PTG GPI-anchored proteins revealed an average of N- and possibly O-glycosylation with higher molar ratios of mannose and GlcNAc in RH GPI-anchored proteins (Table 2). These results suggest that the RH strain possesses a significantly higher degree of N- and possibly O-glycosylation in GPI-anchored proteins than the PTG strain.
Table 2. Carbohydrate composition analysis of protein-free GPIs and GPI-anchored proteins of RH and PTG strains.
| Sample | Ino | Man | GlcNAc | GalNAc | Gal | Glca | Fuc | Xyl |
| RH protein-free GPIs | 1.0±0.1 | 3.9±1.1 | 2.5±0.7 | 1.2±0.3 | 2.6±1.4 | 5.6±2.2 | 2.5±0.3* | 1.2±0.1** |
| PTG protein-free GPIs | 1.0±0.1 | 3.2±0.4 | 1.6±0.3 | 0.7±0.2 | 0.4±0.1 | 2.3±0.7 | 0.8±0.2 | 0.6±0.1 |
| RH GPI-anchored proteins | 1.0±0.1 | 18.9±3.2* | 7.3±0.6* | 1.4±0.3 | 6.3±1.2 | 14.1±3.9 | 7.6±0.1 | 8.0±1.0 |
| PTG GPI-anchored proteins | 1.0±0.1 | 9.3±2.6 | 4.5±1.1 | 1.2±0.1 | 4.1±1.9 | 15.1±4.1 | 5.5±0.5 | 7.2±0.8 |
Glucose is a common contaminant. Values are means of molar ratios ± SD of three replicates.
Values are means of molar ratios ± SD of three replicates.
P<0.005,
P<0.05 RH protein-free GPI and GPI-anchored protein samples compared to PTG protein-free GPI and GPI-anchored protein samples, respectively.
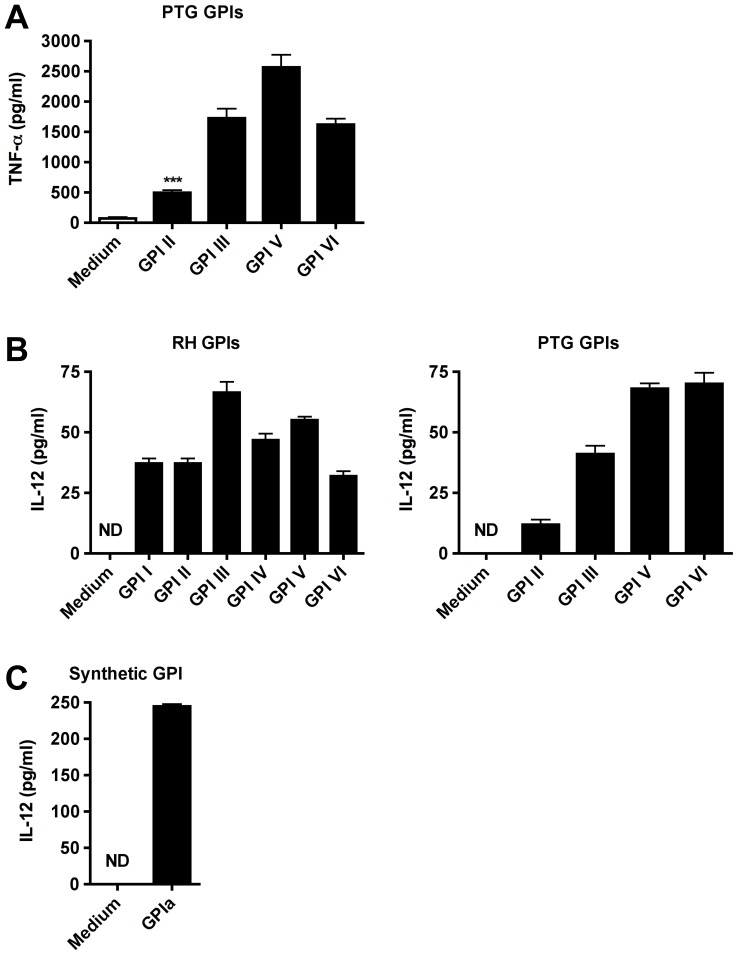
Protein-free GPIs of RH and PTG strains stimulate comparable levels of TNF-α and IL-12 secretion
The six protein-free GPIs of the RH strain have been shown to stimulate macrophages to secrete TNF-α [23]. To measure the biological activity of the four protein-free GPIs of the PTG strain, we individually added them to macrophage cultures. All four PTG GPIs induced significantly the secretion of TNF-α by macrophages (Figure 4A) to a similar extent than those of the RH strain [23]. An early production of IL-12 is essential for the host resistance against T. gondii in mice and only few parasite-derived molecules have been identified to induce IL-12 production [49], [50]. We therefore tested whether the protein-free GPIs of both strains could induce the secretion of IL-12 by macrophages. Figure 4B shows that all GPIs purified from RH (left panel) and PTG (right panel) strain parasites induced significantly the secretion of IL-12 by macrophages. Importantly, glycolipids extracted from uninfected host cells as well as silica alone did not stimulate any cytokine secretion, suggesting that this activity is specific for T. gondii GPIs [23]. To further validate whether the IL-12 production is specifically induced by GPIs and not due to a contaminating molecule, we incubated macrophage cultures with GPIa [(EtN-PO4)-Manα1-2Manα1-6 (Glcα1-4GalNAc β1-4)Manα1-4GlcNα-inositol-PO4], a chemically synthesized structure of RH strain GPI III core glycan. GPIa induced the production of IL-12, thereby demonstrating that the production of IL-12 in response to GPIs is specific and not due to a contaminating molecule (Figure 4C). In sum, these data suggest that RH and PTG GPIs have similar inflammatory capacities due to similar structures.
Figure 4. GPIs of RH and PTG strains induce TNF-α and IL-12p40 secretion by macrophages.
(A) Macrophages were incubated for 24 h with medium alone, or with individual GPIs of the PTG strain extracted from 1×108 parasites and assayed for TNF-α cytokine production. (B) Macrophages were incubated for 24 h with medium alone, or with individual GPIs of the RH (left panel) and the PTG strain (right panel) extracted from 2×108 parasites, respectively, and assayed for IL-12p40 cytokine production. (C) Macrophages were incubated for 24 h with medium alone, or with GPIa (3 mM), a chemically synthesized structure of RH strain GPI III core glycan and assayed for IL-12p40 cytokine production. ***P<0.0005 PTG GPI II compared to medium control. ND, not determined.
Protein-free GPIs of the PTG strain induce TLR4-dependent NF-κB activation
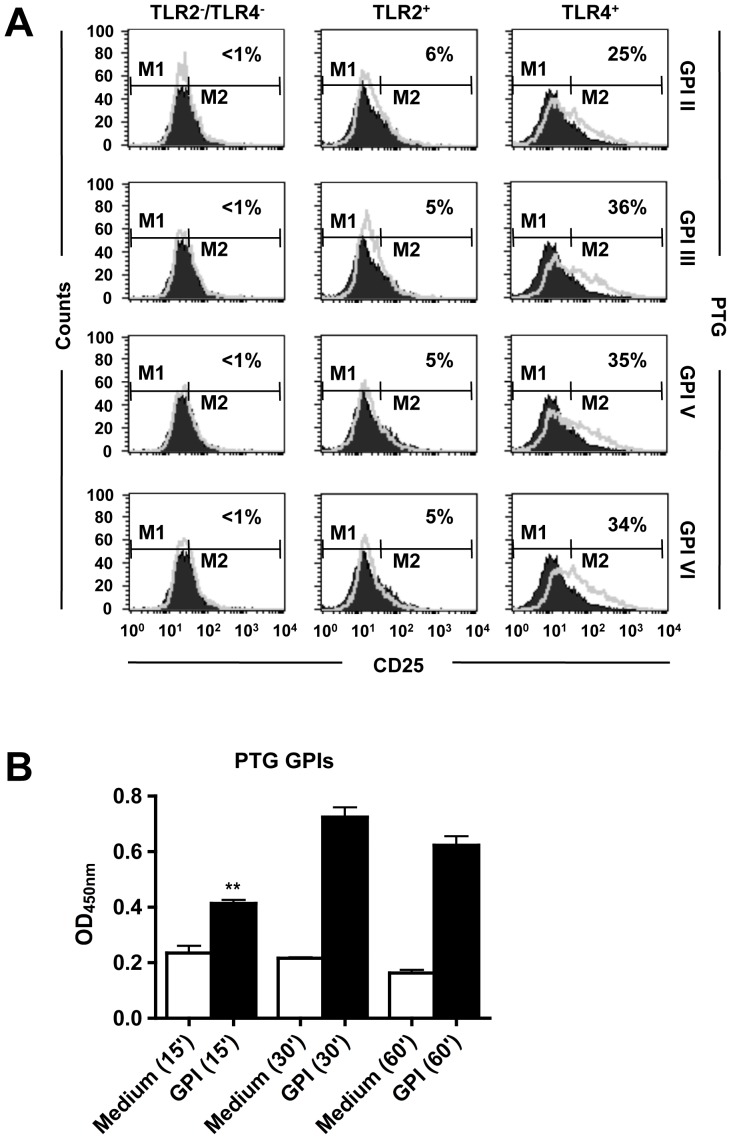
In previous work, we showed that all six RH strain GPIs individually added to CHO/CD14/TLR4 cells induced the expression of CD25 [36]. To determine whether the four GPIs of the PTG strain use similar TLR signaling pathways than those of the RH strain we used two NF-κB reporter cell lines, CHO/CD14/TLR2 and CHO/CD14/TLR4, co-expressing CD14 and TLR2 or TLR4, respectively. No induction of CD25 expression was observed on control cells when stimulated with individual PTG strain GPIs (Figure 5A, left panel). Low CD25 expression levels were measured when PTG strain GPIs were individually incubated with CHO/CD14/TLR2 cells (Figure 5A, middle panel). The expression of CD25 was activated significantly on CHO/CD14/TLR4 cells (Figure 5A, right panel) with magnitudes and kinetics similar to those described for the RH strain GPIs [36]. The GPI-induced TNF-α expression was previously shown to rely on the regulatory control of NF-κB p65 in macrophages [23]. Figure 5B illustrates that the GPIs of the PTG strain were also able to activate NF-κB p65 in macrophages. This result suggests that NF-κB p65 mediates the cytokine production in response to GPIs of the PTG strain. Taken together, these data indicate that RH and PTG strain GPIs have similar inflammatory capacities, mediated by comparable TLR/NF-κB-dependent signaling.
Figure 5. GPIs of the PTG strain activate TLR4/NF-κB signaling.
(A) CHO cells expressing TLR2 (TLR2+), TLR4 (TLR4+), or neither (TLR2−/TLR4−) were either untreated (black line) or exposed to the four different GPIs (II, III, V and VI) extracted from 1×109 parasites (gray line). CD25 expression was measured by FACS analysis 18 h after stimulation. The figure is representative of three independent experiments. Percentage = [percentage of CD25 expression (M2) on GPI-stimulated cells] minus [percentage of CD25 expression (M2) on medium stimulated cells]. (B) Macrophages were incubated for 15, 30, or 60 min with GPI VI extracted from 4×108 PTG strain parasites. Total nuclear protein content was tested using a NF-κB assay as described in Materials and Methods. The figure is a representative experiment with PTG GPI VI and similar results were obtained with GPIs II, III and V of the PTG strain. **P<0.005 compared to medium control.
Discussion
In the present work, we demonstrate that the GPIs of the virulent T. gondii RH and the avirulent PTG strains have similar structures and induce comparable biological effects in macrophages but differ in terms of total GPI-anchor content and degree of GPI-anchored protein N-glycosylation, both were significantly increased in the virulent RH strain. These findings are in line with several earlier studies that have shown that purified GPIs from protozoan parasites are able to activate immune cells of both myeloid and lymphoid origin [51]. In earlier work, we demonstrated that GPIs of RH and PTG strains bind with similar high affinities to human galectin-3 that is required to induce TNF-α secretion by macrophages in response to T. gondii GPIs [37]. Herein we showed that purified and individually tested GPIs from RH and PTG strains induce comparable secretion levels of TNF-α and IL-12 by macrophages, likely as a result of almost identical structures of GPI core glycans and lipid moieties. We showed further that the GPIs of the PTG strain activate TLR4 in a NF-κB CHO/CD14 reporter cell line with comparable magnitudes to those of the RH strain, suggesting that the cytokine induction is due to identical signaling events initiated by macrophage TLRs [36]. The GPIs of both strains have been also shown to stimulate the synthesis of matrix metalloproteinase (MMP)-9 by macrophages in a TNF-α-dependent manner and to degrade extracellular galectin-3 [52]. The GPI-induced degradation of galectin-3 by MMP-9 could have a regulatory function on TLR-signaling in response to GPIs [52]. We conclude that GPIs of RH and PTG strains activate similar signaling events in macrophages due to similar structures.
Structural differences between GPI core glycans of both strains may nevertheless exist since acid-labile residues, such as phosphate, phosphate glycans or phosphoethanolamine, removed by the acidic treatments are not detected. The presence of sn-2 fatty acids (C18∶1) in the T. gondii GPIs found here and elsewhere [44] is reminiscent to the immunogenic GPIs in Plasmodium falciparum mid-schizont stages, which incorporate C18∶1 (88%) and C18∶2 (12%) at the sn-2 position of their diacylglycerolipid [53], and in Trypanosoma cruzi trypomastigotes GPIs, which contain C16∶0 (37%), C18∶1 (31%) along with C18∶2 (21%) at their sn-2 position of their alkylacylglycerolipid [54]. We reasoned that strain-specific differences in the GPI composition could be due to different amounts of protein-free GPIs and/or particular GPI-anchored surface proteins. The comparative analysis of total GPI content demonstrated that the PTG strain had significantly lower quantities of both protein-free GPIs and GPI-anchored proteins, which is likely due to extremely low levels of GPIs I and IV and not because of structural differences in the parasite form between the two strains [55]. The molecular mechanisms and the biological significance underlying the apparent lack of GPIs I and IV in the PTG strain are unknown and have yet to be established. Of further interest would be to analyze the GPI biosynthesis of T. gondii strains other than RH and PTG to verify whether the observed differences in the GPI content represent a general phenomenon between virulent and avirulent strain types. The bulk carbohydrate composition analysis of total GPI-anchored proteins revealed that the molar ratios of Man and GlcNAc are almost twice as high in the RH strain than in those of the PTG strain. Low molar amounts of GalNAc suggest evidence of O-glycosylation in GPI-anchored proteins. Alternatively, the fucose with xylose, galactose and possibly glucose, may form part of a yet unidentified glycan structure that may either be co-purified with the GPI-anchored proteins, or is part of a glycoprotein. In summary, these data suggest that N-linked glycosylation is more extensive in GPI-anchored proteins of the virulent RH strain than in those of the avirulent PTG strain. These results do not, however, demonstrate whether there are specific heavily N-glycosylated GPI-anchored protein(s) missing in the avirulent strain or generally there is less glycosylation of the same GPI-anchored protein(s) (Fauquenoy et al., 2008). Recent work has shown that members of the SRS gene family are polymorphic, codominantly and differentially expressed as distinct, largely nonoverlapping sets of SRS antigens in a parasite strain type- and developmental-stage-specific manner [33]. The biological roles of most SRS and SUSA proteins remain largely understudied, however, some have been reported to function as virulence determinants, involved in parasite adhesion and invasion of host cells (SAG3) [56] and stimulation of host immunity (SAG1) [57], [58]. Recently it has been demonstrated that targeted deletion of the SRS29B (SAG1) and SRS34A (SAG2A) genes in the RH strain led to a specific and substantial increase in SRS29C (SRS2) protein surface expression to a level found on the surface of type 2 strain parasites, resulting in an attenuated virulence phenotype in the mouse model. It was hypothesized that decreased virulence is related to an altered immune response towards fine-tune and/or dampening the inflammatory capacity of other immunoreactive SRS proteins such as SRS29B (SAG1) [33]. Interestingly, our non-quantitative peptide-mass fingerprinting analysis of GPI-anchored proteins revealed that the most abundant parasite-specific SRS proteins, namely SAG1, SAG2, and SRS3 but not SRS2 were present in both strains, whereas the SDS-PAGE analysis of RH and PTG strain GPI-anchored protein fractions clearly showed several differences in the protein distribution pattern, which however could also reflect N-glycosylation differences. Further studies are required to identify and unravel the biological function of these differently expressed proteins in GPI-anchored protein fractions of both strains, including attempts to alter gene expression and targeted gene deletion, followed by testing their relevance in virulence and pathogenesis in the mouse model. The protein-free Glc-GalNAc-substituted GPIs were found equally present and arranged as clusters only in extracellular parasites of both RH and PTG strains. We therefore speculate that protein-free GPIs have dual functions in stimulating host immunity and/or functions in parasite adhesion of target host cells. Several studies have shown that protein-free GPIs or glycoinositolphospholipids (GIPLs) containing ceramides of protozoan parasites elicit a detrimental stimulation of the host innate immune system [51]. We have previously demonstrated that protein-free GPIs containing the Glc-GalNAc side-branch are highly immunogenic in animals and are responsible to elicit an early IgM response in humans [32], [35], [59]. Furthermore, we found that protein-free GPIs are components of DRMs with enrichments of proteins involved in parasite motility and host cell invasion [32]. Further studies are needed, however, to determine whether protein-free Glc-GalNAc-substituted GPIs have indeed dual roles in stimulating the host immunity and functions in host cell invasion as previously proposed for T. gondii profilin [60].
Supporting Information
Protein-free Glc-GalNAc-substituted GPIs are clustered and exclusively present on extracellular parasites. Parasites were collected from cell culture supernatants, while intracellular parasites were mechanically released from host cells with glass beads in a Mixer Mill homogenizer. Parasites were purified, separately fixed on microscope slides with CellTack (BD Biosciences), and stained with the mAb T54 E10 as described in Materials and Methods without initial permeabilization. The mAb T4 1E5, specific for the major T. gondii surface protein SAG1 was used as control. Slides were examined on a Zeiss Axiophot 200M microscope equipped with ApoTome (Carl Zeiss Inc.).
(PDF)
Comparison of T. gondii RH and PTG strain GPI-anchored protein fractions. GPI-anchor protein fraction was obtained after the protein-free GPI extraction from the same parasite pellet by two extractions with water-ethanol-diethylether-pyridine-ammonium hydroxide (15∶15∶5∶1∶0.017, by volume). Extracts were pooled and dried under a stream of nitrogen and solubilized in Laemmli-sample buffer and boiled for 5 min. Equal amounts of GPI-anchored protein fractions of both strains were electrophoresed on 10% (w/v) SDS–PAGE and proteins were stained with Coomassie Blue. The filled arrowhead points to the expected size of SAG1 (P30) protein.
(PDF)
Assignment table of Figure 2C . ES-MS analysis of the PI moieties released by deamination from purified protein-free GPIs from RH and PTG strains. aAll of the molecular species refer to the corresponding peaks in the negative ion mode in Figure 2C. The peaks containing the most abundant species are in bold. bObserved [M-H]-ions, mass over charge from survey scans as described in the experimental procedure section. cPeak identities refer to total number of carbon atoms and double bonds. dOnly the principal component is given. eTheoretical monoisotopic masses were obtained when possible with LIPIDMAPS, otherwise they were calculated using http://www.sisweb.com/referenc/tools/exactmass.htm.
(PDF)
Analysis of RH and PTG strain GPI-anchored proteins. Results obtained from mass-fingerprinting of a fraction containing purified GPI-anchored proteins from both RH and PTG strains. The GPI-anchor protein fraction was obtained after the protein-free GPI extraction from the same parasite pellet by two extractions with water-ethanol-diethylether-pyridine-ammonium hydroxide (15∶15∶5∶1∶0.017, by volume). The supernatant was dried under a stream of nitrogen and proteins were re-suspended in trypsin digestion buffer [12.5 mg/ml modified bovine trypsin (Roche), 0.1% n-octyl glucoside (Calbiochem), 20 mM ammonium bicarbonate]. An equal volume of acetonitrile/0.3% trifluoroacetic acid was added to extract the peptides. MALDI mass spectra were generated using a Voyager DE-STR MALDI-TOF MS system (PerSeptive Biosystems) with delayed extraction in the reflectron mode. Protein identification was from a comparison of peak list data generated from the Data Explorer application (PerSeptive Biosystems) against NCBInr (non-redundant) and Swissprot databases using the Protein-Prospector V3.4.1 software MS-Fit (http://www.prospector.ucsf.edu). Only hits with a high score are given so that a very high confidence level could be maintained, with major surface proteins in bold.
(PDF)
Acknowledgments
The authors thank Dr. Douglas T. Golenbock (University of Massachusetts Medical School) for providing the CHO reporter cell lines, Dr. David Sibley (Washington University School of Medicine) for providing the T. gondii PTG strain, Dr. Jürgen Kimmel and Dr. Hosam Shams-Eldin (Philipps-University Marburg) for research assistance and helpful comments, Dr. Randy Clayton (UPR 9022 du CNRS), Dr. Dominique Ferrandon (UPR 9022 du CNRS), and Dr. R. John MacLeod (Queen's University) for critical reading of the manuscript.
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft (R.T.S.), NIH grant number R01 AI071319 (R.T.G. and R.T.S.) and PROBRAL of Deutscher Akademischer Austausch Dienst/CAPES (R.T.G. and R.T.S.). T.K.S. is supported by a Wellcome Trust Senior Research Fellowship (067441), and Wellcome Trust grants (086658) and (093228). R.T.G. and M.A.C. are research fellows from CNPq. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Montoya JG, Liesenfeld O (2004) Toxoplasmosis. Lancet 363: 1965–1976. [DOI] [PubMed] [Google Scholar]
- 2. Khan A, Dubey JP, Su C, Ajioka JW, Rosenthal BM, et al. (2011) Genetic analyses of atypical Toxoplasma gondii strains reveal a fourth clonal lineage in North America. Int J Parasitol 41: 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khan A, Miller N, Roos DS, Dubey JP, Ajzenberg D, et al. (2011) A monomorphic haplotype of chromosome Ia is associated with widespread success in clonal and nonclonal populations of Toxoplasma gondii. MBio 2: e00228–00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Su C, Khan A, Zhou P, Majumdar D, Ajzenberg D, et al. (2012) Globally diverse Toxoplasma gondii isolates comprise six major clades originating from a small number of distinct ancestral lineages. Proc Natl Acad Sci U S A 109: 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saeij JP, Boyle JP, Boothroyd JC (2005) Differences among the three major strains of Toxoplasma gondii and their specific interactions with the infected host. Trends Parasitol 21: 476–481. [DOI] [PubMed] [Google Scholar]
- 6. Taylor S, Barragan A, Su C, Fux B, Fentress SJ, et al. (2006) A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science 314: 1776–1780. [DOI] [PubMed] [Google Scholar]
- 7. Sibley LD, Boothroyd JC (1992) Virulent strains of Toxoplasma gondii comprise a single clonal lineage. Nature 359: 82–85. [DOI] [PubMed] [Google Scholar]
- 8. Liesenfeld O, Kosek J, Remington JS, Suzuki Y (1996) Association of CD4+ T cell-dependent, interferon-gamma-mediated necrosis of the small intestine with genetic susceptibility of mice to peroral infection with Toxoplasma gondii. J Exp Med 184: 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Melo MB, Jensen KD, Saeij JP (2011) Toxoplasma gondii effectors are master regulators of the inflammatory response. Trends Parasitol 27: 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boothroyd JC (2013) Have it your way: how polymorphic, injected kinases and pseudokinases enable toxoplasma to subvert host defenses. PLoS Pathog 9: e1003296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saeij JP, Boyle JP, Coller S, Taylor S, Sibley LD, et al. (2006) Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science 314: 1780–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Behnke MS, Khan A, Wootton JC, Dubey JP, Tang K, et al. (2011) Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc Natl Acad Sci U S A 108: 9631–9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reese ML, Zeiner GM, Saeij JP, Boothroyd JC, Boyle JP (2011) Polymorphic family of injected pseudokinases is paramount in Toxoplasma virulence. Proc Natl Acad Sci U S A 108: 9625–9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fleckenstein MC, Reese ML, Konen-Waisman S, Boothroyd JC, Howard JC, et al. (2012) A Toxoplasma gondii pseudokinase inhibits host IRG resistance proteins. PLoS Biol 10: e1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fentress SJ, Behnke MS, Dunay IR, Mashayekhi M, Rommereim LM, et al. (2010) Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe 8: 484–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Howard JC, Hunn JP, Steinfeldt T (2011) The IRG protein-based resistance mechanism in mice and its relation to virulence in Toxoplasma gondii. Curr Opin Microbiol 14: 414–421. [DOI] [PubMed] [Google Scholar]
- 17. Behnke MS, Fentress SJ, Mashayekhi M, Li LX, Taylor GA, et al. (2012) The polymorphic pseudokinase ROP5 controls virulence in Toxoplasma gondii by regulating the active kinase ROP18. PLoS Pathog 8: e1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gavrilescu LC, Denkers EY (2001) IFN-gamma overproduction and high level apoptosis are associated with high but not low virulence Toxoplasma gondii infection. J Immunol 167: 902–909. [DOI] [PubMed] [Google Scholar]
- 19. Mordue DG, Monroy F, La Regina M, Dinarello CA, Sibley LD (2001) Acute toxoplasmosis leads to lethal overproduction of Th1 cytokines. J Immunol 167: 4574–4584. [DOI] [PubMed] [Google Scholar]
- 20. Saeij JP, Coller S, Boyle JP, Jerome ME, White MW, et al. (2007) Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature 445: 324–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jensen KD, Wang Y, Wojno ED, Shastri AJ, Hu K, et al. (2011) Toxoplasma polymorphic effectors determine macrophage polarization and intestinal inflammation. Cell Host Microbe 9: 472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aliberti J, Valenzuela JG, Carruthers VB, Hieny S, Andersen J, et al. (2003) Molecular mimicry of a CCR5 binding-domain in the microbial activation of dendritic cells. Nat Immunol 4: 485–490. [DOI] [PubMed] [Google Scholar]
- 23. Debierre-Grockiego F, Azzouz N, Schmidt J, Dubremetz JF, Geyer H, et al. (2003) Roles of glycosylphosphatidylinositols of Toxoplasma gondii. Induction of tumor necrosis factor-alpha production in macrophages. J Biol Chem 278: 32987–32993. [DOI] [PubMed] [Google Scholar]
- 24. Yarovinsky F, Zhang D, Andersen JF, Bannenberg GL, Serhan CN, et al. (2005) TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science 308: 1626–1629. [DOI] [PubMed] [Google Scholar]
- 25. McConville MJ, Ferguson MA (1993) The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem J 294 (Pt 2) 305–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schofield L, Hackett F (1993) Signal transduction in host cells by a glycosylphosphatidylinositol toxin of malaria parasites. J Exp Med 177: 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tachado SD, Schofield L (1994) Glycosylphosphatidylinositol toxin of Trypanosoma brucei regulates IL-1 alpha and TNF-alpha expression in macrophages by protein tyrosine kinase mediated signal transduction. Biochem Biophys Res Commun 205: 984–991. [DOI] [PubMed] [Google Scholar]
- 28. Tachado SD, Gerold P, Schwarz R, Novakovic S, McConville M, et al. (1997) Signal transduction in macrophages by glycosylphosphatidylinositols of Plasmodium, Trypanosoma, and Leishmania: activation of protein tyrosine kinases and protein kinase C by inositolglycan and diacylglycerol moieties. Proc Natl Acad Sci U S A 94: 4022–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boothroyd JC, Hehl A, Knoll LJ, Manger ID (1998) The surface of Toxoplasma: more and less. Int J Parasitol 28: 3–9. [DOI] [PubMed] [Google Scholar]
- 30. Lekutis C, Ferguson DJ, Grigg ME, Camps M, Boothroyd JC (2001) Surface antigens of Toxoplasma gondii: variations on a theme. Int J Parasitol 31: 1285–1292. [DOI] [PubMed] [Google Scholar]
- 31. Jung C, Lee CY, Grigg ME (2004) The SRS superfamily of Toxoplasma surface proteins. Int J Parasitol 34: 285–296. [DOI] [PubMed] [Google Scholar]
- 32. Azzouz N, Shams-Eldin H, Niehus S, Debierre-Grockiego F, Bieker U, et al. (2006) Toxoplasma gondii grown in human cells uses GalNAc-containing glycosylphosphatidylinositol precursors to anchor surface antigens while the immunogenic Glc-GalNAc-containing precursors remain free at the parasite cell surface. Int J Biochem Cell Biol 38: 1914–1925. [DOI] [PubMed] [Google Scholar]
- 33. Wasmuth JD, Pszenny V, Haile S, Jansen EM, Gast AT, et al. (2012) Integrated bioinformatic and targeted deletion analyses of the SRS gene superfamily identify SRS29C as a negative regulator of Toxoplasma virulence. MBio 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wichroski MJ, Ward GE (2003) Biosynthesis of glycosylphosphatidylinositol is essential to the survival of the protozoan parasite Toxoplasma gondii. Eukaryot Cell 2: 1132–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Striepen B, Zinecker CF, Damm JB, Melgers PA, Gerwig GJ, et al. (1997) Molecular structure of the “low molecular weight antigen” of Toxoplasma gondii: a glucose alpha 1–4 N-acetylgalactosamine makes free glycosyl-phosphatidylinositols highly immunogenic. J Mol Biol 266: 797–813. [DOI] [PubMed] [Google Scholar]
- 36. Debierre-Grockiego F, Campos MA, Azzouz N, Schmidt J, Bieker U, et al. (2007) Activation of TLR2 and TLR4 by glycosylphosphatidylinositols derived from Toxoplasma gondii. J Immunol 179: 1129–1137. [DOI] [PubMed] [Google Scholar]
- 37. Debierre-Grockiego F, Niehus S, Coddeville B, Elass E, Poirier F, et al. (2010) Binding of Toxoplasma gondii glycosylphosphatidylinositols to galectin-3 is required for their recognition by macrophages. J Biol Chem 285: 32744–32750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grimwood BG, Hechemy K, Stevens RW (1979) Toxoplasma gondii: purification of trophozoites propagated in cell culture. Exp Parasitol 48: 282–286. [DOI] [PubMed] [Google Scholar]
- 39. Azzouz N, Shams-Eldin H, Schwarz RT (2005) Removal of phospholipid contaminants through precipitation of glycosylphosphatidylinositols. Anal Biochem 343: 152–158. [DOI] [PubMed] [Google Scholar]
- 40. Menon AK, Schwarz RT, Mayor S, Cross GA (1990) Cell-free synthesis of glycosyl-phosphatidylinositol precursors for the glycolipid membrane anchor of Trypanosoma brucei variant surface glycoproteins. Structural characterization of putative biosynthetic intermediates. J Biol Chem 265: 9033–9042. [PubMed] [Google Scholar]
- 41. Tomavo S, Dubremetz JF, Schwarz RT (1992) A family of glycolipids from Toxoplasma gondii. Identification of candidate glycolipid precursor(s) for Toxoplasma gondii glycosylphosphatidylinositol membrane anchors. J Biol Chem 267: 11721–11728. [PubMed] [Google Scholar]
- 42. Yamashita K, Mizuochi T, Kobata A (1982) Analysis of oligosaccharides by gel filtration. Methods Enzymol 83: 105–126. [DOI] [PubMed] [Google Scholar]
- 43. Macrae JI, Ferguson MA (2005) A robust and selective method for the quantification of glycosylphosphatidylinositols in biological samples. Glycobiology 15: 131–138. [DOI] [PubMed] [Google Scholar]
- 44. Smith TK, Kimmel J, Azzouz N, Shams-Eldin H, Schwarz RT (2007) The role of inositol acylation and inositol deacylation in the Toxoplasma gondii glycosylphosphatidylinositol biosynthetic pathway. J Biol Chem 282: 32032–32042. [DOI] [PubMed] [Google Scholar]
- 45. Tomavo S, Couvreur G, Leriche MA, Sadak A, Achbarou A, et al. (1994) Immunolocalization and characterization of the low molecular weight antigen (4–5 kDa) of Toxoplasma gondii that elicits an early IgM response upon primary infection. Parasitology 108 (Pt 2) 139–145. [DOI] [PubMed] [Google Scholar]
- 46. Pekari K, Tailler D, Weingart R, Schmidt RR (2001) Synthesis of the fully phosphorylated GPI anchor pseudohexasaccharide of Toxoplasma gondii. J Org Chem 66: 7432–7442. [DOI] [PubMed] [Google Scholar]
- 47. Delude RL, Yoshimura A, Ingalls RR, Golenbock DT (1998) Construction of a lipopolysaccharide reporter cell line and its use in identifying mutants defective in endotoxin, but not TNF-alpha, signal transduction. J Immunol 161: 3001–3009. [PubMed] [Google Scholar]
- 48. Lien E, Sellati TJ, Yoshimura A, Flo TH, Rawadi G, et al. (1999) Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J Biol Chem 274: 33419–33425. [DOI] [PubMed] [Google Scholar]
- 49. Pollard AM, Knoll LJ, Mordue DG (2009) The role of specific Toxoplasma gondii molecules in manipulation of innate immunity. Trends Parasitol 25: 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hunter CA, Sibley LD (2012) Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat Rev Microbiol 10: 766–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gazzinelli RT, Denkers EY (2006) Protozoan encounters with Toll-like receptor signalling pathways: implications for host parasitism. Nat Rev Immunol 6: 895–906. [DOI] [PubMed] [Google Scholar]
- 52. Niehus S, Elass E, Coddeville B, Guerardel Y, Schwarz RT, et al. (2012) Glycosylphosphatidylinositols of Toxoplasma gondii induce matrix metalloproteinase-9 production and degradation of galectin-3. Immunobiology 217: 61–64. [DOI] [PubMed] [Google Scholar]
- 53. Naik RS, Branch OH, Woods AS, Vijaykumar M, Perkins DJ, et al. (2000) Glycosylphosphatidylinositol anchors of Plasmodium falciparum: molecular characterization and naturally elicited antibody response that may provide immunity to malaria pathogenesis. J Exp Med 192: 1563–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Almeida IC, Camargo MM, Procopio DO, Silva LS, Mehlert A, et al. (2000) Highly purified glycosylphosphatidylinositols from Trypanosoma cruzi are potent proinflammatory agents. Embo J 19: 1476–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dubey JP, Lindsay DS, Speer CA (1998) Structures of Toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts. Clin Microbiol Rev 11: 267–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dzierszinski F, Mortuaire M, Cesbron-Delauw MF, Tomavo S (2000) Targeted disruption of the glycosylphosphatidylinositol-anchored surface antigen SAG3 gene in Toxoplasma gondii decreases host cell adhesion and drastically reduces virulence in mice. Mol Microbiol 37: 574–582. [DOI] [PubMed] [Google Scholar]
- 57. Khan IA, Eckel ME, Pfefferkorn ER, Kasper LH (1988) Production of gamma interferon by cultured human lymphocytes stimulated with a purified membrane protein (P30) from Toxoplasma gondii. J Infect Dis 157: 979–984. [DOI] [PubMed] [Google Scholar]
- 58. Rachinel N, Buzoni-Gatel D, Dutta C, Mennechet FJ, Luangsay S, et al. (2004) The induction of acute ileitis by a single microbial antigen of Toxoplasma gondii. J Immunol 173: 2725–2735. [DOI] [PubMed] [Google Scholar]
- 59. Striepen B, Tomavo S, Dubremetz JF, Schwarz RT (1992) Identification and characterisation of glycosyl-inositolphospholipids in Toxoplasma gondii. Biochem Soc Trans 20: 296S. [DOI] [PubMed] [Google Scholar]
- 60. Plattner F, Yarovinsky F, Romero S, Didry D, Carlier MF, et al. (2008) Toxoplasma profilin is essential for host cell invasion and TLR11-dependent induction of an interleukin-12 response. Cell Host Microbe 3: 77–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein-free Glc-GalNAc-substituted GPIs are clustered and exclusively present on extracellular parasites. Parasites were collected from cell culture supernatants, while intracellular parasites were mechanically released from host cells with glass beads in a Mixer Mill homogenizer. Parasites were purified, separately fixed on microscope slides with CellTack (BD Biosciences), and stained with the mAb T54 E10 as described in Materials and Methods without initial permeabilization. The mAb T4 1E5, specific for the major T. gondii surface protein SAG1 was used as control. Slides were examined on a Zeiss Axiophot 200M microscope equipped with ApoTome (Carl Zeiss Inc.).
(PDF)
Comparison of T. gondii RH and PTG strain GPI-anchored protein fractions. GPI-anchor protein fraction was obtained after the protein-free GPI extraction from the same parasite pellet by two extractions with water-ethanol-diethylether-pyridine-ammonium hydroxide (15∶15∶5∶1∶0.017, by volume). Extracts were pooled and dried under a stream of nitrogen and solubilized in Laemmli-sample buffer and boiled for 5 min. Equal amounts of GPI-anchored protein fractions of both strains were electrophoresed on 10% (w/v) SDS–PAGE and proteins were stained with Coomassie Blue. The filled arrowhead points to the expected size of SAG1 (P30) protein.
(PDF)
Assignment table of Figure 2C . ES-MS analysis of the PI moieties released by deamination from purified protein-free GPIs from RH and PTG strains. aAll of the molecular species refer to the corresponding peaks in the negative ion mode in Figure 2C. The peaks containing the most abundant species are in bold. bObserved [M-H]-ions, mass over charge from survey scans as described in the experimental procedure section. cPeak identities refer to total number of carbon atoms and double bonds. dOnly the principal component is given. eTheoretical monoisotopic masses were obtained when possible with LIPIDMAPS, otherwise they were calculated using http://www.sisweb.com/referenc/tools/exactmass.htm.
(PDF)
Analysis of RH and PTG strain GPI-anchored proteins. Results obtained from mass-fingerprinting of a fraction containing purified GPI-anchored proteins from both RH and PTG strains. The GPI-anchor protein fraction was obtained after the protein-free GPI extraction from the same parasite pellet by two extractions with water-ethanol-diethylether-pyridine-ammonium hydroxide (15∶15∶5∶1∶0.017, by volume). The supernatant was dried under a stream of nitrogen and proteins were re-suspended in trypsin digestion buffer [12.5 mg/ml modified bovine trypsin (Roche), 0.1% n-octyl glucoside (Calbiochem), 20 mM ammonium bicarbonate]. An equal volume of acetonitrile/0.3% trifluoroacetic acid was added to extract the peptides. MALDI mass spectra were generated using a Voyager DE-STR MALDI-TOF MS system (PerSeptive Biosystems) with delayed extraction in the reflectron mode. Protein identification was from a comparison of peak list data generated from the Data Explorer application (PerSeptive Biosystems) against NCBInr (non-redundant) and Swissprot databases using the Protein-Prospector V3.4.1 software MS-Fit (http://www.prospector.ucsf.edu). Only hits with a high score are given so that a very high confidence level could be maintained, with major surface proteins in bold.
(PDF)