Abstract
Background
In the X-ray repair cross-complementing group 1 (XRCC1) gene, a polymorphism, Arg399Gln (rs25487), has been shown to change neoconservative amino acid and thus result in alternation of DNA repair capacity. Numerous studies have investigated the association between Arg399Gln and breast cancer risk in the American population, but yielding inconsistent results. This study aimed to clarify the role of this polymorphism in susceptibility to breast cancer.
Methods
Literatures were searched in multiple databases including PubMed, Springer Link, Ovid, EBSCO and ScienceDirect databases up to April 2013. A comprehensive meta-analysis was conducted to estimate the overall odds ratio (OR), by integrating data from 18 case control studies of 10846 cases and 11723 controls in the American population.
Results
Overall, significant association was observed between the Arg399Gln polymorphism and breast cancer risk under the random-effects model (OR for dominant model = 1.12, 95% CI: 1.02–1.24, P heterogeneity = 0.003; OR for additive model = 1.07, 95% CI: 1.01–1.14, P heterogeneity = 0.017). Further sensitivity analysis supported the robust stability of this current result by showing similar ORs before and after removal of a single study.
Conclusions
This meta-analysis suggests that the XRCC1 Arg399Gln polymorphism may significantly contribute to susceptibility of breast cancer in the American population.
Introduction
Breast cancer is the most common cancer and a predominate cause of cancer related-death in female population worldwide [1]. In 2013, an estiamted 232,340 new cases in women were expected to occurred and 39,620 women were expected to die from breast cancer in the USA [2]. Breast cancer is a complex trait caused by environmental and genetic factors. Multiple environmental factors for breast cancer have been identified, including age at first birth, menarche and menopause, and family history, but the underlying genetic basis remained largely unknown [3].
Base-excision repair (BER), an important DNA repair pathway, is responsible for the repair of base damage resulting from exposure to X-rays, oxygen radicals, and alkylating agents [4], [5], [6]. In the BER pathway, the X-ray repair cross-complementing group 1 (XRCC1) gene, encoding a scaffolding protein, involved in the repair of single-strand breaks, the most common lesions in cellular DNA [7]. Molecular studies showed if lacking the XRCC1 active cell would be hypersensitive to DNA damage. In the XRCC1 gene, a functional polymorphism, Arg399Gln (rs25487) has been extensively investigated in many cancers [8], [9], [10], [11]. Regarding breast cancer, multiple studies have been conducted to explore the association of this polymorphism and the disease risk in the USA [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26]; however, results were inconsistent. For instance, Duell et al. suggested that the variant of Arg399Gln might confer increased risk of breast cancer [12], whereas Dawei Bu et al. reported no association of this polymorphism and breast cancer [18]. Based on previously published studies, four meta-analysis have been conducted on the Arg399Gln and breast cancer risk [27], [28], [29], [30], but not special in the American population. Maybe due to heterogeneity across different countries, no conclusion has been drawn yet. Unfortunately, in the two most recent meta-analysis [27], [29], some errors in the data extraction have introduced the incorrect results. Herein, we believed that it is essential to conduct an update comprehensive meta-analysis including studies published since 2001 to provide a more precise assessment of the association between the Arg399Gln in XRCC1 and breast cancer risk in the American population.
Materials and Methods
Literature Search
Relevant articles published before April 1st, 2013 were identified through a electronically search in the PubMed, Springer Link, Ovid, EBSCO and ScienceDirect databases using the combination of key words: ‘XRCC1’, ‘polymorphism’, ‘Arg399Gln’, ‘SNP’, ‘variant’, ‘BC’ and ‘breast cancer’. References of retrieved publications were also screened. Disagreements were resolved through discussions between the two authors (Yang Peng and Yong Sun).
Inclusion and Exclusion Criteria
In our meta-analysis, studies were included if they met the all of the following criteria: (a) case-control studies investigated the relationship between XRCC1 Arg399Gln and breast cancer risk; (b) patients should be confirmed with histologically breast cancer; (c) studies should provided data about the frequencies of alleles or genotypes. (d) American population is meant that all the inhabitants of America. Meta-analysis, letters, reviews or editorial articles were excluded. If studies shared the same participants, only the one with the largest population or the most complete information was included. If more than one ethnical population were included in one publication, each population was considered separetly. The meta-analysis was conducted according to the guidelines of Preferred Reporting Items for Systemic Reviews and Meta-Analyses statement (PRISMA) [31], as shown in Checklist S1 (http://www.prisma-statement.org).
Data Extraction
The following data from included studies were extracted independently by two authors (Li Zhao and Yang Peng) into a standardized form: the first author's name, year of publication, ethnicity of participants, study design, sample size, pre- and postmenopausal status, genotyping method, allele and genotype frequencies, study population, sample materials of study participants and evidence of Hardy-Weinberg equilibrium (HWE) in controls. In case of conflicting evaluations, disagreements were resolved through discussions between the authors.
Quality Assessment of Included Studies
Two authors independently assessed the quality of included studies according to the 9-star Newcastle-Ottawa Scale. The study quality was assessed by the 9-star Newcastle-Ottawa Scale. A full score is 9 stars, and a score ≥6 stars is considered to be high quality. The quality of case-control studies was assessed as follows: adequate definition of cases, representativeness of cases, selection of controls, definition of control, control for the most important factor or the second important factor, exposure assessment, same method of ascertainment for all subjects, and non-response rate. The score of each individual publications was shown in Table S1.
Statistical analysis
For each study, odds ratios (ORs) and their 95% confidence intervals (CIs) as the metrics of effect size were recalculated for additive, dominant [(Gln/Gln+Arg/Gln) versus Arg/Arg] and recessive [Gln/Gln versus (Arg/Gln + Arg/Arg)] genetic models. For additive model, common homozygotes, heterozygotes, and rare homozygotes were assigned as scores of 0, 1, and 2, respectively, and then ORs per unit score were calculated by comparing between cases and controls in logistic regression model. The χ2 based Cochran's Q statistic test was employed to test between-study heterogeneity, and heterogeneity was considered significant when P<0.1 for Q statistic. Heterogeneity was quantified by I2 statistic examining the percentage of heterogeneity (I2 = 0–25%, no heterogeneity; I2 = 25–50%, moderate heterogeneity; I2 = 50–75%, large heterogeneity; I2 = 75–100%, extreme heterogeneity) [32]. For pooling ORs and 95% CIs. A random-effects model using the DerSimonian and Laird's method was applied, with significant evidence of heterogeneity; otherwise, a fixed-effects with Mantel-Haenszel's method was utilized. Furthermore, subgroup analyses were performed by ethnicity, menopausal status, genotyping method and control source, to explore the source of heterogeneity. Sensitivity analysis was also conducted to assess influence of single study on the overall estimate, by sequential removal of individual studies. Publication bias was estimated by funnel plot and Egger's test [33]. All analyses were carried out by using the Stata 12.0 software.
Results
Characteristics of included studies
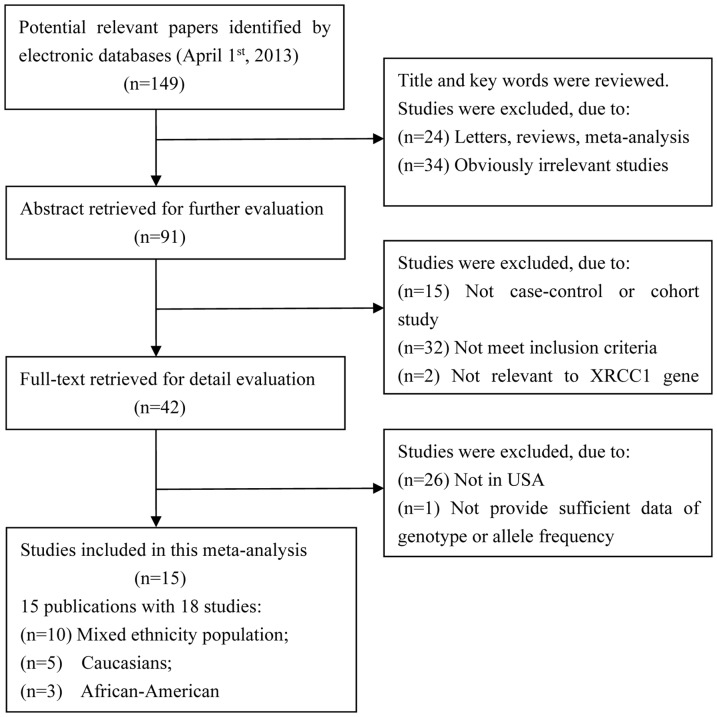
Figure 1 shows the procedure of study selection. A total of 15 publications with 18 case-control studies of 10846 breast cancer cases and 11723 controls were finally included in this meta-analysis. Among them, 10 studies were conducted in mixed ethnicity population, 3 studies were in the African-American, and 5 studies in Caucasians. The characteristics of individual studies are summarized in Table 1.
Figure 1. Flow chart of literature search and selection in the meta-analysis.
Table 1. Characteristics of included studies in this meta-analysis.
| First author | Year | Ethnicity | Case | Control | Population based | HWE | Sample material | Genotyping methods | ||||
| AA | AG | GG | AA | AG | GG | |||||||
| Duell [12] | 2001 | African | 164 | 82 | 7 | 198 | 64 | 4 | Population | Y | Blood | PCR-RFLP |
| Duell [12] | 2001 | Caucasian | 162 | 175 | 49 | 164 | 58 | 56 | Population | Y | Blood | PCR-RFLP |
| Smith [13] | 2003 | Mixed | 99 | 122 | 30 | 115 | 123 | 29 | Hospital | Y | Blood | PCR-RFLP |
| Smith [15] | 2003 | Caucasian | 70 | 72 | 20 | 119 | 150 | 31 | Hospital | Y | Peripheral lymphocyte | PCR-RFLP |
| Han [14] | 2003 | Mixed | 391 | 460 | 135 | 545 | 616 | 176 | Population | Y | Blood | Pyrosequencing |
| Shen [17] | 2005 | Mixed | 412 | 539 | 116 | 444 | 536 | 130 | Population | Y | Blood | PCR-RFLP |
| Patel [16] | 2005 | Mixed | 196 | 195 | 61 | 194 | 202 | 56 | Population | Y | Buffy coat | TaqMan Real Time PCR |
| Bu [18] | 2006 | Mixed | 84 | 84 | 22 | 42 | 43 | 10 | Hospital | Y | Blood | PCR-RFLP |
| Zhang [21] | 2006 | Caucasian | 392 | 1433 | 1214 | 360 | 1173 | 1054 | Population | Y | Mouthwash cytobrush | PCR-RFLP |
| Brewster [20] | 2006 | Mixed | 108 | 159 | 38 | 126 | 135 | 49 | Population | Y | Blood | PCR-RFLP |
| Thyagarajan [19] | 2006 | Mixed | 57 | 76 | 60 | 135 | 140 | 47 | Population | Y | Blood,normal tissue | PCR-RFLP |
| Pachkowski [22] | 2006 | African | 536 | 203 | 22 | 493 | 172 | 11 | Population | Y | Blood | TaqMan Real Time PCR |
| Pachkowski [22] | 2006 | Caucasian | 504 | 581 | 159 | 480 | 494 | 148 | Population | Y | Blood | TaqMan Real Time PCR |
| Ali [23] | 2008 | Mixed | 11 | 16 | 13 | 21 | 20 | 7 | Population | Y | Normal tissues | PCR-RFLP |
| Smith [24] | 2008 | Caucasian | 135 | 141 | 36 | 179 | 181 | 46 | Population | Y | Blood | MassARRAY Sequenome |
| Smith [24] | 2008 | African | 38 | 13 | 1 | 58 | 15 | 1 | Population | Y | Blood | MassARRAY Sequenome |
| Zipprich [25] | 2010 | Mixed | 126 | 115 | 30 | 139 | 141 | 43 | Population | Y | Blood | SYBR Green PCR |
| Roberts [26] | 2011 | Mixed | 104 | 361 | 417 | 164 | 772 | 814 | Hospital | Y | Blood, mouthwash | MassARRAY Sequenome |
Overall meta-analysis
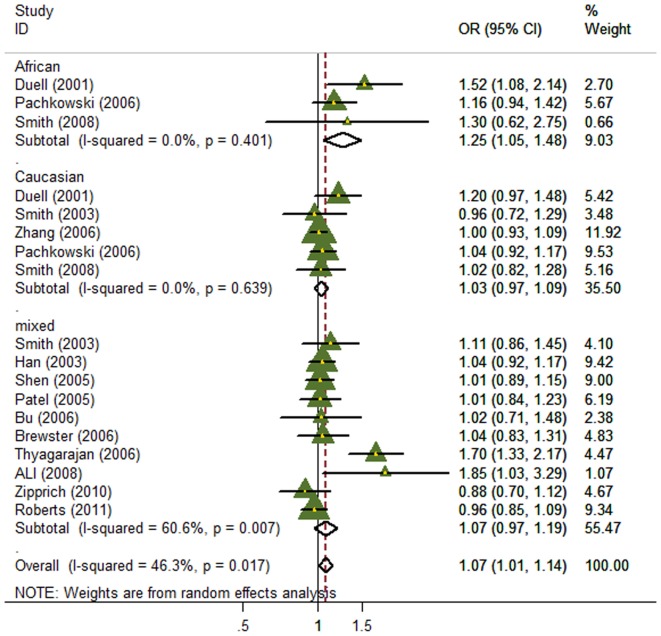
In the overall meta-analysis, significant between-study heterogeneity were observed for all genetic models (P for heterogeneity = 0.003, 0.003 and 0.017 for dominant, recessive and additive models, respectively), and thus the random-effects model was employed. Significant associations were observed between the XRCC1 Arg399Gln and breast cancer risk in both of the dominant and additive models (OR for dominant model = 1.12, 95% CI: 1.02–1.24; OR for additive model = 1.07, 95% CI: 1.01–1.14; Figure 2–3), but no association was found in recessive model (OR = 0.95, 95% CI: 0.84–1.08; Figure S1).
Figure 2. Forest plot of the association between the XRCC1 Arg399Gln and breast cancer risk for the dominant model.
Figure 3. Forest plot of the association between the XRCC1 Arg399Gln polymorphism and breast cancer risk for the additive model.
Subgroup meta-analysis
When subgroup analysis was performed by ethnical populations, for the dominant model, only the subgroup with mixed population showed significant association of the Arg399Gln without evidence of heterogeneity (OR = 1.10, 95% CI:1.01–1.20; Figure 2), whereas heterogeneity still existed and no associations were found for both subgroups of African-Americans and Caucasians, possibly due to their relatively small sample size and the moderate effect of this polymorphism under the dominant model. For the additive model, heterogeneity was effectively removed in African-Americans and Caucasians, but only the African-American population showed significant association (OR = 1.25, 95% CI: 1.05–1.48; Figure 3). For recessive model, heterogeneity was effectively removed in African-Americans and Caucasians, but there was still no association in any subgroups.
3 studies provided data according to premenopausal or postmenopausal status (Table S2) [17], [21], [26]. Heterogeneity was effectively removed in postmenopausal subgroup (Figure S2, S3, S4), but no significant association was detected. We considered that based on current limited data, it may lack of sufficient power to detect the real effect of this polymorphism according to premenopausal or postmenopausal status.
When stratified by the genotyping method, the significant was effectively removed in TaqMan Real Time PCR and MassARRAY Sequenome subgroup, but no association was found. In the PCR-RFLP subgroup, heterogeneity was seen (for dominant model: P = 0.005, I2 = 62.0%; for additive model: P = 0.001 I2 = 62.2%), possibly due to the different sources of controls and ethnicity. Significant association was also seen in this subgroup, with ORs for dominant model and additive model were 1.27 (95% CI = 1.08–1.49) and 1.15 (95% CI = 1.02–1.29), respectively.
Subgroup analysis was also performed by sources of controls (Table 2). The population based subgroup showed significant association, but with evidence of heterogeneity (for dominant model: P = 0.001, I2 = 63.4%; for additive model: P = 0.006, I2 = 55.5%). No heterogeneity and no significant association were seen in the hospital based subgroup.
Table 2. Results of overall analysis and subgroup analysis in this meta-analysis.
| Group | Dominant model | Additive model | Recessive model |
| Ethnicity | |||
| Caucasian | 1.13 (0.90–1.41) | 1.02 (0.96–1.08) | 1.08 (0.92–1.27) |
| African-American | 1.09 (0.75–1.58) | 1.24 (1.05–1.47) | 0.56 (0.30–1.03) |
| Mixed | 1.10 (1.01–1.21) | 1.07 (0.96–1.18) | 0.95 (0.84–1.09) |
| Menopausal status | |||
| Premenopausal | 1.90 (0.73–4.91) | 1.08 (0.98–1.19) | 2.24 (0.48–10.35) |
| Postmenopausal | 0.98 (0.86–1.12) | 0.93 (0.86–1.01) | 1.02 (0.87–1.21) |
| Genotyping method | |||
| PCR-RFLP | 1.27 (1.08–1.49) | 1.15 (1.02–1.29) | 0.90 (0.70–1.17) |
| TaqMan Real Time PCR | 1.08 (0.96–1.22) | 1.04 (1.00–1.13) | 0.93 (0.72–1.20) |
| MassARRAY Sequenome | 0.83 (0.68–1.02) | 0.98 (0.88–1.09) | 0.97 (0.83–1.13) |
| Control source | |||
| Population | 1.14 (1.02–1.28) | 1.09 (1.01–1.18) | 0.95 (0.80–1.13) |
| Hospital | 1.03 (0.83–1.29) | 1.00 (0.89–1.10) | 0.95 (0.82–1.10) |
| Overall OR | 1.12 (1.02–1.24) | 1.07 (1.01–1.14) | 0.95 (0.84–1.09) |
Sensitivity Analysis and Publication Bias
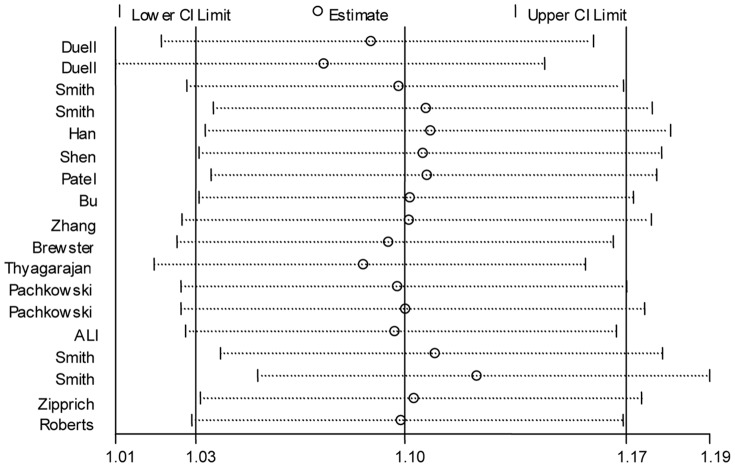
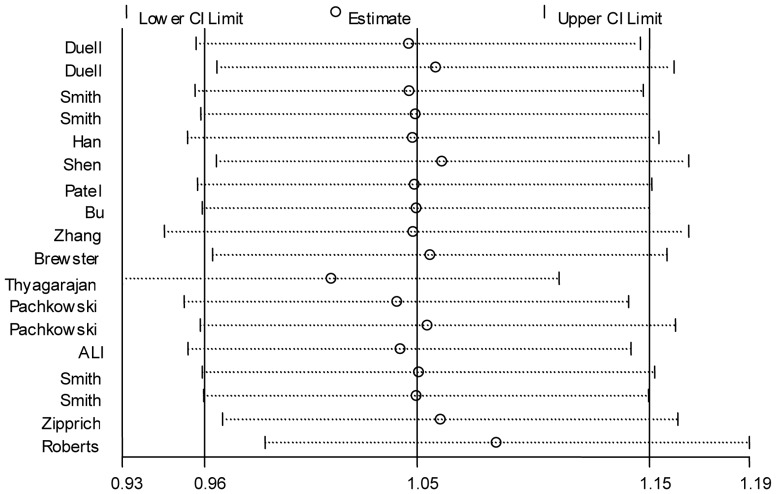
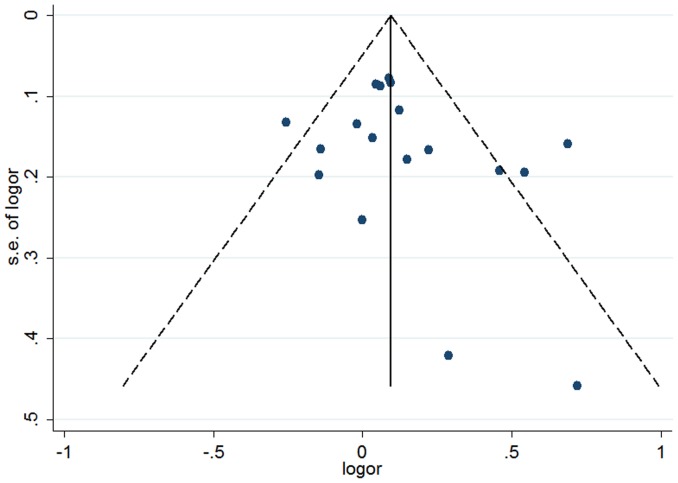
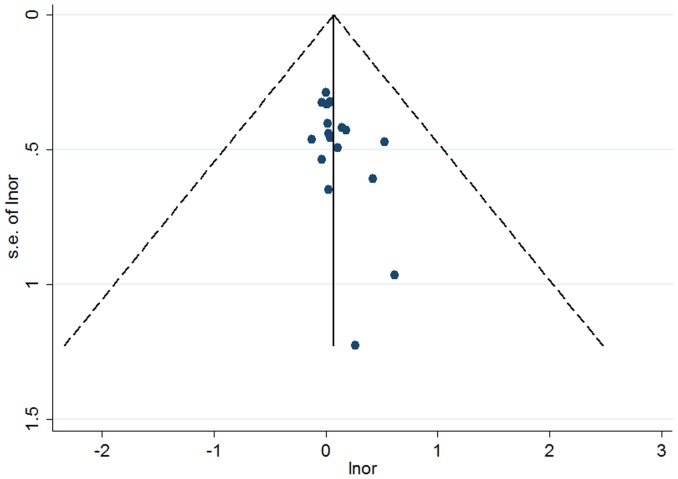
Given the significant between-study heterogeneity for the Arg399Gln polymorphism, we performed a sensitivity analysis to assess the effects of single study on pooled ORs under a random-effects model (Figure 4–5, Figure S5). The pooled ORs were similar before and after removal of each study, suggesting no single study significantly changes the pooled ORs. As reflected by funnel plots (Figure 6–7 and Figure S6) and Egger's tests, there was no publication bias in the dominant and recessive models (P for Egger's test >0.10). For the additive model, a borderline significant publication bias was observed (P for Egger's test = 0.04).
Figure 4. Sensitivity analysis of the association between the XRCC1 Arg399Gln and breast cancer risk for the dominant model.
Figure 5. Sensitivity analysis of the association between the XRCC1 Arg399Gln and breast cancer risk for the additive model.
Figure 6. Funnel plot of the association between the XRCC1 Arg399Gln and breast cancer risk for the dominant model.
Figure 7. Funnel plot of the association between the XRCC1 Arg399Gln and breast cancer risk for the additive model.
Discussion
This meta-analysis incorporated 18 studies of 10846 breast cancer cases and 11723 controls concerning the Arg399Gln in XRCC1. The Arg399Gln variant presented significant association breast cancer risk in the American population. Further sensitivity analysis suggested the stability of the current results, by showing similar ORs before and after sequential removal of single study. This meta-analysis, based on updated published data, has further increased sample size and enlarged the statistical power to reflect the precision effect of the Arg399Gln in breast cancer in the American population.
XRCC1 plays an important role in the BER pathway, which has been thought of as the predominant DNA-damage repair pathway for the processing of small base lesions derived from oxidation and alkylation damage [34]. The major significance of XRCC1 in maintaining genomic stability has been raised by high frequency of chromosome deletions or aberrations in the gene mutant cells, and thus the XRCC1 gene has been posed as a candidate gene for many cancer susceptibility. In the coding region of XRCC1, the nonsynonymous polymorphism, Arg399Gln, has caught much attention in breast cancer risk for years. This polymorphism is located in the critical COOH-terminal side of PARP-binding BRCT-domain [35], [36]. The amino acid substitution caused by this variant in the BRCT domian has been shown to completely disrupt the function of XRCC1, and thus may result in reduction of DNA repair capacity [37]. In view of its functional significance, it is biologically possible that the Arg399Gln polymorphism may modulate the risk of breast cancer. As expected, this meta-analysis provides an obvious evidence that the XRCC1 Arg399Gln polymorphism is significant associated with of breast cancer in the American population. Intriguingly, the significant association was presented in the dominant and additive models, which is inconsistent with the most recent meta-analysis [27], [29]. In these previous meta-analysis, the authors have wrongly extract the control's AA frequency. In the original article by Patel AV et al., the AA genotype frequency in controls was 194; however, in the meta-analysis by Huang Y et al. and Wu K et al., it changed to 280, which would influence the accuracy of the pooled analysis. Additionally, Caucasian and African-American assessed in the previous meta-analysis were distinct with our meta-analysis, possibly resulting in the inconsistent result with our meta-analysis.
The association of XRCC1 with breast cancer has been investigated in many other countries. In China, Liu L et al. reported XRCC1 -77T>C may be a genetic determinant for developing breast cancer [38]. For lung cancer, Liu L et al. find that XRCC1 T-77C could be genetic determinant for prognosis of advanced non-small-cell lung cancer patients treated with platinum-based chemotherapy [39]. Thus, we believed that XRCC1 Arg399Gln polymorphism maybe also associate with breast cancer.
Nevertheless, significant between-study heterogeneity was seen in this meta-analysis. To explore the source of heterogeneity, we performed subgroup analysis. After stratified by premenopausal or postmenopausal status, heterogeneity was significant removed, indicating that the premenopausal or postmenopausal status may be one source of heterogeneity. According to ethnicity, we found that for recessive and additive models, heterogeneity was effectively removed in Africans and Caucasians, whereas for dominant model, it retained in Africans and Caucasians, but in the mixed population, no evidence of heterogeneity was shown, suggesting ethnical population may also partly explained the heterogeneity of this meta-analysis. With regard to the control source, heterogeneity was removed in hospital-based subgroup, but was detected in population-based subgroup. Furthermore, the results of PCR-RFLP subgroup analysis were similar to population based subgroup analysis. Heterogeneity was also partly explained by population based and genotpying method of this meta-analysis. Moreover, all the included studies showed high quality (≥6 stars) by the 9-star Newcastle-Ottawa Scale, and no publications bias was observed in dominant and recessive models.
Several limitations in this meta-analysis should be figured out. First, in the subgroup analysis by ethnicity and premenopausal/postmenopausal status, the sample size was relatively small and the statistical power might be insufficient. Second, potential sources of heterogeneity in this meta-analysis could include other factors, such as family history of breast cancer, staging of breast cancer, history of begin breast disease. However, due to the limited data, we failed to further explore these factors in the current meta-analysis. Finally, multiple epidemiological studies have demonstrated gene-gene or gene-enviroment interactions may play more important role in cancer development as compared with genetic factors [40], [41]. However, gene-gene interactions and gene-environment interactions could not be appraised in this meta-analysis owing to a lack of special data.
In conclusion, this meta-analysis provided evidence that the XRCC1 Arg399Gln polymorphism was significantly associated with risk of breast cancer in the American population. Nevertheless, in the future, well-designed studies with large sample sizes will be warranted in diverse populations.
Supporting Information
The PRISMA 2009 Checklist.
(DOC)
Forest plot of the association between the XRCC1 Arg399Gln and breast cancer risk for the recessive model.
(TIF)
Forest plot of the association between the XRCC1 Arg399Gln and breast cancer risk of menopausal subgroup for the dominant model.
(TIF)
Forest plot of the association between the XRCC1 Arg399Gln and breast cancer risk of menopausal subgroup for the recessive model.
(TIF)
Forest plot of the association between the XRCC1 Arg399Gln and breast cancer risk of menopausal subgroup for the additive model.
(TIF)
Sensitivity analysis of the association between the XRCC1 Arg399Gln and breast cancer risk the recessive model.
(TIF)
Funnel plot of the association between the XRCC1 Arg399Gln and breast cancer risk for the recessive model.
(TIF)
Quality assessment of case–control studies included in this meta-analysis.
(DOC)
Data of premenopausal or postmenopausal studies.
(DOC)
Funding Statement
This work was supported by Medical Scientific Research Foundation of Guangdong Province B2012176 and Foundation for Distinguished Young Talents in Higher Education of Guangdong 2012LYM_0082 to Li Liu. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Parkin DM, Bray F, Ferlay J, Pisani P (2001) Estimating the world cancer burden: Globocan 2000. International journal of cancer 94: 153–156. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Cancer Facts & Figures 2013. Atlanta: American Cancer Society; 2013. [Google Scholar]
- 3. Espinosa E, G A Mez-Pozo A, S A Nchez-Navarro I, Pinto A, Casta N Eda CA, et al. (2012) The present and future of gene profiling in breast cancer. Cancer and Metastasis Reviews 31: 41–46. [DOI] [PubMed] [Google Scholar]
- 4. Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374. [DOI] [PubMed] [Google Scholar]
- 5. Wood RD, Mitchell M, Sgouros J, Lindahl T (2001) Human DNA repair genes. Science 291: 1284–1289. [DOI] [PubMed] [Google Scholar]
- 6. Goode EL, Ulrich CM, Potter JD (2002) Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiology Biomarkers & Prevention 11: 1513–1530. [PubMed] [Google Scholar]
- 7. Tudek B (2007) Base excision repair modulation as a risk factor for human cancers. Molecular aspects of medicine 28: 258–275. [DOI] [PubMed] [Google Scholar]
- 8. Huang G, Cai S, Wang W, Zhang Q, Liu A (2013) Association between XRCC1 and XRCC3 Polymorphisms with Lung Cancer Risk: A Meta-Analysis from Case-Control Studies. PLOS ONE 8: e68457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo S, Li X, Gao M, Li Y, Song B, et al. (2013) The relationship between XRCC1 and XRCC3 gene polymorphisms and lung cancer risk in northeastern Chinese. PloS one 8: e56213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Y, Liu F, Tan S, Wang Y, Li S (2012) X-ray repair cross-complementing group 1 (XRCC1) genetic polymorphisms and cervical cancer risk: a huge systematic review and meta-analysis. PLoS One 7: e44441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yuan P, Liu L, Wu C, Zhong R, Yu D, et al. (2010) No association between XRCC1 polymorphisms and survival in non-small-cell lung cancer patients treated with platinum-based chemotherapy. Cancer biology \& therapy 10: 854–859. [DOI] [PubMed] [Google Scholar]
- 12. Duell EJ, Millikan RC, Pittman GS, Winkel S, Lunn RM, et al. (2001) Polymorphisms in the DNA repair gene XRCC1 and breast cancer. Cancer Epidemiol Biomarkers Prev 10: 217–222. [PubMed] [Google Scholar]
- 13. Smith TR, Levine EA, Perrier ND, Miller MS, Freimanis RI, et al. (2003) DNA-repair genetic polymorphisms and breast cancer risk. Cancer Epidemiol Biomarkers Prev 12: 1200–1204. [PubMed] [Google Scholar]
- 14. Han J, Hankinson SE, De Vivo I, Spiegelman D, Tamimi RM, et al. (2003) A prospective study of XRCC1 haplotypes and their interaction with plasma carotenoids on breast cancer risk. Cancer Res 63: 8536–8541. [PubMed] [Google Scholar]
- 15. Smith TR, Miller MS, Lohman K, Lange EM, Case LD, et al. (2003) Polymorphisms of XRCC1 and XRCC3 genes and susceptibility to breast cancer. Cancer Lett 190: 183–190. [DOI] [PubMed] [Google Scholar]
- 16. Patel AV, Calle EE, Pavluck AL, Feigelson HS, Thun MJ, et al. (2005) A prospective study of XRCC1 (X-ray cross-complementing group 1) polymorphisms and breast cancer risk. Breast Cancer Res 7: R1168–R1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shen J, Gammon MD, Terry MB, Wang L, Wang Q, et al. (2005) Polymorphisms in XRCC1 modify the association between polycyclic aromatic hydrocarbon-DNA adducts, cigarette smoking, dietary antioxidants, and breast cancer risk. Cancer Epidemiol Biomarkers Prev 14: 336–342. [DOI] [PubMed] [Google Scholar]
- 18. Bu D, Tomlinson G, Lewis CM, Zhang C, Kildebeck E, et al. (2006) An intronic polymorphism associated with increased XRCC1 expression, reduced apoptosis and familial breast cancer. Breast Cancer Res Treat 99: 257–265. [DOI] [PubMed] [Google Scholar]
- 19. Thyagarajan B, Anderson KE, Folsom AR, Jacobs DJ, Lynch CF, et al. (2006) No association between XRCC1 and XRCC3 gene polymorphisms and breast cancer risk: Iowa Women's Health Study. Cancer Detect Prev 30: 313–321. [DOI] [PubMed] [Google Scholar]
- 20. Brewster AM, Jorgensen TJ, Ruczinski I, Huang HY, Hoffman S, et al. (2006) Polymorphisms of the DNA repair genes XPD (Lys751Gln) and XRCC1 (Arg399Gln and Arg194Trp): relationship to breast cancer risk and familial predisposition to breast cancer. Breast Cancer Res Treat 95: 73–80. [DOI] [PubMed] [Google Scholar]
- 21. Zhang Y, Newcomb PA, Egan KM, Titus-Ernstoff L, Chanock S, et al. (2006) Genetic polymorphisms in base-excision repair pathway genes and risk of breast cancer. Cancer Epidemiol Biomarkers Prev 15: 353–358. [DOI] [PubMed] [Google Scholar]
- 22. Pachkowski BF, Winkel S, Kubota Y, Swenberg JA, Millikan RC, et al. (2006) XRCC1 genotype and breast cancer: functional studies and epidemiologic data show interactions between XRCC1 codon 280 His and smoking. Cancer Res 66: 2860–2868. [DOI] [PubMed] [Google Scholar]
- 23. Ali MF, Meza JL, Rogan EG, Chakravarti D (2008) Prevalence of BER gene polymorphisms in sporadic breast cancer. Oncol Rep 19: 1033–1038. [PubMed] [Google Scholar]
- 24. Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, et al. (2008) Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis 29: 2132–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zipprich J, Terry MB, Brandt-Rauf P, Freyer GA, Liao Y, et al. (2010) XRCC1 polymorphisms and breast cancer risk from the New York Site of the Breast Cancer Family Registry: A family-based case-control study. J Carcinog 9: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roberts MR, Shields PG, Ambrosone CB, Nie J, Marian C, et al. (2011) Single-nucleotide polymorphisms in DNA repair genes and association with breast cancer risk in the web study. Carcinogenesis 32: 1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang Y, Li L, Yu L (2009) XRCC1 Arg399Gln, Arg194Trp and Arg280His polymorphisms in breast cancer risk: a meta-analysis. Mutagenesis 24: 331–339. [DOI] [PubMed] [Google Scholar]
- 28. Li H, Ha TC, Tai BC (2009) XRCC1 gene polymorphisms and breast cancer risk in different populations: a meta-analysis. Breast 18: 183–191. [DOI] [PubMed] [Google Scholar]
- 29. Wu K, Su D, Lin K, Luo J, Au WW (2011) XRCC1 Arg399Gln gene polymorphism and breast cancer risk: a meta-analysis based on case-control studies. Asian Pac J Cancer Prev 12: 2237–2243. [PubMed] [Google Scholar]
- 30. Saadat M, Ansari-Lari M (2009) Polymorphism of XRCC1 (at codon 399) and susceptibility to breast cancer, a meta-analysis of the literatures. Breast cancer research and treatment 115: 137–144. [DOI] [PubMed] [Google Scholar]
- 31. Moher D, Liberati A, Tetzlaff J, Altman DG (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Annals of internal medicine 151: 264–269. [DOI] [PubMed] [Google Scholar]
- 32. Higgins J, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Statistics in medicine 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 33. Egger M, Smith GD, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. Bmj 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chou W, Wang H, Wong F, Ding S, Wu P, et al. (2008) Chk2-dependent phosphorylation of XRCC1 in the DNA damage response promotes base excision repair. The EMBO journal 27: 3140–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hung RJ, Hall J, Brennan P, Boffetta P (2005) Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. American journal of epidemiology 162: 925–942. [DOI] [PubMed] [Google Scholar]
- 36. Shen MR, Jones IM, Mohrenweiser H (1998) Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer research 58: 604–608. [PubMed] [Google Scholar]
- 37. Masson M, Niedergang C, Schreiber VER, Muller S, Menissier-de Murcia J, et al. (1998) XRCC1 is specifically associated with poly (ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Molecular and Cellular Biology 18: 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu L, Yuan P, Liu L, Wu C, Zhang X, et al. (2011) A functional -77T>C polymorphism in XRCC1 is associated with risk of breast cancer. Breast Cancer Res Treat 125: 479–487. [DOI] [PubMed] [Google Scholar]
- 39. Liu L, Yuan P, Wu C, Zhang X, Wang F, et al. (2011) Assessment of XPD Lys751Gln and XRCC1 T-77C polymorphisms in advanced non-small-cell lung cancer patients treated with platinum-based chemotherapy. Lung Cancer 73: 110–115. [DOI] [PubMed] [Google Scholar]
- 40. Zhong R, Liu L, Zou L, Sheng W, Zhu B, et al. (2013) Genetic variations in the TGFbeta signaling pathway, smoking and risk of colorectal cancer in a Chinese population. Carcinogenesis 34: 936–942. [DOI] [PubMed] [Google Scholar]
- 41. Liu L, Wu C, Wang Y, Zhong R, Wang F, et al. (2011) Association of candidate genetic variations with gastric cardia adenocarcinoma in Chinese population: a multiple interaction analysis. Carcinogenesis 32: 336–342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The PRISMA 2009 Checklist.
(DOC)
Forest plot of the association between the XRCC1 Arg399Gln and breast cancer risk for the recessive model.
(TIF)
Forest plot of the association between the XRCC1 Arg399Gln and breast cancer risk of menopausal subgroup for the dominant model.
(TIF)
Forest plot of the association between the XRCC1 Arg399Gln and breast cancer risk of menopausal subgroup for the recessive model.
(TIF)
Forest plot of the association between the XRCC1 Arg399Gln and breast cancer risk of menopausal subgroup for the additive model.
(TIF)
Sensitivity analysis of the association between the XRCC1 Arg399Gln and breast cancer risk the recessive model.
(TIF)
Funnel plot of the association between the XRCC1 Arg399Gln and breast cancer risk for the recessive model.
(TIF)
Quality assessment of case–control studies included in this meta-analysis.
(DOC)
Data of premenopausal or postmenopausal studies.
(DOC)