Abstract
Diffusion tensor imaging (DTI) is achieved by collecting a series of diffusion-weighted images (DWIs). Signal averaging of multiple repetitions can be performed in the k-space (k-avg) or in the image space (m-avg) to improve the image quality. Alternatively, one can treat each acquisition as an independent image and use all of the data to reconstruct the DTI without doing any signal averaging (no-avg). To compare these three approaches, in this study, in vivo DTI data was collected from five normal mice. Noisy data with signal-to-noise ratios (SNR) that varied between five and 30 (before averaging) were then simulated. The DTI indices, including relative anisotropy (RA), trace of diffusion tensor (TR), axial diffusivity (λ║), and radial diffusivity (λ┴), derived from the k-avg, m-avg, and no-avg, were then compared in the corpus callosum white matter, cortex gray matter, and the ventricles. We found that k-avg and m-avg enhanced the SNR of DWI with no significant differences. However, k-avg produced lower RA in the white matter and higher RA in the gray matter, compared to the m-avg and no-avg, regardless of SNR. The latter two produced similar DTI quantifications. We concluded that k-avg is less preferred for DTI brain imaging.
Introduction
Diffusion tensor imaging (DTI) is an emerging image modality, which quantifies water molecular diffusion and is sensitive to the microstructural changes (1,2). During the past decades, DTI has been used successfully to produce non-invasive nerve fiber tracking (3,4) and white matter segmentation (5). DTI-derived quantities, such as the axial diffusivity (λ║), radial diffusivity (λ⊥), and diffusion anisotropy, have been proposed to serve as potential markers for various white matter pathologies (6-9). Recent studies also demonstrated that the microstructural changes involved in neuroplasticity might also be detectable by DTI (10-12).
Despite the wide ranges of the applications using DTI in the nervous system, the fundamental data acquisition of DTI usually involves a series of diffusion-weighted images (DWI). Because diffusion-weighting factors attenuate the MRI signals, DWIs are inherently prone to low signal-to-noise ratios (SNR). Data averaging among the repetitions of DWIs in the k-space is a widely used approach to overcome the low SNR of DWIs (13,14). The averaged DWIs were then used to reconstruct the diffusion tensor, D, via Eq. [1], here named “k-avg” DTI.
| [1] |
where S is the signal intensity of DWI; b is the diffusion weighting b value; u is the gradient encoding unit vector; and S0 is the signal intensity without diffusion weighting (2).
Alternatively, the DWI of each repetition could be produced, via a Fourier transformation. The DWIs with the same diffusion-encoding vector were averaged, and then used to reconstruct D via Eq. [1], here named “m-avg” DTI. As a second alternative, a Fourier transformation can be executed on each repetition individually, without averaging, followed by applying these repetitions into Eq. [1] to reconstruct the diffusion tensor. Since there are more equations than unknowns, the ordinary-least-squares method was used to find an approximate solution to this over-determined system. Here we called it the “no-avg” DTI.
Theoretically, the k-avg is preferred over the m-avg, because the noise in the k-space are complex numbers with a zero-mean Gaussian probability distribution (15). However, k-space data is not always available, especially within some clinical settings. For applications in fiber tracking, studies may collect data with a large number of diffusion encoding directions with no repetition (16-18). As such, a no-avg DTI would be performed. It is unknown whether m-avg or no-avg approaches would be a more practical solution, especially with data that has low SNR.
In this study, we performed mouse brain in vivo DTI. The data from a set of directive DWIs is acquired with three repetitions. Simulated noise was added in the k-space to produce seven sets of data with various SNRs ranging between 4.5 and 33.3 (measured in cortex before averaging). The effects of k-avg, m-avg, and no-avg algorithms were compared side-by-side on the DWIs and reconstructed DTI maps. DTI indices of the gray matter, white matter, and ventricles of mouse brains were quantitatively compared among these three algorithms.
Materials and Methods
All animal procedures were done in accordance with National Institutes of Health guidelines. The experimental procedures were approved by the Institutional Animal Care and Use Committee of Loma Linda University.
Five C57BL/6 female mice, eight weeks of age, were anesthetized by 1.5% isoflurane/oxygen using an isoflurane vaporizer (VetEquip, Pleasanton, CA). The core body temperature was maintained using a warm-water-circulating pad. The transmitter was a 7-cm inner diameter Bruker linear RF coil, and the receiver was a 2-cm surface coil. DTI was performed in a Bruker 4.7T BioSpec animal scanner using a spin echo imaging sequence with a TR of 3s, TE of 26 ms, with a duration between a diffusion gradient pair (Δ) of 10 ms, diffusion gradient duration (δ) of 6 ms, field of view of 1.5cm, slice thickness of 0.5mm, 19 interleaved slices, and data matrix of 96 × 96 (with zero-padding to 256 × 256), and six-direction diffusion scheme with the b-value of 0.850 ms/μm2 along encoding directions [x, y, z] = [0.707, 0.707, 0], [0.707, 0, 0.707], [0, 0.707, 0.707], [−0.707, 0.707, 0], [−0.707, 0, 0.707], and [0, −0.707, 0.707], plus one image with no diffusion weighting (B0). The acquisition was not gated. Data was acquired over three repetitions. The total acquisition time was about three hours.
Simulated noise was introduced by adding random values from the standard normal distribution in both real and imaginary parts of the raw data in k-space using Matlab (MathWorks, Natick, MA, USA). The data was then processed through three different algorithms:
Averaging k-space data across three repetitions (k-avg), and then converting them to the image space via a Fourier transformation, followed by Eq. [1] to generate the diffusion tensor D.
Averaging the image-space data across three repetitions (m-avg), followed by tensor D calculation using Eq. [1].
No averaging across three repetitions but applying the image-space data in Eq. [1] to derive the diffusion tensor D (no-avg).
Following each algorithm, the eigenvalues (λ1, λ2, and λ3) were derived from the diffusion tensor D The axial diffusivity (λ║), radial diffusivity (λ⊥), relative anisotropy (RA), and the trace of the diffusion tensor (TR) were defined by the following equations (2,19):
| [2] |
| [3] |
| [4] |
| [5] |
All of these processes were accomplished with customized software written in Matlab (MathWorks, Natick, MA, USA).
The regions of interest (ROIs) were selected in cortex, corpus callosum, and lateral ventricle to evaluate the DTI quantification in white matter, gray matter, and ventricles respectively (Figure 1). The SNR of B0 images was calculated as the mean intensity of the selected region divided by the standard deviation of the background. For statistical analysis, multiple comparisons to compare DTI indices among k-avg, m-avg, and no-avg were performed using one-way analysis of variance (ANOVA) followed by the post-hoc Student-Newman-Keuls test using Sigma Plot 11.0 (San Jose, CA), and p < 0.05 was considered significant.
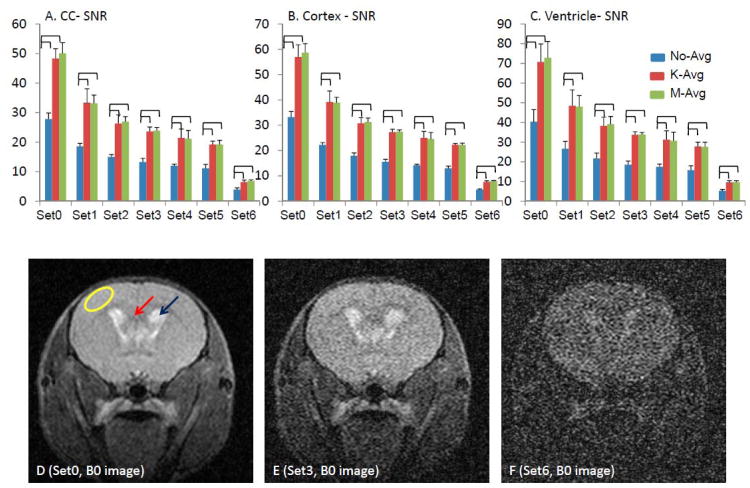
Figure 1.
Noise was added to simulate seven sets of DWIs with various SNR levels. The SNR of the white matter, gray matter, and ventricles were measured in the corpus callosum (red arrow in D), cortex (yellow oval in D), and lateral ventricle (blue arrow in D), respectively, on B0 images. The SNR at each region of each data set was summarized in A (corpus callosum, CC), B (cortex), and C (ventricle). The brackets indicated p < 0.05. The SNR of m-avg and k-avg were significantly higher than the SNR of no-avg, while there was no significant difference between m-avg and k-avg. Examples of B0 images from the Set0, Set3, and Set6 were shown in D – E, respectively.
Results
The SNR was 27.8 ± 2.1 in the corpus callosum, 33.3 ± 2.2 in the cortex, and 40.3 ± 6.2 in the ventricles of the collected B0 images. Noise was added in k-space, and the noise levels were gradually increased to produce a total of 7 sets of images with the lowest SNR down to 3.9 ± 0.6 in the corpus callosum, 4.5 ± 0.4 in the cortex, and 5.2 ± 0.7 in the ventricles (B0 images). Both k-avg and m-avg significantly enhanced SNR 1.72 – 1.77 times from the non-averaged B0 images (Figure 1). There was no significant SNR difference on the averaged B0 images between k-avg and m-avg.
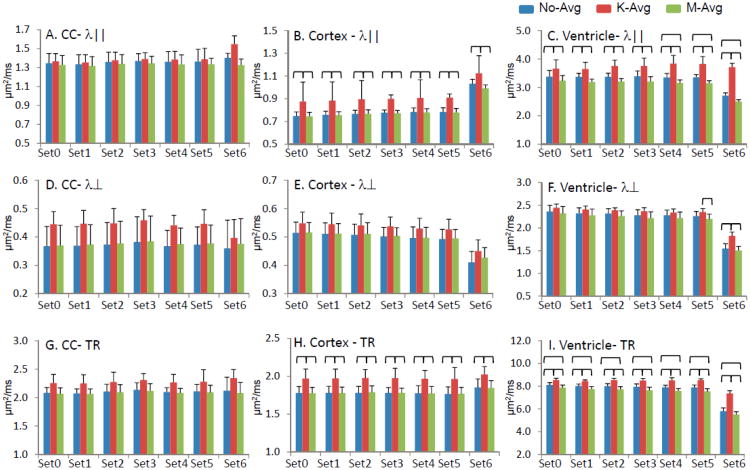
While k-avg and m-avg enhanced SNR at similar levels, the DTI indices produced by k-avg and m-avg were different (Figures 2 and 3). In the corpus callosum (CC), a white matter highly anisotropic region, k-avg led to a ∼20% increase in λ⊥, a ∼9% increase in TR, and a ∼10% decrease in RA, compared to those derived from the m-avg and no-avg. However, these differences did not reach a statistically significant level. In the cortex, a low anisotropic gray matter region, k-avg led to a ∼16% increase in λ‖ (p < 0.05), a ∼7% increase λ⊥ (not statistically significant), a ∼10% increase TR (p < 0.05), and a ∼20% increase in RA (not statistically significant). In the ventricle, an isotropic region, k-avg led to a 13 – 48% increase in λ‖ (p < 0.05), a 5 – 20% increase λ⊥ (p < 0.05 in Set5 and Set6 but not in Set0 - Set4), 9 – 30% increase TR (p < 0.05), and a 21 – 38% increase in RA (p < 0.05 in Set4 - Set6 but not in Set0 - Set3), compared to the m-avg and no-avg.
Figure 2.
The λ‖ (A – C), λ⊥ (D – F), and TR (G – I) diffusivities derived from no-avg, k-avg, and m-avg measured in corpus callosum (CC, A, D, G), cortex (B, E, H), and ventricle (C, F, I). The brackets indicated p < 0.05. The k-avg produced a significant increase of λ‖ and TR in the cortex and ventricle, compared to m-avg and no-avg.
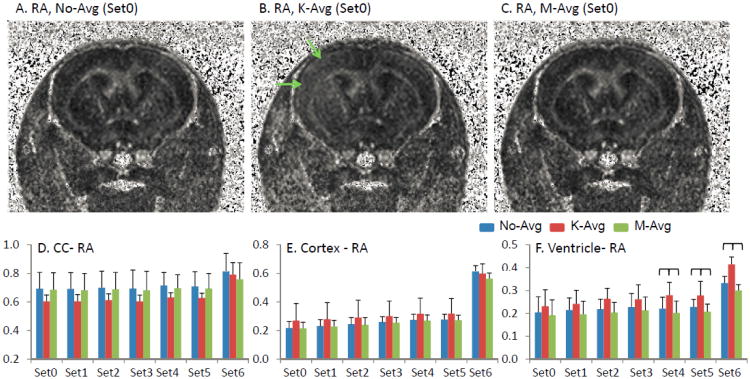
Figure 3.
RA maps derived from no-avg, k-avg, and m-avg. An example of the RA maps of the same dataset derived from no-avg, k-avg, and m-avg was shown in A – C, respectively. The RA maps derived from no-avg (A) and m-avg (C) were indistinguishable. However, in k-avg, artificially increased RA was seen in the cortex (green arrows). The measurements of RA from all data sets were summarized in corpus callosum (CC, D), cortex (E), and ventricle (F). In CC, the RA derived from k-avg was usually lower than the RA derived from m-avg and no-avg. In contrast, in the cortex and ventricles, the RA derived from k-avg was usually higher than the RA derived from m-avg and no-avg. However, most of these differences did not reach a statistically significant level. Only in the ventricles with low SNR (Set4 - Set6), the differences were statistically significant (p < 0.05) and were marked with brackets.
We also investigated the DTI reconstructed from data with and without an averaging process. As shown in Figure 3, the RA maps of no-avg are highly similar to the one produced via the m-avg. In k-avg, however, a reduced RA in white matter and an increased RA in gray matter led to a reduced contrast of image to identify white matter on the RA maps. Statistical analysis showed no significant difference between DTI indices derived from m-avg and no-avg in DTI measurements in the corpus callosum and cortex. In the ventricles, there was no difference of DTI indices (except TR) between no-avg and m-avg in data with SNR > 20 (before averaging). In the lower SNR situations (Set4 – Set6), the m-avg gradually produced a reduced λ‖, compared to those derived from no-avg (p < 0.05). As for TR, m-avg produced a ∼2% decrease of TR in ventricles, compared to no-avg (p < 0.05), regardless of the SNR.
Discussion
In this study, DTI quantifications produced by k-avg, m-avg, and no-avg were compared using mouse brains in vivo. The SNR of the simulated data were ranging from a conventional level (∼30, before averaging) to a much noisier situation (∼5, before averaging). There were three important findings in this study:
Both k-avg and m-avg were equally good at enhancing the SNR from repeatedly acquired B0 images (no significant difference). Both averaging processes enhanced SNR 1.72 – 1.77 times, which is equivalent to the theoretical square root of three (repetition amount), 1.73.
The DTI indices derived through no-avg were indistinguishable from those derived from m-avg.
Several DTI indices derived from k-avg were significantly different from those derived from no-avg or m-avg. The reduced RA in the white matter and the increased RA in the gray matter, provided via the k-avg, led to a reduced contrast of the image to identify white matter on RA maps, compared to no-avg and m-avg.
Data averaging is a widely adopted strategy to improve the image SNR in MRI (13,14). However, the success of using averaging to improve image SNR relies on the assumption of the identical signals of repetitions, with the exception of the noise. However, for the DTI with its longer scanning time, the motion of the subjects (20), the physiological fluctuation (21), eddy-current effects, and the increase of the electrical circuit/coil temperature (22) during the scan may alter signals. Because MRI signals were acquired as a complex number in k-space, the variations would appear in phase and magnitude of MRI signals. While both no-avg and m-avg discard the phase portions of MRI signals after the Fourier transform, k-avg uses the full k-space complex numbers for averaging. The variation in phase can cause signal cancelation during the k-avg process, adding errors in DTI quantifications.
Phase variation has been recognized as an issue for data averaging in MRI. One solution is to use navigator echoes to identify the corrupted measurements and reacquire those measurements when the anatomy returns to the baseline position (23,24). A more time-efficient method is to extract information of in-plane and through-plane displacements from the navigator echoes so that k-space data can be retrospectively corrected (25-28). However, navigator displacement measurements require a priori knowledge of the type of motion so that the navigator can be tailored to the specific type of motion. Alternatively, the corrupted phase encoding views could be detected from the difference between different sets of k-space data. The corrupted data are then replaced either with synthesized data (29) or nearest pre-average images (30). However, only those errors which are large enough (usually through a threshold setting) were identified and corrected. The phase errors smaller than the threshold would be missed. In this study, we explored the alternatives using m-avg and no-avg without any phase correction process. We found m-avg produced SNR enhancements similar to k-avg. Both m-avg and no-avg avoided the phase-induced signal canceling and produced highly reproducible DTI quantifications regardless the noise levels.
Because the DTI produced by no-avg is indistinguishable from those produced via m-avg, the averaging process to improve SNR of DWIs for DTI reconstruction may not be as critical as previously considered. If the SNR of the original DWI is not a concern, the precious scanning time may not need to be used for multiple repetitions, but rather to acquire DWIs at various diffusion encoding directions. This finding supported the notions of choosing a large number of encoding directions over the SNR to acquire DWIs for a better DTI quantification (16-18).
In this study, a spin-echo imaging sequence was used. Despite the widely used echo-planer imaging (EPI) sequences in clinical MRI, the spin-echo imaging is still dominantly used for small animal imaging. Because of the short scanning time in EPI, the motion-induced phase errors may be less pronounced, compared to the spin-echo imaging. Similarly, when a large number of diffusion gradient encoding directions are used, the number of repetitions for data averaging is usually reduced in order to maintain the same total scanning time. With a reduced averaging process, the DTI errors caused by k-avg would be less pronounced but may still be noticeable. The findings of this study are still applicable in these situations.
In conclusion, we demonstrated that no-avg and m-avg avoided the phase variation during data acquisition and produced similar DTI indices. In contrast, k-avg tended to produce a higher RA in gray matter and ventricles and a lower RA in white matter, compared to m-avg or no-avg. Thus, the m-avg or no-avg should be considered to achieve a better reliable DTI quantification, instead of using k-avg. Although the data presented in this study were collected based on in vivo mouse brain imaging, the findings are applicable to clinical MRI for human brain imaging.
Acknowledgments
This study was partly supported by NIH R01 NS062830.
Grant support: NIH R01 NS062830.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pierpaoli C, Jezzard P, Basser PJ, Barnett A, Di Chiro G. Diffusion tensor MR imaging of the human brain. Radiology. 1996;201(3):637–648. doi: 10.1148/radiology.201.3.8939209. [DOI] [PubMed] [Google Scholar]
- 2.Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994;66(1):259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. In vivo fiber tractography using DT-MRI data. Magn Reson Med. 2000;44(4):625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 4.Danielian LE, Iwata NK, Thomasson DM, Floeter MK. Reliability of fiber tracking measurements in diffusion tensor imaging for longitudinal study. Neuroimage. 2010;49(2):1572–1580. doi: 10.1016/j.neuroimage.2009.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun SW, Song SK, Hong CY, Chu WC, Chang C. Directional correlation characterization and classification of white matter tracts. Magn Reson Med. 2003;49(2):271–275. doi: 10.1002/mrm.10362. [DOI] [PubMed] [Google Scholar]
- 6.Sun SW, Liang HF, Cross AH, Song SK. Evolving Wallerian degeneration after transient retinal ischemia in mice characterized by diffusion tensor imaging. Neuroimage. 2008;40(1):1–10. doi: 10.1016/j.neuroimage.2007.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q, Vlkolinsky R, Xie M, Obenaus A, Song SK. Diffusion tensor imaging detected optic nerve injury correlates with decreased compound action potentials after murine retinal ischemia. Invest Ophthalmol Vis Sci. 2012;53(1):136–142. doi: 10.1167/iovs.11-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song SK, Sun SW, Ju WK, Lin SJ, Cross AH, Neufeld AH. Diffusion tensor imaging detects and differentiates axon and myelin degeneration in mouse optic nerve after retinal ischemia. Neuroimage. 2003;20(3):1714–1722. doi: 10.1016/j.neuroimage.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Sun SW, Liang HF, Trinkaus K, Cross AH, Armstrong RC, Song SK. Noninvasive detection of cuprizone induced axonal damage and demyelination in the mouse corpus callosum. Magn Reson Med. 2006;55(2):302–308. doi: 10.1002/mrm.20774. [DOI] [PubMed] [Google Scholar]
- 10.Sagi Y, Tavor I, Hofstetter S, Tzur-Moryosef S, Blumenfeld-Katzir T, Assaf Y. Learning in the fast lane: new insights into neuroplasticity. Neuron. 2012;73(6):1195–1203. doi: 10.1016/j.neuron.2012.01.025. [DOI] [PubMed] [Google Scholar]
- 11.Blumenfeld-Katzir T, Pasternak O, Dagan M, Assaf Y. Diffusion MRI of structural brain plasticity induced by a learning and memory task. PLoS One. 2011;6(6):e20678. doi: 10.1371/journal.pone.0020678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Groof G, Verhoye M, Van Meir V, Tindemans I, Leemans A, Van der Linden A. In vivo diffusion tensor imaging (DTI) of brain subdivisions and vocal pathways in songbirds. Neuroimage. 2006;29(3):754–763. doi: 10.1016/j.neuroimage.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 13.Sarlls JE, Pierpaoli C. Diffusion-weighted radial fast spin-echo for high-resolution diffusion tensor imaging at 3T. Magn Reson Med. 2008;60(2):270–276. doi: 10.1002/mrm.21639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agosta F, Kostic VS, Davidovic K, Kresojevic N, Sarro L, Svetel M, Stankovic I, Comi G, Klein C, Filippi M. White matter abnormalities in Parkinson's disease patients with glucocerebrosidase gene mutations. Mov Disord. 2013 doi: 10.1002/mds.25397. [DOI] [PubMed] [Google Scholar]
- 15.Edelstein WA, Glover GH, Hardy CJ, Redington RW. The intrinsic signal-to-noise ratio in NMR imaging. Magn Reson Med. 1986;3(4):604–618. doi: 10.1002/mrm.1910030413. [DOI] [PubMed] [Google Scholar]
- 16.Tuch DS, Reese TG, Wiegell MR, Makris N, Belliveau JW, Wedeen VJ. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn Reson Med. 2002;48(4):577–582. doi: 10.1002/mrm.10268. [DOI] [PubMed] [Google Scholar]
- 17.Hope T, Westlye LT, Bjornerud A. The effect of gradient sampling schemes on diffusion metrics derived from probabilistic analysis and tract-based spatial statistics. Magn Reson Imaging. 2012;30(3):402–412. doi: 10.1016/j.mri.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Jones DK. The effect of gradient sampling schemes on measures derived from diffusion tensor MRI: a Monte Carlo study. Magn Reson Med. 2004;51(4):807–815. doi: 10.1002/mrm.20033. [DOI] [PubMed] [Google Scholar]
- 19.Sun SW, Liang HF, Xie M, Oyoyo U, Lee A. Fixation, not death, reduces sensitivity of DTI in detecting optic nerve damage. Neuroimage. 2009;44(3):611–619. doi: 10.1016/j.neuroimage.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 20.Jiang H, Golay X, van Zijl PC, Mori S. Origin and minimization of residual motion-related artifacts in navigator-corrected segmented diffusion-weighted EPI of the human brain. Magn Reson Med. 2002;47(4):818–822. doi: 10.1002/mrm.10102. [DOI] [PubMed] [Google Scholar]
- 21.Le Bihan D, Poupon C, Amadon A, Lethimonnier F. Artifacts and pitfalls in diffusion MRI. J Magn Reson Imaging. 2006;24(3):478–488. doi: 10.1002/jmri.20683. [DOI] [PubMed] [Google Scholar]
- 22.Doty FD, Entzminger G, Kulkarni J, Pamarthy K, Staab JP. Radio frequency coil technology for small-animal MRI. NMR Biomed. 2007;20(3):304–325. doi: 10.1002/nbm.1149. [DOI] [PubMed] [Google Scholar]
- 23.Sachs TS, Meyer CH, Irarrazabal P, Hu BS, Nishimura DG, Macovski A. The diminishing variance algorithm for real-time reduction of motion artifacts in MRI. Magn Reson Med. 1995;34(3):412–422. doi: 10.1002/mrm.1910340319. [DOI] [PubMed] [Google Scholar]
- 24.Crowe LA, Keegan J, Gatehouse PD, Mohiaddin RH, Varghese A, Symmonds K, Cannell TM, Yang GZ, Firmin DN. 3D volume-selective turbo spin echo for carotid artery wall imaging with navigator detection of swallowing. J Magn Reson Imaging. 2005;22(4):583–588. doi: 10.1002/jmri.20424. [DOI] [PubMed] [Google Scholar]
- 25.Anderson AW, Gore JC. Analysis and correction of motion artifacts in diffusion weighted imaging. Magn Reson Med. 1994;32(3):379–387. doi: 10.1002/mrm.1910320313. [DOI] [PubMed] [Google Scholar]
- 26.Ordidge RJ, Helpern JA, Qing ZX, Knight RA, Nagesh V. Correction of motional artifacts in diffusion-weighted MR images using navigator echoes. Magn Reson Imaging. 1994;12(3):455–460. doi: 10.1016/0730-725x(94)92539-9. [DOI] [PubMed] [Google Scholar]
- 27.Anderson AG, 3rd, Velikina J, Block W, Wieben O, Samsonov A. Adaptive retrospective correction of motion artifacts in cranial MRI with multicoil three-dimensional radial acquisitions. Magn Reson Med. 2012 doi: 10.1002/mrm.24348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Ehman RL. Retrospective adaptive motion correction for navigator-gated 3D coronary MR angiography. J Magn Reson Imaging. 2000;11(2):208–214. doi: 10.1002/(sici)1522-2586(200002)11:2<208::aid-jmri20>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 29.Bydder M, Larkman DJ, Hajnal JV. Generalized SMASH imaging. Magn Reson Med. 2002;47(1):160–170. doi: 10.1002/mrm.10044. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Kholmovski EG, Guo J, Parker DL. TSE with average-specific phase encoding ordering for motion detection and artifact suppression. J Magn Reson Imaging. 2007;25(6):1271–1282. doi: 10.1002/jmri.20908. [DOI] [PubMed] [Google Scholar]