Summary
Inhibition of DAF-2 (IGF-1 receptor) or RSKS-1 (S6K), key molecules in the insulin/IGF-1 signaling (IIS) and target of rapamycin (TOR) pathways respectively, extends lifespan in C. elegans. However it has not been clear how they interact with each other and in which tissues to modulate longevity. Here we demonstrate that mutations in daf-2 and rsks-1 when combined produce a nearly five-fold increase in longevity that is much greater than the sum of single mutations. This synergistic lifespan extension requires positive feedback regulation of DAF-16 (FOXO) via the AMP-activated protein kinase (AMPK) complex. We further identify germ line as the key tissue for the synergistic longevity. Moreover, germline-specific inhibition of rsks-1 activates DAF-16 in the intestine. Together, our findings highlight the importance of the germ line in significantly prolonged longevity by daf-2 rsks-1, which provides important implications for interactions between the two major conserved longevity pathways in more complex organisms.
Keywords: C. elegans, daf-2, rsks-1, daf-16, AMPK, germ line, synergistic lifespan extension
Introduction
Aging can be modulated by both genetic and environmental factors. Alterations in insulin/insulin-like growth factor (IGF-1) signaling (IIS), target of rapamycin (TOR) pathway, signals from the reproductive system, and dietary restriction (DR) significantly affect lifespan (Kenyon, 2005; Kenyon, 2010). The highly conserved TOR kinase serves as a nutrient sensor to promote growth and proliferation via regulation of mRNA translation, ribosomal biogenesis, metabolism, and autophagy (Kapahi et al., 2010; Wullschleger et al., 2006). TOR promotes mRNA translation largely through the downstream ribosomal S6 kinase (S6K) and translation initiation factor eIF-4E-binding protein (4E-BP). Inhibition of TOR or S6K significantly extends lifespan in multiple species (Hansen et al., 2007; Harrison et al., 2009; Jia et al., 2004; Kaeberlein et al., 2005; Kapahi et al., 2004; Pan et al., 2007; Vellai et al., 2003). The mechanisms are overlapping with those by DR, an environmental manipulation that extends lifespan and slows age-related pathologies (Kapahi et al., 2010). rsks-1 encodes the C. elegans ortholog of S6K. In addition to lifespan extension, rsks-1 mutants also show delayed development and reduced fertility (Hansen et al., 2007; Korta et al., 2012; Pan et al., 2007; Selman et al., 2009). The longevity phenotype of rsks-1 requires PHA-4, a FOXA transcription factor, and AAK-2, a catalytic subunit of the 5′ adenosine monophosphate-activated protein kinase (AMPK) (Selman et al., 2009; Sheaffer et al., 2008). AMPK is a key cellular energy homeostasis regulator that is also partially required for lifespan extension by reduced IIS (Apfeld et al., 2004).
Inhibition of IIS results in prolonged longevity in worms, flies, mice and probably humans (Clancy et al., 2001; Holzenberger et al., 2003; Kenyon et al., 1993). In C. elegans, loss-of-function mutations in daf-2, which encodes the insulin/IGF-1 receptor homolog, lead to more than doubled adult lifespan as well as significant changes in development, metabolism and increased stress resistance (Gems et al., 1998; Kenyon et al., 1993; Kimura et al., 1997). The significantly prolonged longevity of daf-2 is totally dependent upon the downstream DAF-16 (FOXO) transcription factor (Lin et al., 1997; Ogg et al., 1997). Functional genomics studies identified DAF-16 target genes, which are involved in stress response, metabolism and detoxification (Lee et al., 2003; McElwee et al., 2004; Murphy et al., 2003). DAF-16 acts in specific tissues to modulate lifespan. Restoring the DAF-16 activity in the intestine (adipose tissue) substantially increases the lifespan of daf-16; daf-2 double mutants (Libina et al., 2003). On the other hand, DAF-16 functions through different factors to regulate the expression of downstream genes both cell-autonomously and -non-autonomously (Zhang et al., 2013). These findings suggest that IIS functions in an endocrine-like manner to modulate aging in C. elegans.
Signals from the reproductive system regulate lifespan in worms, flies and potentially in mice (Flatt et al., 2008; Hsin and Kenyon, 1999). In C. elegans, removal of the germ line significantly extends lifespan through activating DAF-16 in the intestine via a steroid hormone signaling (Berman and Kenyon, 2006; Hsin and Kenyon, 1999). Lifespan extension by germline loss requires DAF-16-mediated regulation of fat metabolism (McCormick et al., 2012; O'Rourke et al., 2009; Wang et al., 2008) and proteasome activity (Vilchez et al., 2012). Interestingly, removal of the germ line in certain daf-2 mutants synergistically enhances the prolonged longevity phenotype, suggesting there might be regulatory interactions between IIS and signals from the reproductive system (Hsin and Kenyon, 1999).
Despite the well-characterized roles of daf-2 and rsks-1 in aging and their apparently overlapping functions, it has not been clear whether and how they might interact with each other to affect longevity. To address this important question, we constructed a daf-2 rsks-1 double mutant, which displayed a synergistic effect on longevity. This nearly five-fold lifespan extension is mediated by positive feedback regulation of DAF-16 via AMPK. Further analyses identified germ line as the key tissue for RSKS-1, DAF-16 and AMPK to modulate the synergistically prolonged longevity. Furthermore, inhibition of rsks-1 in the germ line non-autonomously activates DAF-16 in the intestine. Collectively, our findings demonstrated a novel interaction between IIS and S6K in specific tissues that leads to significantly extended lifespan.
Results
Synergistic lifespan extension by daf-2 rsks-1 requires DAF-16
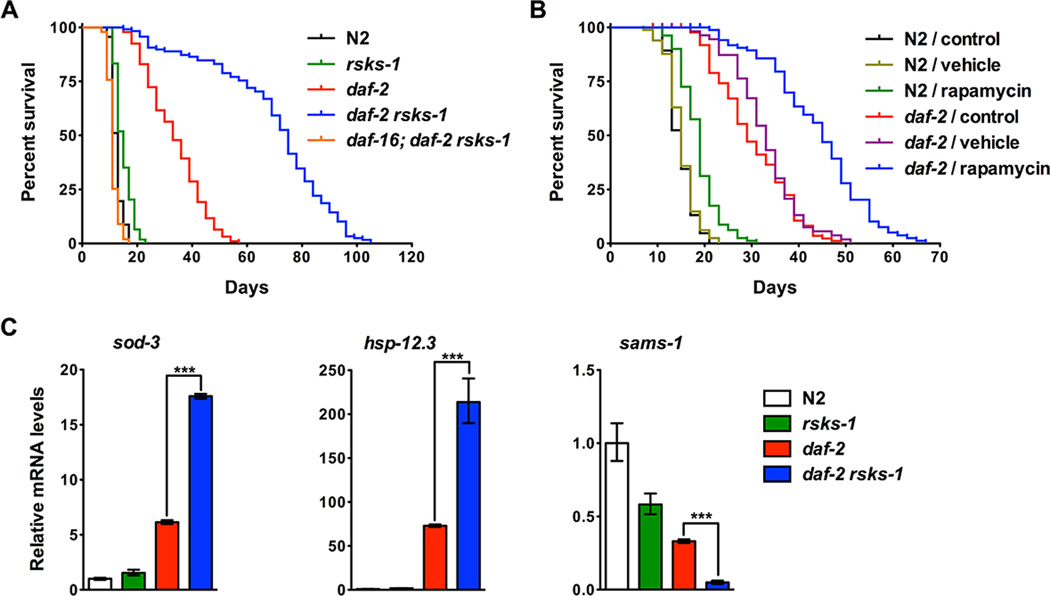
To examine the genetic interaction between daf-2 and rsks-1, we constructed a double mutant that carries the daf-2(e1370) strong loss-of-function allele and rsks-1(ok1255) deletion allele. The double mutant is viable, fertile and does not arrest at the diapause dauer stage under standard culture conditions, which allowed us to characterize the adult lifespan phenotypes. Since the daf-2 mutation is temperature-sensitive, animals were grown at the permissive temperature (15°C or 20°C) until the late L4 larval stage and then transferred to the restrictive temperature (25°C) during adulthood for survival assays. The rsks-1 and daf-2 single mutants increased mean lifespan by 20% and 169%, respectively; whereas the daf-2 rsks-1 double mutant showed a synergistic lifespan extension by 454% compared to the wild-type N2 (Figure 1A and Table S1). This lifespan extension phenotype was not due to unknown mutations in the background since all mutants were backcrossed with the same wild-type N2 for a minimum of six times. Furthermore, similar lifespan extension phenotypes were observed using another deletion rsks-1(tm1714) allele (Figure S1A), another point mutation daf-2(e1391) allele (Figure S1B) or when animals were cultured at the intermediate temperature (20°C) throughout life (Figure S1C). We also performed lifespan assays with animals treated with rapamycin to inhibit TOR, the upstream activator of RSKS-1. Rapamycin mildly extended lifespan of N2 by 26%, while it extended lifespan of the daf-2 mutant by 45% (Figure 1B).
Figure 1. Double mutations in daf-2 and rsks-1 lead to synergistically prolonged longevity that requires DAF-16.
(A) The daf-2 rsks-1 double mutant showed synergistically prolonged longevity (454% extension compared to N2) that is dependent on DAF-16. (B) Inhibition of TOR by rapamycin led to increased lifespan extension in daf-2 compared to N2. Rapamycin (100 μM) extended N2 and daf-2 lifespan by 26% and 45%, respectively (log-rank, p < 0.0001). Animals treated with the vehicle (DMSO) alone did not show significantly affected lifespan (log-rank, p > 0.05). Quantitative data and statistical analyses are included in Table S1. (C) daf-2 rsks-1 animals showed significantly increased DAF-16 transcriptional activity. mRNA levels of DAF-16 targets that are either activated (sod-3 and hsp-12.3) or inhibited (sams-1) by DAF-16 were quantified using qRT-PCR. Asterisks indicate statistical differences using two-tailed t tests: ***, p < 0.001.
The longevity phenotype of daf-2 is dependent on the downstream DAF-16 transcription factor (Lin et al., 1997; Ogg et al., 1997). To test the role of DAF-16 in the daf-2 rsks-1- mediated synergistic longevity, we constructed a daf-16; daf-2 rsks-1 triple mutant using the daf-16(mgDf47) null allele. The daf-16 deletion fully suppressed the prolonged longevity phenotype by daf-2 rsks-1 (Figure 1A and Table S1). We then examined DAF- 16 transcriptional activity by performing quantitative RT-PCR (qRT-PCR) to measure mRNA levels of genes that are regulated by DAF-16. sod-3 and hsp-12.3 are activated by DAF-16, while sams-1 is repressed by DAF-16 at the transcription level. Compared with daf-2, the daf-2 rsks-1 double mutant showed a synergistic enhancement of DAF-16 transcription activity indicated by significantly increased expression of sod-3 and hsp- 12.3, and significantly decreased expression of sams-1 (Figure 1C). Therefore, deletion of rsks-1 in daf-2 leads to synergistically extended lifespan by significantly increasing DAF-16 activity.
Previous studies have identified transcription factors that are essential for the prolonged longevity by daf-2 or rsks-1 single mutant. hsf-1 encodes the C. elegans heat-shock transcription factor ortholog and is required for daf-2-medaited lifespan extension (Hsu et al., 2003). Inhibition of hsf-1 by RNAi decreased lifespan in all four genetic backgrounds tested (Figure S2). hsf-1 inhibition suppressed the lifespan of daf-2 rsks-1 at a higher level (73%) than it did in other backgrounds (40% in N2, 57% in rsks-1, and 65% in daf-2), and the lifespan extension by daf-2 rsks-1 was almost completely suppressed by the hsf-1 RNAi treatment (Figure S2). skn-1 encodes the Nrf transcription factor ortholog that regulates oxidative stress response. Mutations in skn-1 suppress lifespan extension by certain daf-2 alleles (Tullet et al., 2008). However, inhibition of skn-1 did not suppress the synergistic lifespan extension by daf-2 rsks-1 (Figure S2D). pha-4 encodes a FOXA transcription factor that is required for rsks-1 and DR-mediated lifespan extension (Panowski et al., 2007; Sheaffer et al., 2008). Inhibition of pha-4 shortened N2 and rsks-1 lifespan, but had no effect on daf-2 lifespan (Figure S2A-C). Surprisingly, pha-4 knock-down did not affect the lifespan of daf-2 rsks-1 (Figure S2D), supporting the notion that the daf-2 rsks-1 double mutant extends lifespan by engaging different mechanisms than the single mutants alone.
Effects of daf-2 rsks-1 on development, reproduction, stress resistance and DR
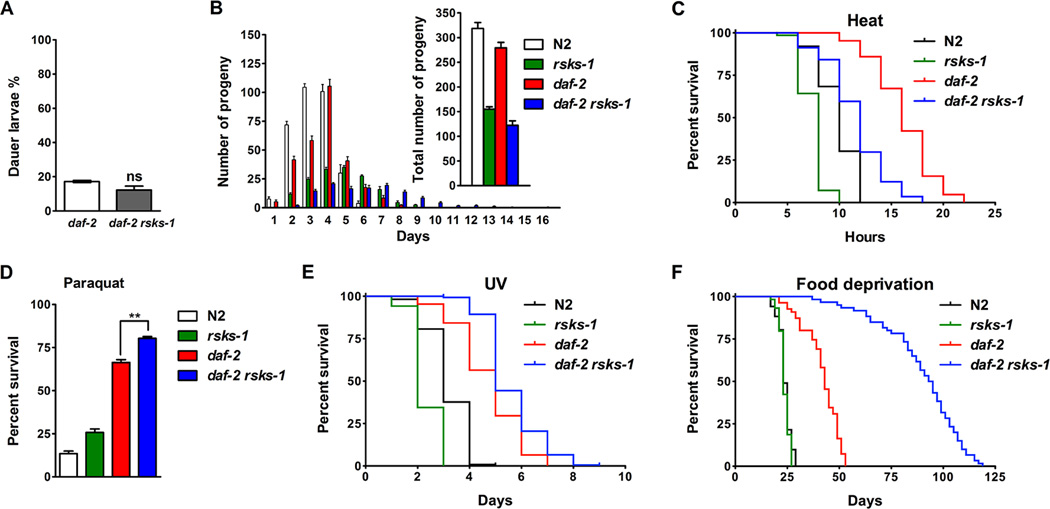
In order to further investigate the daf-2 rsks-1 mutant, we examined other phenotypes associated with extended lifespan. In addition to prolonged longevity, mutations in daf- 2 also affect development, reproduction and stress resistance. Under harsh conditions, C. elegans may arrest development entering a diapause stage called dauer for extended survival (Riddle and Albert, 1997). Inhibition of IIS leads to constitutive dauer arrest in a temperature-sensitive manner, with complete penetrance at the restrictive temperature 25°C. Some daf-2 animals transiently arrest as dauer at the intermediate temperature 22.5°C, which provides a condition to test whether rsks-1 plays a role in the IIS-dependent dauer formation. We found that rsks-1 had no significant effect on daf-2 dauer formation at 22.5°C (Figure 2A), suggesting an uncoupling of mechanisms for the synergistic longevity and dauer formation in daf-2 rsks-1.
Figure 2. Effects of daf-2 rsks-1 on development, reproduction, stress resistance and dietary restriction.
(A) rsks-1 did not affect daf-2 dauer formation at 22.5°C. ns, not significant. (B) daf-2 rsks-1 animals showed delayed, prolonged and overall reduced reproduction. (C) daf-2 rsks-1 animals were more sensitive to heat stress (35°C) than daf-2 (log-rank, p < 0.0001). (D) daf-2 rsks-1 animals were more resistant to oxidative stress by paraquat compared to daf-2 (**, p < 0.01, t - test). (E) daf-2 rsks-1 animals were more resistant to UV stress (2,000 J/m2) than daf-2 (log-rank, p < 0.0001). (F) daf-2 rsks-1 animals showed significantly increased survival under DR by bacterial food deprivation compared to daf-2 (log-rank, p < 0.0001).
Previous studies have demonstrated that both daf-2 and rsks-1 regulate germline proliferation and reproduction (Korta et al., 2012; Michaelson et al., 2010). We examined the reproduction profiles of N2, rsks-1, daf-2 and daf-2 rsks-1. The experiments were performed at 15°C because at higher temperatures, both daf-2 and daf-2 rsks-1 mutants show high incidence of embryo retention and thus internal hatching, which leads to death and inaccurate measurement of reproduction. We found that compared to daf-2, the daf-2 rsks-1 mutant showed a delayed and prolonged reproduction period and overall reduced brood size (Figure 2B). This is consistent with the recent studies showing that daf-2 and rsks-1 act in parallel to regulate germline stem cell proliferation and differentiation (Korta et al., 2012). The reproductive profile of daf-2 rsks-1 was similar to that of rsks-1, with slightly more severe phenotypes. The lack of correlation between lifespan and reproduction suggests that the trade-off between longevity and fertility is unlikely to be a major cause of the synergistic lifespan extension by daf-2 rsks-1.
The extended lifespan of daf-2 mutants has also been correlated with the activation of stress response genes and increased stress resistance (Murphy et al., 2003; Samuelson et al., 2007). We performed various stress assays to determine whether the synergistic longevity phenotype also correlates with increased stress resistance. Surprisingly, we found that daf-2 rsks-1 animals were more sensitive to heat stress (35°C), with the mean survival decreased by 34% compared to the daf-2 single mutant (Figure 2C). However, daf-2 rsks-1 animals were slightly more resistant to oxidative stress induced by paraquat (Figure 2D) and ultraviolet (UV) stress (Figure 2E). Therefore, increased resistance stress may not be the main causes of the synergistic longevity phenotype.
DR is a robust environmental manipulation that slows down aging. Since both IIS and TOR pathways are regulated by nutrients, we examined whether DR regulates the synergistic longevity by daf-2 rsks-1 using a modified bacterial food deprivation DR regimen (DR-FD) (Kaeberlein et al., 2006; Lee et al., 2006). Unlike animals growing under ad libitum (AL) conditions, the rsks-1 single mutant under DR-FD did not show lifespan extension (log-rank, p > 0.05). However, the daf-2 rsks-1 double mutant still showed robust lifespan extension by 114% compared to daf-2 (Figure 2F), suggesting the synergistic longevity phenotype is not dependent on nutrients.
Positive feedback regulation of DAF-16 via AMPK mediates the synergistic longevity by daf-2 rsks-1
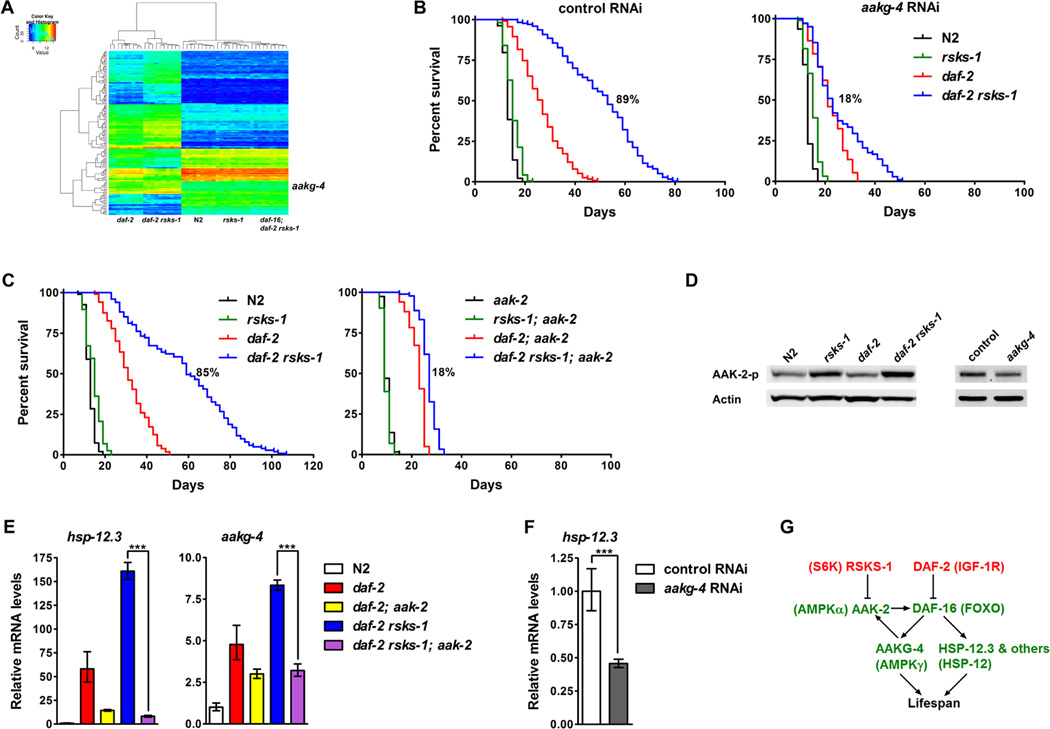
To characterize the molecular mechanisms of the synergistic longevity by daf-2 rsks-1, we compared gene expression profiles in N2, rsks-1, daf-2, daf-2 rsks-1 and daf-16; daf- 2 rsks-1 by microarrays. Genes that are differentially expressed in daf-2 rsks-1 to a greater extent than in daf-2 in a DAF-16 dependent manner were chosen for analysis (Figure 3A and Table S2). Gene Ontology (GO) analysis indicated that “aging” and “determination of adult lifespan” are among the most significant GO terms from this group of genes. We chose 42 genes based on their expression patterns and identities to perform a genetic screen by RNAi to identify genes that mediate the synergistic lifespan extension. We found 11 genetic suppressors of daf-2 rsks-1, inhibition of which decreases lifespan in daf-2 rsks-1 to a greater extent than in N2, rsks-1 and daf-2 (Table S3).
Figure 3. The synergistic longevity by daf-2 rsks-1 is mediated by positive feedback regulation of DAF-16 via AMPK.
(A) Genes that are differentially expressed in daf-2 rsks-1 were identified by microarrays analyses. (B) Identification of aakg-4 as a strong suppressor of daf-2 rsks-1. rsks-1-mediated lifespan extension in daf-2 (daf-2 vs. daf-2 rsks-1): 89% (control RNAi), 18% (aakg-4 RNAi). (C) A deletion in aak-2 suppressed the synergistic longevity by daf-2 rsks-1. rsks-1-mediated lifespan extension in daf-2 (daf-2 vs. daf-2 rsks-1): 85% (with aak-2), 18% (without aak-2). Quantitative data and statistical analyses are included in Table S1. (D) The daf-2 rsks-1 double mutant showed further increased phosphorylation of AAK-2. Inhibition of aakg-4 significantly decreased AAK-2 phosphorylation in daf-2 rsks-1. (E) The aak-2 deletion suppressed the significantly increased DAF-16 transcriptional activity in daf-2 rsks-1. ***, p < 0.001. (F) Inhibition of aakg-4 in daf-2 rsks-1 reduced DAF-16 transcriptional activity. ***, p < 0.001. (G) A model depicting the synergistic lifespan extension through positive feedback regulation of DAF-16 via AMPK.
aakg-4, which encodes the γ regulatory subunit of AMPK, showed the strongest suppression of daf-2 rsks-1 upon inhibition (Figure 3B and Table S1). In control RNAitreated animals, the daf-2 rsks-1 double mutant showed an 89% (synergistic) lifespan extension compared to the daf-2 single mutant (Figure 3B left panel). In aakg-4 RNAitreated animals, daf-2 rsks-1 only showed an 18% (additive) lifespan extension compared to daf-2 (Figure 3B right panel). qRT-PCR experiments confirmed that aakg-4 mRNA levels were further up-regulated in daf-2 rsks-1 (Figure S3A), and this regulation requires DAF-16 (Figure S3B). Since chromatin profiling studies by DNA adenine methyltransferase identification (DamID) demonstrated binding of DAF-16 to the aakg-4 promoter (Schuster et al., 2010), aakg-4 is likely to be a direct target of DAF-16 that is critical for the synergistic longevity by daf-2 rsks-1. Consistently, a deletion in aak-2, which encodes the α catalytic subunit of AMPK, also suppressed the synergistic longevity (Figure 3C and Table S1). In the presence of aak-2, the rsks-1 deletion synergistically extended the mean lifespan of daf-2 by 85% (Figure 3C left panel), whereas without aak-2, rsks-1 only additively extended the mean lifespan of daf-2 by 18% (Figure 3C right panel).
AMPK serves as an important energy homeostasis regulator by activating catabolic pathways to promote ATP generation upon energy starvation. It is a hetero-trimer that consists of a catalytic α subunit and regulatory β and γ subunits. AMP competes with ATP to bind the γ subunit, which stimulates AMPK by keeping the phosphorylation state of a highly conserved Threonine in the α subunit (Hardie, 2011). Consistent with previous studies (Selman et al., 2009), the rsks-1 single mutant showed increased AMPK activation as indicated by elevated phosphorylation of AAK-2 (Figure 3D). The daf-2 rsks- 1 double mutant showed further increase in AAK-2 phosphorylation compared with the rsks-1 single mutant (Figure 3D). This phenotype is AAKG-4-dependent, as knockingdown aakg-4 significantly reduced AAK-2 phosphorylation in daf-2 rsks-1 (Figure 3D). Previous studies demonstrated that AAK-2 directly phosphorylates and activates DAF-16 in C. elegans (Greer et al., 2007). We found that inhibition of AMPK either by the aak-2 deletion or aakg-4 RNAi blocked the significantly elevated DAF-16 transcriptional activity in daf-2 rsks-1, indicated by reduced mRNA levels of hsp-12.3 and aakg-4 (Figure 3E and F). The fact that AAK-2 promotes the expression of its own activator AAKG-4 suggests a positive feedback mechanism. In the daf-2 rsks-1 double mutant, AAK-2 is activated to increase DAF-16 activity, promoting the expression of lifespan determinant genes such as aakg-4, which in turn further activates AAK-2 and DAF-16 to form a positive feedback loop that eventually leads to significantly increased DAF-16 activity and lifespan extension (Figure 3G).
Tissue-specific regulation of the synergistic longevity by daf-2 rsks-1
In multiple cellular organisms, different tissues coordinately modulate physiology and lifespan at the whole organism level in response to genetic and environmental manipulations. The IIS pathway plays an endocrine role to modulate lifespan in a cellnon- autonomous manner. Genetic mosaics lacking daf-2 in neuronal precursor cells are long-lived (Apfeld and Kenyon, 1998), and neuronal expression of daf-2 rescues the daf- 2 mutant-mediated lifespan extension (Wolkow et al., 2000). However, the essential IIS downstream transcription factor DAF-16 functions mainly in the intestine to modulate longevity (Libina et al., 2003). Although RSKS-1 regulates many cellular processes globally, its tissue-specific role in lifespan determination has not been characterized. To better understand the endocrine functions of daf-2 and rsks-1, we examined tissue-specific involvement of rsks-1, daf-16, hsf-1 and aak-2 in the daf-2 rsks-1-mediated synergistic longevity by tissue-specific RNAi. RDE-1 is a member of the PIWI/STING/Argonaute family of proteins that is an essential component of the RNAi machinery (Tabara et al., 1999). Tissue-specific RNAi can be achieved using transgenic rde-1 mutants complemented with tissue-specific promoters driving rde-1 cDNA expression (Espelt et al., 2005; Qadota et al., 2007). Additionally, mutation of rrf-1, which encodes an RNA-directed RNA polymerase, allows RNAi to be functional exclusively in the germ line but not in somatic tissues (Sijen et al., 2001).
Using these tissue-specific RNAi tools, we identified the tissues in which rsks-1 RNAi synergistically extended lifespan of daf-2 animals. Knocking-down rsks-1 in the daf-2 single mutant extended the mean lifespan by 54%. Germline- and hypodermis-specific rsks-1 RNAi extended daf-2 lifespan by 41% and 39%, respectively. Intestine-specific rsks-1 RNAi in daf-2 caused a moderate lifespan extension of 21%. Body wall muscle-specific rsks-1 RNAi did not extend daf-2 lifespan significantly (Figure 4A, Figure S4 and Table S1). Thus, rsks-1 loss-of-function in the germ line and hypodermis are likely to play an important role in the synergistic longevity.
Figure 4. Tissue-specific regulation of the synergistically prolonged longevity by daf-2 rsks-1.
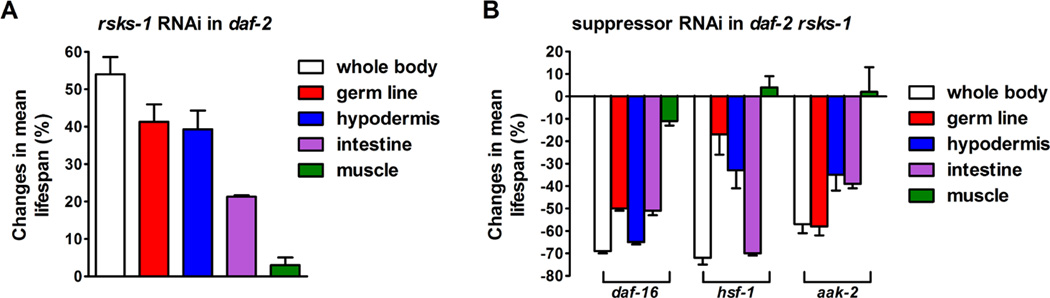
(A) Mean lifespan extension by rsks-1 RNAi knocking-down in different tissues of daf-2 animals. (B) Changes in mean lifespan relative to the control RNAi-treated animals by tissue-specific RNAi knocking-down of daf-16, hsf-1 and aak-2 in daf-2 rsks-1. Data from three independent experiments are shown. Survival curves are included in Figure S4. Quantitative data and statistical analyses are included in Table S1.
Next, we tested tissue-specific involvement of the key suppressors of daf-2 rsks-1, including daf-16, hsf-1, and aak-2. Knocking-down daf-16, hsf-1 and aak-2 by RNAi in daf-2 rsks-1 suppressed the mean lifespan by 69%, 72% and 58%, respectively. Upon inhibition of these genes in the germ line, the synergistic longevity was significantly suppressed by daf-16 RNAi (49%) and aak-2 RNAi (58%), but only mildly affected by hsf- 1 RNAi (18%). Intestine-specific knocking-down daf-16, hsf-1 and aak-2 significantly suppressed daf-2 rsks-1 lifespan by 50%, 69% and 39%, respectively. Additionally, inhibition of daf-16 in the hypodermis also significantly decreased daf-2 rsks-1 lifespan by 63%, whereas knocking-down aak-2 and hsf-1 in the hypodermis moderately reduced the lifespan by 35% and 36%, respectively. In the body wall muscle, knocking-down daf- 16, aak-2 and hsf-1 had little effect on longevity (Figure 4B, Figure S4 and Table S1). In summary, it is likely that DAF-16 functions in the germ line, intestine and hypodermis, AAK-2 functions in the germ line and intestine, while HSF-1 mainly functions in the intestine to modulate the longevity of daf-2 rsks-1. These results demonstrated that in addition to the intestine, other tissues especially the germ line also play an important role in aging.
Germline signaling modulates the synergistic longevity by daf-2 rsks-1
Previous studies have indicated that signals from the reproductive system modulate aging in multiple species. Signals from the germ line shorten lifespan, whereas signals from the somatic gonad extend lifespan (Hsin and Kenyon, 1999). Prolonged longevity via removal of germline precursor cells requires DAF-16 and DAF-12 (nuclear hormone receptor). Hence, we examined whether germline signaling modulates the synergistic longevity by daf-2 rsks-1.
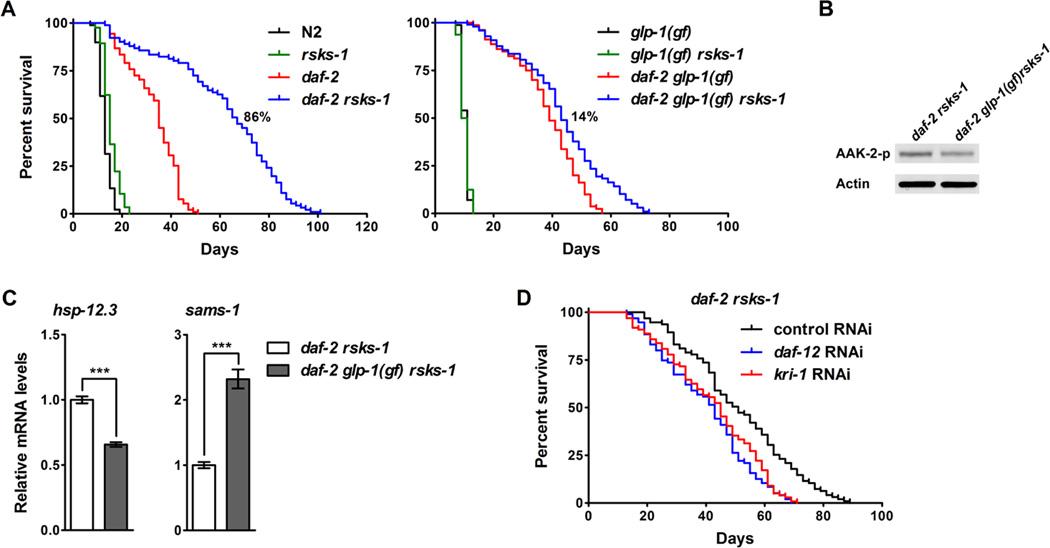
glp-1 encodes a Notch family receptor that is essential for mitotic proliferation of germline cells (Austin and Kimble, 1987; Priess et al., 1987). The long-lived glp-1 loss-offunction (lf) mutant serves as a genetic mimic of germline removal. Similar to the rsks-1 mutant, glp-1(lf) animals showed significantly increased phosphorylation of AAK-2 (Figure S5A). Consistently, inhibition of glp-1 by RNAi extended lifespan in N2, but not in the aak-2 deletion mutant (Figure S5B). glp-1(ar202) is a gain-of-function (gf) allele exhibiting germline over-proliferation and shortened adult lifespan. We found that glp-1(gf) suppressed the synergistic lifespan extension by daf-2 rsks-1. Without glp-1(gf), the rsks-1 deletion synergistically extended the mean lifespan of daf-2 by 86% (Figure 5A left panel), whereas with glp-1(gf), rsks-1 only additively extended the mean lifespan of daf-2 by 14% (Figure 5A right panel). Consistently, glp-1(gf) also significantly decreased AAK-2 phosphorylation (Figure 5B) and DAF-16 transcriptional activity (Figure 5C) in daf-2 rsks-1. DAF-12, a nuclear hormone receptor, and KRI-1, an ankyrin repeats containing protein, were identified as important regulators of germline loss-mediated longevity (Berman and Kenyon, 2006; Hsin and Kenyon, 1999). Inhibition of daf-12 or kri-1 by RNAi decreased the lifespan of daf-2 rsks-1 by 24% and 19%, respectively (Figure 5D and Table S1). Together, these results support the idea that germline signals play an important role in daf-2 rsks-1-mediated activation of AMPK and DAF-16 and synergistic lifespan extension.
Figure 5. Germline signaling modulates the daf-2 rsks-1-mediated synergistic lifespan extension through AMPK and DAF-16.
(A) The glp-1(gf) mutation suppressed the synergistic longevity by daf-2 rsks-1. rsks-1-mediated lifespan extension in daf-2 (daf-2 vs. daf-2 rsks-1): 86% [without glp-1(gf)], 14% [with glp-1(gf)]. (B) The glp-1(gf) mutation decreased phosphorylation of AAK-2 in daf-2 rsks-1. (C) The glp-1(gf) mutation suppressed the significantly increased DAF-16 transcriptional activity in daf-2 rsks-1. ***, p < 0.001. (D) Inhibition of DAF-12 or KRI-1, essential mediators of the germline signaling, significantly suppressed the synergistic longevity by daf-2 rsks-1 (log-rank, p < 0.0001). Quantitative data and statistical analyses are included in Table S1.
Cell-non-autonomous activation of DAF-16 by knocking-down of rsks-1 in the germ line
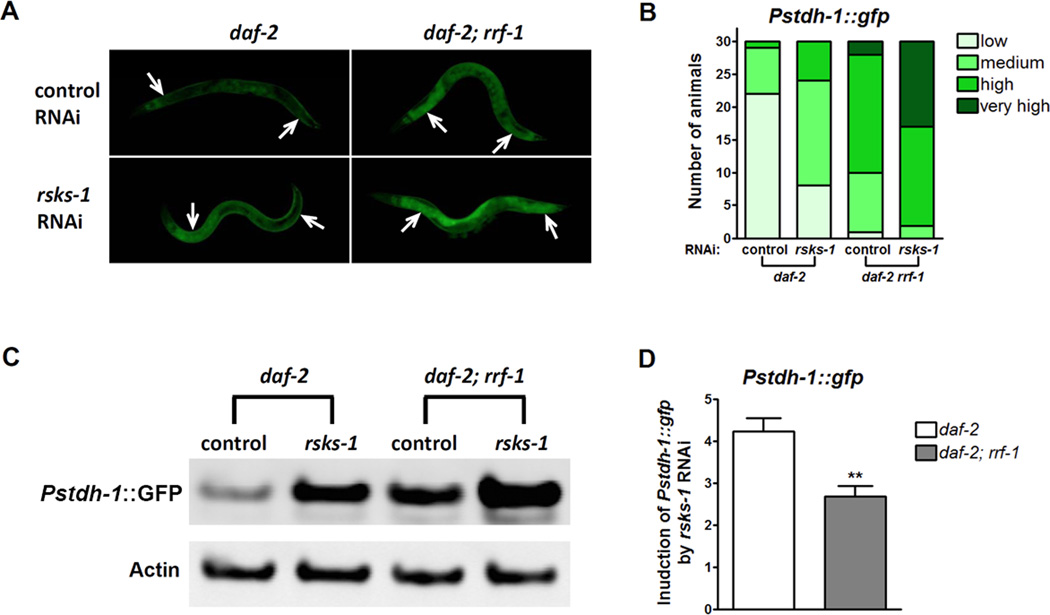
Since germline signaling has endocrine properties to regulate DAF-16 cell-non-autonomously in the intestine (Berman and Kenyon, 2006), we decided to examine whether inhibition of rsks-1 in the germ line affects downstream genes cell-autonomously or -non-autonomously. To answer this question, we crossed an integrated gfp reporter driven by the stdh-1 promoter (Pstdh-1::gfp) into daf-2 and daf- 2; rrf-1 to examine stdh-1 expression patterns upon rsks-1 RNAi treatment. stdh-1 is transcriptionally activated by DAF-16. It is widely expressed in neurons, muscles and intestine, allowing us to monitor DAF-16 activities in various tissues. In the daf-2 background, rsks-1 RNAi activated stdh-1 expression in the intestine. In daf-2; rrf-1, which allows RNAi to be functional only in the germ line, rsks-1 RNAi led to a significant induction of stdh-1 expression in the intestine (Figure 6A, B). To better quantify the induction of Pstdh-1::gfp by rsks-1 RNAi, we performed Western blotting to measure GFP levels from whole worm lysates since majority of the Pstdh-1::gfp expression was from the intestine. Consistent with the imaging results, both regular and germline-specific rsks-1 RNAi increased Pstdh-1::gfp expression (Figure 6C). We then quantified the intensities of GFP bands relative to those of Actin, which serves as the internal control for equal loading. The fold induction of Pstdh-1::gfp by rsks-1 RNAi compared to the control RNAi was calculated. Although the germline-specific RNAi mutant rrf-1 increased the basal levels of stdh-1 expression, the induction of stdh-1 expression by rsks-1 RNAi is higher in daf-2 than in daf-2; rrf-1 (Figure 6D). Thus, inhibition of rsks-1 in the germ line non-autonomously activates DAF-16 in the intestine.
Figure 6. Cell-non-autonomous activation of DAF-16 by rsks-1 RNAi knocking-down in the germ line.
(A) Activation of the DAF-16 target stdh-1 by rsks-1 RNAi in daf-2 and daf- 2; rrf-1. rsks-1 RNAi knocking-down in the germ line of daf-2 animals (daf-2; rrf-1) significantly increased the intestinal expression of GFP driven by the stdh-1 promoter. Arrows indicate the anterior and posterior intestine. (B) Quantification of GFP expression driven by the stdh-1 promoter. Thirty animals were examined for each treatment. (C) Measurement of Pstdh-1::gfp expression by Western blots using anti-GFP antibodies. (D) Quantification of GFP expression from Western blots. Relative GFP levels were calculated by normalizing the intensities of GFP bands to Actin. Fold increase in Pstdh-1::gfp expression induced by rsks-1 RNAi was calculated through dividing the relative GFP levels in rsks-1 RNAi-treated animals by those in control RNAi-treated animals. **, p < 0.01. The quantification was performed with four biological replicates.
Discussion
IIS and TOR pathways play conserved roles in modulating lifespan in multiple species. However, it is unclear how they might interactively modulate aging. We set out to address this question by constructing a daf-2 rsks-1 double mutant, which has reduced function of IIS and an important branch of the TOR pathway. Surprisingly, the daf-2 rsks- 1 double mutant shows a nearly 5-fold lifespan extension (Figure 1A and Table S1). We defined this phenotype as synergistic lifespan extension based on the observation that longevity of the daf-2 rsks-1 double mutant is beyond the combined effects of rsks-1 and daf-2 single mutants. This synergistic longevity phenotype cannot be explained by the hypothesis that daf-2 and rsks-1 function in parallel to modulate lifespan independently since an additive effect will be expected under such an assumption.
The synergistic longevity phenotype is different from what we previously reported that rsks-1 RNAi further extended daf-2 lifespan by 24% (Pan et al., 2007). One major change in the experimental procedures was that in the previous study, daf-2 animals were treated with rsks-1 RNAi only during adulthood in contrast to this work, in which the double mutant carries the putative null allele of rsks-1 throughout life. When we treated daf-2 animals with rsks-1 RNAi for two generations resulting in a more complete reduction in rsks-1 mRNA levels, we observed a 54% further lifespan extension (Figure S4A and Table S1). These results suggest that inhibition of rsks-1 during development is critical for the synergistic longevity phenotype. Consistently, inhibition of the RSKS-1 upstream activator LET-363/CeTOR in daf-2 during adulthood led to 17% additive lifespan extension (Figure S6). Since let-363 is an essential gene, inhibition of which during development leads to larval arrest, we used a pharmaceutical approach to inhibit let-363 by treating animals with rapamycin as previously reported (Robida-Stubbs et al., 2012). Rapamycin treatment throughout life extended lifespan of N2 and daf-2 animals by 26% and 45%, respectively (Figure 1B). There are multiple possible reasons why rapamycin treatment could not extend lifespan of daf-2 animals as much as the rsks-1 deletion mutant does. One possibility is rapamycin treatment did not fully block RSKS-1, which is required for the synergistic longevity. Another possibility is rapamycin treatment at this dosage has been shown to inhibit both TOR complex 1 and complex 2 activities (Robida-Stubbs et al., 2012), and there might be other lifespan determinant genes affected by the drug. Nevertheless, these results are consistent with the idea that inhibiting rsks-1 in daf-2 during development leads to synergistic lifespan extension.
Previous studies showed that null mutants of age-1, which encodes a catalytic subunit of the phosphatidylinositol-3-kinase (PI3K) in the IIS pathway, exhibit exceptional lifespan extension in a DAF-16-dependent manner (Ayyadevara et al., 2008). Since the daf-2 mutations we used in this study are not null alleles, one possible explanation for the synergistic longevity by daf-2 rsks-1 is that the rsks-1 deletion makes daf-2 mutant phenotypes more severe. We think this is unlikely to be true because there are many aging-related phenotypes of daf-2 not enhanced by the rsks-1 deletion. As shown in Figure 2, rsks-1 does not affect daf-2-mediated dauer arrest, and rsks-1 has a minor or even opposite effect on most stress resistance. Understanding why these phenotypes are uncoupled from the synergistically prolonged longevity by daf-2 rsks-1 will help to understand the basic mechanisms of aging.
TOR plays a conserved role in DR-mediated lifespan extension (Kapahi et al., 2010). We tested the effect of nutrients on the synergistic longevity using the DR-FD regimen (Figure 2F). The rsks-1 single mutant did not show lifespan extension under DR, which is consistent with the idea that DR and reduced TOR signaling function through overlapping mechanisms to extend lifespan. Interestingly, the synergistic longevity by daf-2 rsks-1 is nutrient independent, suggesting rsks-1 functions through novel mechanisms to further extend lifespan of daf-2 animals.
To better understand the molecular mechanisms of the synergistic longevity by daf-2 rsks-1, we set out to identify critical mediators by testing known regulators of IIS or rsks- 1. The heat-shock factor HSF-1 is critical for daf-2-mediated lifespan extension. Inhibition of hsf-1 almost completely abolished the lifespan extension by daf-2 rsks-1 (Figure S2). Lifespan extension via genetic or pharmaceutical inhibition of TOR requires the IIS downstream transcription factor SKN-1 (Robida-Stubbs et al., 2012). Surprisingly, inhibition of skn-1 by RNAi had little effect on the synergistic longevity by daf-2 rsks-1 (Figure S2). Similarly, inhibition of PHA-4, a FOXA transcription factor that is required for the rsks-1 single mutant-mediated lifespan extension, did not affect lifespan of daf-2 rsks-1 (Figure S2). This is further evidence that the mechanism of the synergistic longevity in the daf-2 rsks-1 double mutant is distinct from the lifespan extension by the single mutants.
We then performed microarray studies and identified genes that are differentially expressed in daf-2 rsks-1 (Figure 3A and Table S2). A genetic screen using RNAi helped to identify the AMPK complex as the key mediator of the synergistic longevity by daf-2 rsks-1 (Figure 3B, C, Table S1 and Table S3). Quantitative analysis of the lifespan data indicated that suppression of daf-2 rsks-1 lifespan by inhibiting AMPK was not due to general sickness. Instead, inhibition of AMPK suppressed the synergy part of the lifespan extension. Further analysis identified positive feedback regulation of DAF-16 via AMPK in the daf-2 rsks-1 mutant (Figure 3D-G). AMPK plays important roles in various cellular functions (Hardie, 2011). Under energy-starved conditions, AMPK is activated to promote catabolism and thus ATP production. Further characterization of the role of AMPK in metabolism will aid in the understanding of the synergistic longevity by daf-2 rsks-1.
Both IIS and signals from the reproductive system have endocrine functions. Modulation of these pathways in one tissue leads to non-autonomous activation of DAF-16 in the intestine (Berman and Kenyon, 2006; Libina et al., 2003). To better understand how aging is coordinately modulated across multiple tissues, we tested the involvement of key regulators of the daf-2 rsks-1-mediated synergistic longevity by tissue-specific RNAi. We found that rsks-1, daf-16 and aak-2 function in the germ line to regulate the synergistic lifespan extension (Figure 4), which can also be suppressed by a genetic mutation that causes germ line over-proliferation and by inhibiting key mediators of the germline signaling (Figure 5). In addition, inhibiting rsks-1 in the germ line leads to non-autonomous activation of DAF-16 in the intestine (Figure 6). Previous studies on the tissue-specific requirements of key longevity determinants, including DAF-16, mainly employed transgenic rescue approaches. However, the traditional microinjection method creates transgenic lines with high copy number of transgenes, which will be silenced in the germ line. Our results indicate the germ line as an important tissue to integrate signals from the IIS pathway and S6K for lifespan determination.
Similar to the rsks-1 single mutant, daf-2 rsks-1 animals showed significantly delayed, prolonged and overall reduced reproduction (Figure 2B). This is consistent with a recent study showing that RSKS-1 acts in parallel with the IIS pathway to play an essential role in the establishment of the germline stem cell/progenitor pool (Korta et al., 2012). Interestingly, RSKS-1 functions cell-autonomously to regulate the germline progenitor establishment. This effect is independent of its known suppressors in the regulation of lifespan (Korta et al., 2012). These findings suggest that the synergistic longevity of daf-2 rsks-1 cannot simply be linked with its functions in germline development and reproduction.
In C. elegans, the intestine carries out multiple nutrient-related functions, and it is the site for food digestion and absorption, fat storage, and immune response. DAF-16 is one of the essential transcription factors that function in the intestine to modulate lifespan. We found that intestinal-specific inhibition of daf-16, aak-2 or hsf-1 largely abolishes the synergistic lifespan extension of daf-2 rsks-1 (Figure 4B). However, knocking-down of rsks-1 in the intestine only has an additive effect on daf-2 lifespan (Figure 4A), suggesting that rsks-1 may function through non-autonomous mechanisms to activate DAF-16.
The hypodermis is considered as part of the epithelial system in C. elegans. It is involved in basic body plan establishment, cell fate specification, axon migration, apoptotic cells removal, and fat storage. We found that hypodermis-specific knocking-down of rsks-1 in daf-2 also leads to synergistic lifespan extension, and that hypodermis-specific knocking-down of daf-16 significantly reduces the synergistic lifespan extension (Figure 4). Our results provide evidence for the important role of the hypodermis in lifespan determination. In future studies, it will be interesting to examine which biological functions of the hypodermis are involved in regulating the synergistic longevity by daf-2 rsks-1.
Previous studies showed that muscle decline is one of the major physiological causes of aging in C. elegans (Herndon et al., 2002). Neither rsks-1 nor the downstream regulators daf-16, hsf-1 and aak-2 seem to function in the muscle to modulate the synergistic lifespan extension (Figure 4). However, we cannot rule out the possibility that these regulators may function in other tissues to non-autonomously regulate muscle functions in daf-2 rsks-1. Characterization of age-dependent muscle decline in daf-2 rsks-1 will help to understand whether muscle functions are important for the synergistic lifespan extension.
There are limitations for assessing tissue-specific involvement of key regulators in lifespan determination by RNAi such as uncertainty of knock-down efficiency and potential leakiness. It has been reported that in rrf-1 mutants, RNAi can be processed in certain somatic tissues including the intestine at least for the genes tested (Kumsta and Hansen, 2012). However, the critical function of rsks-1 in the germ line is unlikely to be an artifact as rsks-1 knock-down in the intestine of daf-2 animals did not lead to synergistic lifespan extension. Moreover, inhibition of certain strong suppressors of daf-2 rsks-1 such as hsf-1 in the intestine but not in the germ line significantly decreased the synergistic lifespan extension by daf-2 rsks-1. Further analyses by single-copied, isoform-specific transgenic rescue will help to quantitatively determine the tissue-specific involvement of key regulators in the synergistic lifespan extension by daf-2 rsks-1.
It has not been clear whether DAF-16 is quantitatively more active or it is uniquely activated in certain tissues such as the germ line of daf-2 rsks-1. Though we identified the AMPK-mediated positive feedback regulation of DAF-16 based on genes that are expressed to a greater extent in daf-2 rsks-1 animals, we speculate that the double mutant has some unique properties as shown in dauer formation and various stress tolerance assays. Our data with the phenotypic analysis of the double mutant and epistasis analysis of tissue requirement of DAF-16 suggests that with the rsks-1 deletion, DAF-16 plays a more important role in certain tissues like the germ line to further extend lifespan of daf-2. Characterization of the genes that are uniquely up-regulated in daf-2 rsks-1 or those that are regulated independently of DAF-16 will help distinguish these models.
In conclusion, we found that the daf-2 rsks-1 double mutant shows synergistic lifespan extension, which is achieved through positive feedback regulation of DAF-16 by AMPK. Tissue-specific epistasis analysis suggests that this enhanced activation of DAF-16 is initiated by signals from the germ line and that the germ line tissue may play a key role in integrating the interactions between daf-2 and rsks-1 to cause synergistic lifespan extension. Since DAF-2, RSKS-1, AMPK and DAF-16 are highly conserved molecules, similar regulation may also exist in mammals. Further characterization of the daf-2 rsks- 1-mediated synergistic longevity will contribute to a better understanding of the molecular mechanisms of aging and age-related diseases.
Experimental Procedures
Lifespan assays
Animals were maintained at 15°C or 20°C until late L4 stages and then transferred to 25°C. The first day of adulthood is Day 1 in survival curves. Animals were scored as alive, dead or lost every 2–3 days.
qRT-PCR assays
The SYBR Green dye (Quanta) was used for qRT-PCR reactions performed on an LC480 machine (Roche). Relative-fold changes were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). qRT-PCR experiments were performed three times with consistent results using three independent RNA preparations.
Microarray analysis
Microarray hybridization was performed at the Buck Institute Genomics Core using the NimbleGen 12-Plex Gene Expression Arrays and arrays were quantified using the NimbleScan2 software.
Western blotting
Western blotting for phosphorylated AAK-2 (Cell Signaling, 1:300), Actin (Cell Signaling, 1:1,000) and GFP (UC Davis/NIH NeuroMab Facility, 1:1,000) were performed using the LICOR system. Band intensities were quantified using the Odyssey V3.0 software.
Supplementary Material
daf-2 rsks-1 double mutant shows synergistic lifespan extension in C. elegans
AMPK mediates positive feedback regulation of DAF-16 in daf-2 rsks-1
Germ line is a key tissue in modulating the synergistic longevity of daf-2 rsks-1
Inhibiting rsks-1 in the germ line leads to cell-non-autonomous activation of DAF-16
Acknowledgments
We thank K. Felkey for microarrays experiments, Dr. J.M. Tullet for advice on Western blotting, J. Graham and N. Naude for technical assistance, Drs. D. Bailie, K. Blackwell, M. Hansen, E.J.A. Hubbard, K. Kaibuchi, C. Kenyon, D.Z. Korta, K. Strange, H. Tissenbaum and S. Tuck for strains and unpublished results, Drs. L. Barrett, G. Du, M. McGee, and members of the Kapahi lab, Lithgow lab and Gill lab for discussions and comments on the manuscript. Some nematode strains were provided by the C. elegans Genetics Center, funded by NIH National Center for Research Resources and by the Mitani lab, Tokyo Women's Medical University, School of Medicine, Japan. This work was supported by grants from the American Foundation for Aging Research, Hillblom Foundation, a Nathan Shock Startup award, Genomics Core support from the Nathan Shock Center (NIH P30AG025708 ), and the NIH (R01AG038688, RL1AAG032113 & 3RL1AG032113- 03S1) to P.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
The GEO accession number for the microarray data reported in this paper is XXXXXX.
References
- Apfeld J, Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- Apfeld J, O'Connor G, McDonagh T, DiStefano PS, Curtis R. The AMPactivated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J, Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- Ayyadevara S, Alla R, Thaden JJ, Shmookler Reis RJ. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell. 2008;7:13–22. doi: 10.1111/j.1474-9726.2007.00348.x. [DOI] [PubMed] [Google Scholar]
- Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Espelt MV, Estevez AY, Yin X, Strange K. Oscillatory Ca2+ signaling in the isolated Caenorhabditis elegans intestine: role of the inositol-1,4,5-trisphosphate receptor and phospholipases C beta and gamma. J Gen Physiol. 2005;126:379–392. doi: 10.1085/jgp.200509355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T, Min KJ, D'Alterio C, Villa-Cuesta E, Cumbers J, Lehmann R, Jones DL, Tatar M. Drosophila germ-line modulation of insulin signaling and lifespan. Proc Natl Acad Sci U S A. 2008;105:6368–6373. doi: 10.1073/pnas.0709128105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kaeberlein TL, Smith ED, Tsuchiya M, Welton KL, Thomas JH, Fields S, Kennedy BK, Kaeberlein M. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006;5:487–494. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, Kockel L. With TOR less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Korta DZ, Tuck S, Hubbard EJ. S6K links cell fate, cell cycle and nutrient response in C. elegans germline stem/progenitor cells. Development. 2012;139:859–870. doi: 10.1242/dev.074047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C, Hansen M. C. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS One. 2012;7:e35428. doi: 10.1371/journal.pone.0035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, Ingram DK, Zou S. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–524. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McCormick M, Chen K, Ramaswamy P, Kenyon C. New genes that extend Caenorhabditis elegans' lifespan in response to reproductive signals. Aging Cell. 2012;11:192–202. doi: 10.1111/j.1474-9726.2011.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- Michaelson D, Korta DZ, Capua Y, Hubbard EJ. Insulin signaling promotes germline proliferation in C. elegans. Development. 2010;137:671–680. doi: 10.1242/dev.042523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- O'Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Priess JR, Schnabel H, Schnabel R. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Qadota H, Inoue M, Hikita T, Koppen M, Hardin JD, Amano M, Moerman DG, Kaibuchi K. Establishment of a tissue-specific RNAi system in C. elegans. Gene. 2007;400:166–173. doi: 10.1016/j.gene.2007.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle DL, Albert PS. C elegans II. Plainview NY: Cold Spring Harbor Laboratory Press; 1997. Genetic and Environmental Regulation of Dauer Larva Development; pp. 739–768. [PubMed] [Google Scholar]
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell TK. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012;15:713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson AV, Carr CE, Ruvkun G. Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev. 2007;21:2976–2994. doi: 10.1101/gad.1588907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster E, McElwee JJ, Tullet JM, Doonan R, Matthijssens F, Reece-Hoyes JS, Hope IA, Vanfleteren JR, Thornton JM, Gems D. DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol. 2010;6:399. doi: 10.1038/msb.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer KL, Updike DL, Mango SE. The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol. 2008;18:1355–1364. doi: 10.1016/j.cub.2008.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues AP, Manning G, Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- Wang MC, O'Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Zhang P, Judy M, Lee SJ, Kenyon C. Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab. 2013;17:85–100. doi: 10.1016/j.cmet.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.