Abstract
Among insect taxa, ants exhibit one of the most variable chromosome numbers ranging from n = 1 to n = 60. This high karyotype diversity is suggested to be correlated to ants diversification. The karyotype evolution of ants is usually understood in terms of Robertsonian rearrangements towards an increase in chromosome numbers. The ant genus Mycetophylax is a small monogynous basal Attini ant (Formicidae: Myrmicinae), endemic to sand dunes along the Brazilian coastlines. A recent taxonomic revision validates three species, Mycetophylax morschi, M. conformis and M. simplex. In this paper, we cytogenetically characterized all species that belongs to the genus and analyzed the karyotypic evolution of Mycetophylax in the context of a molecular phylogeny and ancestral character state reconstruction. M. morschi showed a polymorphic number of chromosomes, with colonies showing 2n = 26 and 2n = 30 chromosomes. M. conformis presented a diploid chromosome number of 30 chromosomes, while M. simplex showed 36 chromosomes. The probabilistic models suggest that the ancestral haploid chromosome number of Mycetophylax was 17 (Likelihood framework) or 18 (Bayesian framework). The analysis also suggested that fusions were responsible for the evolutionary reduction in chromosome numbers of M. conformis and M. morschi karyotypes whereas fission may determines the M. simplex karyotype. These results obtained show the importance of fusions in chromosome changes towards a chromosome number reduction in Formicidae and how a phylogenetic background can be used to reconstruct hypotheses about chromosomes evolution.
Introduction
Chromosomes are the units of inheritance contained in the nuclei of eukaryotic cells. They can have different sizes, shapes and DNA compositions and there is ample evidence that chromosome changes may promote speciation [1]. A large number of ant species have been studied cytogenetically and they exhibit an enormous diversity of chromosome number, varying from n = 1 to n = 60 (reviewed in [2]). This marked variation aroused attention over 35 years ago [3], such that several evolutionary mechanisms and the manner in which this diversity has evolved have been proposed. Among the evolutionary mechanism proposed, rearrangements involving Robertsonian fissions stands out (see [4]–[5]) in the so–called “minimum interaction theory”. According to this theory, the chromosome evolution in ants generally tends towards an increase in chromosome number in order to reduce the risk of deleterious rearrangements [5]. Nevertheless, it has been thought that the typical trend in chromosome increase probably respects certain limits [2].
The growing number of cytogenetic studies in ants have highlighted that chromosome evolution has accompanied genus and species diversification [2]. Thus, karyotype descriptions and their comparative analyses is an important independent tool for taxonomy and understanding chromosome evolution, particularly when relying on phylogenetic tree construction [6]. The ready availability of DNA sequences and advances in molecular phylogenetic analysis has allowed researchers to infer the relationships of ants that can be used in an integrative cytogenetic approach. In addition, sequences of protein–coding nuclear genes have been shown to be useful for resolving phylogenetic relationships within genera and between related species in ants [7].
The Attini tribe belongs to the Myrmicinae subfamily and comprises ants that are known to engage in a symbiosis with a Basidiomycota fungus, which serves as their main food source. They are restricted to the New World and are primarily distributed in the Neotropics, where they achieve their greatest diversity. Currently, the tribe comprises more than 230 described species grouped into 14 genera [8]–[9]. Although some systematic studies on Attini tribe have been conducted, information is scarce regarding the majority of the groups and the taxonomy of many species still requires revision. Indeed, recent revisionary studies have permitted the identification of sibling species [10], description of new species [11], and even the description of a new genus [9].
The genus Mycetophylax is a small monogynous basal Attini that has recently gained more attention [9], [13]–[15]. About 20 species, subspecies and varieties were coded to the genus Mycetophylax [9], although Kempf [16] lists 15 species living in Brazil. Following the taxonomic revision based on morphological systematics, the majority of those species were synonymized and some others were included into Mycetophylax, originally belonging to the Attini genus Cyphomyrmex. Currently the genus Mycetophylax is composed of three valid species, M. conformis (Mayr, 1884), M. morschi (Emery, 1888) and M. simplex (Emery, 1888). However, some issues concerning the occurrence of sibling species within Mycetophylax and the status of M. morschi belonging to the genus still remain under discussion (Mayhé–Nunes pers. Com.). Analysis of wingless and long–wave rhodopsin led to the recent molecular phylogenetic hypothesis of the Mycetophylax genus [17], which was in agreement with the morphological features. Recently, the nuclear content of the three species were estimated by flow cytometry, data that provided noteworthy information at a higher level than the species level [14], but information concerning the Mycetophylax karyotype is no longer available.
Thus, the aim of this study was to provide the first characterization of Mycetophylax species karyotype, including chromosome number, morphology, heterochromatin location and chromatin AT/GC richness. We discuss the evolutionary dynamics of the karyotypes within the genus in the light of the recently published phylogeny. Additionally, lineage–specific rearrangements leading to different chromosome numbers in Mycetophylax were tested using ancestral state reconstruction. For this we used two different, recently developed approaches by Mayrose et al. [18] based on Maximum Likelihood and Bayesian methods in order to propose insights concerning chromosome evolution in ants.
Materials and Methods
Biological material and chromosome preparation
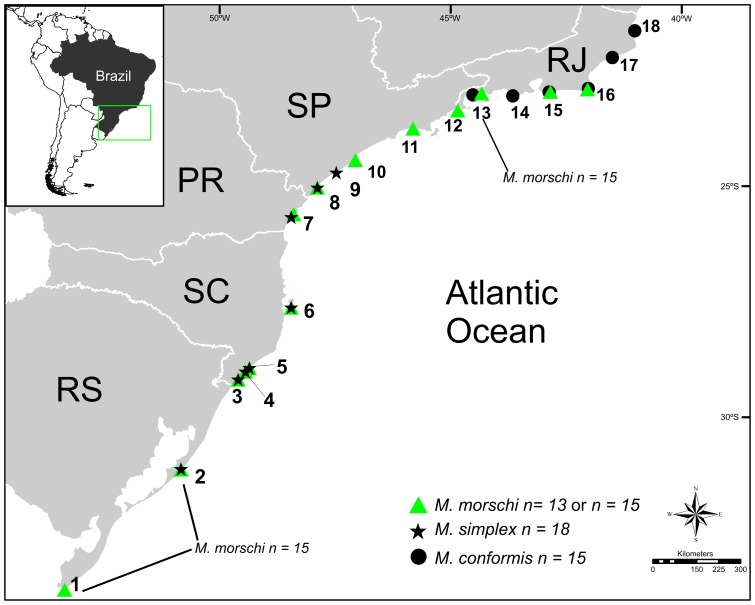
Colonies of the three species were collected from sand dunes throughout their occurrence area along the Brazilian Atlantic coast, from Rio Grande do Sul State to Rio de Janeiro State between December 2009 and March 2011. The colonies of M. simplex (19 colonies) were collected on beaches in the States of Rio Grande do Sul, Santa Catarina, Paraná and Rio de Janeiro. M. conformis (21 colonies) were collected on beaches in the States of Rio de Janeiro and São Paulo, while M. morschi (38 colonies) were collected in all the states mentioned (Figure 1; Table S1 for sampling details). Following collection, the colonies were transported to the laboratory and reared according to the protocol described by Cardoso et al. [13] until brooding occurred. When available, at least ten individuals from each colony were used in cytogenetic analyses. All the ants collected were preserved in ethanol and confirmation of species identification was performed by Rodrigo Feitosa, at the Museum of Zoology of the University of São Paulo (Museu de Zoologia da Universidade de São Paulo, MUZSP), where vouchers were also deposited. All species' collections were authorized by the Brazilian Institute for Biodiversity Conservation (ICMBio) by means of a special permit (number 24869–2) recorded by SISBio. Collecting permit was issued to Danon Clemes Cardoso in Brazil.
Figure 1. Cytogenetically characterized populations of Mycetophylax species across the distribution of the genus along Atlantic coast.
Sampling sites and karyotype results. Populations of M. morschi with haploid number of 15 chromosomes are indicated. Numbers represent sampling localities as given in the Table S1.
Metaphase spreads were prepared from the cerebral ganglia of post–defecant larvae, according to protocol proposed by Imai et al. [19]. The cerebral ganglion was dissected in colchicine–hypotonic solution (0.005%) under a stereoscopic microscope, transposed to a new drop of same solution and incubated under light protection for one hour until slide preparation (see reference [19] for detailed procedure). The slide with metaphases were examined under a phase contrast microscope and stained with 4% Giemsa solution in Sorensen's buffer, pH 6.8, to determine chromosome number and morphology. We classified the chromosomes following a modified nomenclature based on the proposed by Levan et al. [20], which is based on four types of centromeric position: acrocentric (A), subtelocentric (ST), submetacentric (SM) and metacentric (M).
C–banding and Fluorochrome staining
In order to determine the distribution pattern of heterochromatin, the BSG (barium hydroxide/saline/Giemsa) banding technique was performed, essentially following the method described by Sumner [21], with modifications in the duration of treatment with Ba(OH)2, as proposed by Pompolo and Takahashi [22]. Sequential fluorochrome staining with chromomycin A3/distamycin A/4′–6′–diamindino–2–phenylindole (CMA3/DA/DAPI) was conducted according to Schweizer [23] in order to characterize CG and AT richness region on chromosomes. The slides were analyzed under an epifluorescence microscope (Olympus BX 60) equipped with a digital camera system (Q color 3 Olympus®). The fluorescent signals were analyzed with different filters: WB filter (450 to 480 nm) for the fluorochrome CMA3 and WU filter (330 to 385 nm) for the fluorochrome DAPI. At least nine and six slides of each species with well-spread metaphases were submitted to C-banding technique and fluorochrome staining, respectively.
Chromosome evolution analysis
In order to infer and support the patterns and processes underpinning chromosomal evolution in Mycetophylax an integrative cytogenetic and molecular phylogeny study was conducted. To determine the direction of chromosomal changes (i.e. fusion versus fission) that occurred in the genus Mycetophylax, Attini species with known karyotypes were used as an out–group. Thus, two different methods were performed with the purpose of outlining a chromosome evolution hypothesis for this genus. The software ChromEvol 1.3 [18] was used to infer the chromosome evolution model and haploid ancestral states (chromosome numbers) by Maximum Likelihood and Bayesian methods, relying on a previously phylogenetic hypotheses published by our group [17].
The molecular phylogenetic tree for Mycetophylax, on which the haploid ancestral states were inferred in this work, was based on the wingless and long–wave rhodopsin matrix of Cardoso et al. [17]. Sequences of wingless and long–wave rhodopsin genes were downloaded from GenBank (Table S2) and aligned using Mega 5.0 [24]. Therefore, we reconstructed the Bayesian tree from that study using selected taxa that were cytogenetically characterized or that have their karyotype known, with the same setting parameters and substitution model HKY+G for long–wave rhodopsin and GTR+G for wingless to run MrBayes 3.2.2 [25]. ChromEvol 1.3 was carried out and relied on this reconstructed tree. The program allows the evaluation of eight models of chromosome evolution taking into account: gains and losses of single chromosomes; duplications, whole–genome duplication; and demi–duplication, a mechanism that facilitates the transition from an n chromosome to 1.5 n. The last feature was not evaluated in our analysis, since it is only widespread and common in plants. Therefore, four models of chromosome evolution that did not consider demi–duplication were carried out. All the parameters were adjusted to the data following the recommendation of Mayrose et al. [18]. The models and their null hypotheses were analyzed with 10,000 simulations and the one that best fit the data set was selected under the Akaike information criterion (AIC).
Subsequently, to define a more complete chromosome evolution hypothesis, the node ages of Mycetophylax were estimated. This analysis was performed to determine when the possible splits between lineages occurred, to assess the information of chromosome changes in a geological and evolutionary context. Thus, molecular dating of Mycetophylax lineages were estimated using previously reported nuclear clock calibrations for Attini ants [8]. In a matrix for the genes wingless and long–wave rhodopsin downloaded from the NCBI GenBank (see Table S2), we included sequences of cytogenetically characterized Mycetophylax species. The nuclear genes matrix was analyzed under a Bayesian framework and uncorrelated lognormal–relaxed clock model in BEAST v. 1.6.1. [26] as described by Rabeling et al. [27].
Results
Karyotype analysis and chromosome banding
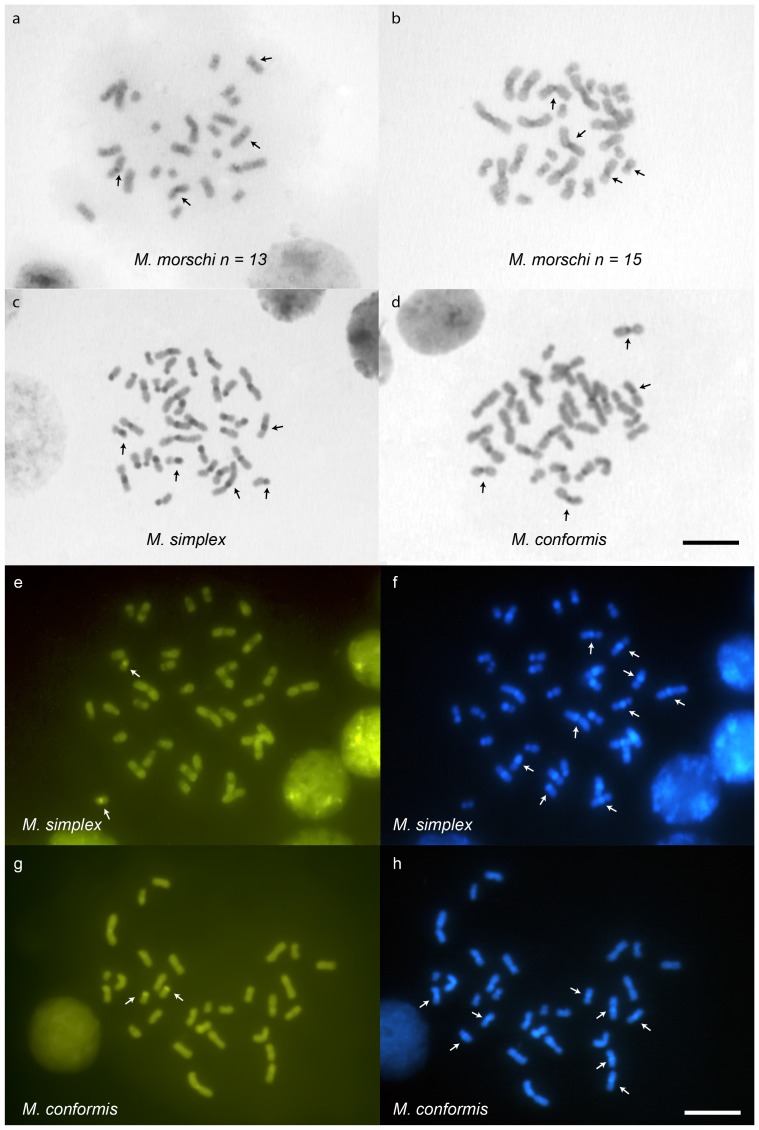
The three species of the genus Mycetophylax investigated present different chromosome numbers and karyotype morphologies (Figure 2). M. morschi showed populations with two distinct chromosome sets. Populations from some beaches of Rio Grande do Sul and Rio de Janeiro State presented diploid chromosome numbers equal to 2n = 30 (n = 15) or 2n = 26 (n = 13), whereas all the populations from Santa Catarina, Paraná and São Paulo State showed 2n = 26 (Figure 1, 2). No hybrids were found. M. morschi presented an almost bimodal karyotype that consisted of seven large and six small chromosome pairs for populations with 2n = 26 chromosomes, and six large and nine small pairs in populations with 2n = 30 chromosomes (Figure 2a and b). Both M. morschi karyotypes displayed nine metacentric pairs and one acrocentric pair; however M. morschi 2n = 26 showed only three submetacentric pairs, whereas specimens with 2n = 30 showed five. In karyotypes with 2n = 26, one strongly and two weakly stained regions were observed in the terminal portion of the long arm of chromosome 3 and chromosomes 2 and 5, respectively. These regions may correspond to secondary constrictions. Similarly, a secondary constriction on chromosome 3 was observed in the karyotype 2n = 30.
Figure 2. Conventional staining of mitotic cells of Mycetophylax.
Species images, metaphases and diploid karyotypes of Mycetophylax morschi 2n = 26 (a) and 2n = 30 (b), Mycetophylax conformis (c) and Mycetophylax simplex (d). M = metacentric, SM = submetacentric and A = acrocentric.
In all of the metaphases analyzed, M. conformis presented 2n = 30 (n = 15) and a chromosome complement composed of eleven metacentric and four submetacentric pairs (Figure 2c). A secondary constriction was observed on the second pair of metacentric chromosomes.
M. simplex showed the largest chromosome number of the genus, 2n = 36 (n = 18). It was composed by ten metacentric and eight submetacentric pairs (Figure 2d). The first metacentric and submetacentric chromosomes (pairs 1 and 11) were large and the remainders were from medium to small in size.
The results of chromosome banding and staining are shown in Table 1. The species showed different patterns of heterochromatin distribution (Figure 3). In M. morschi karyotypes, the heterochromatin is quite evident and can be distinguished in a few chromosomes in the centromeric region (Figure 3a and b, dark grey as indicated by the arrows). Moreover, M. conformis and M. simplex showed conspicuous heterochromatin blocks in the centromeric and pericentromeric regions (Figure 3c and d, dark grey as indicated by the arrows). The sequential fluorochrome staining revealed positive GC rich blocks (CMA3 +) in only one pair, on the telomeric region, in M. conformis and on the pericentromeric region in M. simplex (Figure 3e, g – white arrows). DAPI showed general banding pattern coincident with the C–bands, indicating that the heterochromatin is AT rich (Fig. 3f and h – shiny blue as indicated by white arrows). M. morschi did not show any GC or AT rich regions in either karyotype, since the chromosomes were stained uniformly (data not shown).
Table 1. Cytogenetic data of the Mycetophylax species. Summary of chromosome.
| Chromosome number (n) | Chromosome morphology | C–banding positive blocks | Fluorochrome staining | ||||||
| M | SM | A | C | PC | SA | AT+ bands | GC+ bands | ||
| M. simplex | 18 | 10 | 8 | no | yes | yes | yes | yes | yes |
| M. conformis | 15 | 11 | 4 | no | yes | yes | yes | yes | yes |
| M. morschi | 15 | 9 | 5 | 1 | yes | no | no | no | no |
| M. morschi | 13 | 9 | 3 | 1 | yes | no | no | no | no |
number, chromosome morphology and banding patterns of observed karyotypes of all the specimens analyzed here.
M: metacentric; SM: submetacentric; A: acrocentric.
C: centromeric; PC: pericentromeric; SA: Short arm.
Figure 3. Mycetophylax metaphases submitted to C–banding technique and stained with fluorochromes.
(a) C – banding in the worker metaphase of M. morschi 2n = 26, (b) M. morschi 2n = 30, (c) M. simplex, (d) M. conformis denoting the heterochromatic positive bands (dark grey as indicated by the arrows). Metaphase of (e, f) M. simplex and (g, h) M. conformis stained with fluorochromes CMA3 and DAPI, respectively. The white arrows indicate the positive staining for CMA3. DAPI positivity was in agreement with the C – banding pattern (some positive bands are indicated by white arrows). Bar = 5 µm.
Chromosome evolution
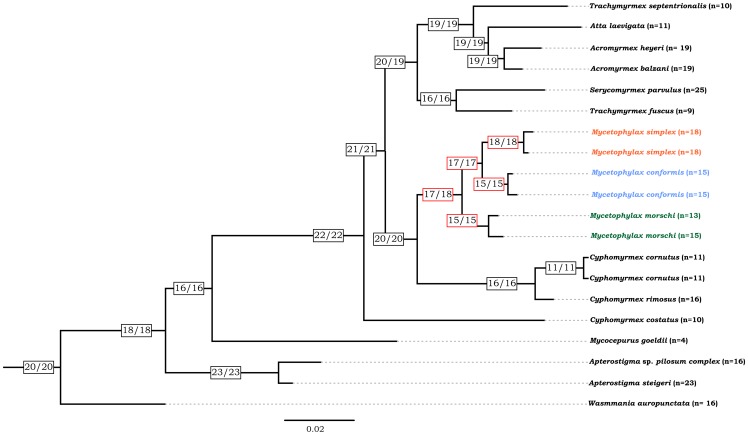
The results obtained in the analysis of chromosome evolution suggested that the best supported model of the process underpinning chromosome change was the hypothesis with constant gain, loss and duplication (Table 2). The rate parameters estimated in the best model were 16.52 for loss (δ), 7.01 for gain (λ) and 0.40 for duplication (ρ). The total inferred chromosome loss events were 181.41, gain 72.91 and duplication 1.70. These results revealed the occurrence of polyploidization events and suggested that whole karyotype duplication could have occurred during the chromosome evolution of these species. The main events inferred were loss (fusion) and gain (fission), which showed PP>0.5. In the Bayesian analysis, the haploid chromosome number at the most recent common ancestor (MRCA) of Mycetophylax with highest posterior probability (PP) was n = 17 and in the ML analysis the most likely haploid number was n = 18 (Figure 4, see Figure S1 for details).
Table 2. AIC scores and likelihood estimates for the data set analyzed for each model implemented by ChromEvol software.
| Models | Log-likelihood | AIC scores |
| Gain, Loss and Duplication constant* | −56.91 | 119.8 |
| Gain and Loss constant, no duplication | −58.01 | 120 |
| Gain, Loss and duplication constant, Gain and Loss depend linearly on the current chromosome number | −55.67 | 121.3 |
| Gain and Loss constant, Gain and Loss depend linearly on the current chromosome number, no duplication | −57.37 | 122.7 |
*Best fitting model.
Figure 4. Chromosome number evolution and inferred ancestral chromosome state in the genus Mycetophylax (red boxes) inferred under Bayesian and Maximum likelihood optimization, including other Attini ants and an outgroup.
Boxes at the nodes present the inferred ancestral haploid chromosome number for each node by Bayesian and ML analysis, respectively. Numbers at the tips are the known haploid chromosome numbers of species.
To describe an evolutionary scenario for chromosome evolution inferred by ChromEvol to Mycetophylax, we focused on the haploid chromosome numbers estimated in the Bayesian method, since this method provides posterior probabilities (PP) as a statistical parameter. From the MRCA of the Mycetophylax species, the chromosome number decreased, becoming n = 15 (PP = 0.43) and subsequently, n = 13 in the branch leading to M. morschi. Likewise, in the branch leading to M. conformis, the chromosome number decreased to n = 15 (PP = 0.63). In the case of M. simplex, the haploid chromosome number increased to n = 18 (PP = 0.74).
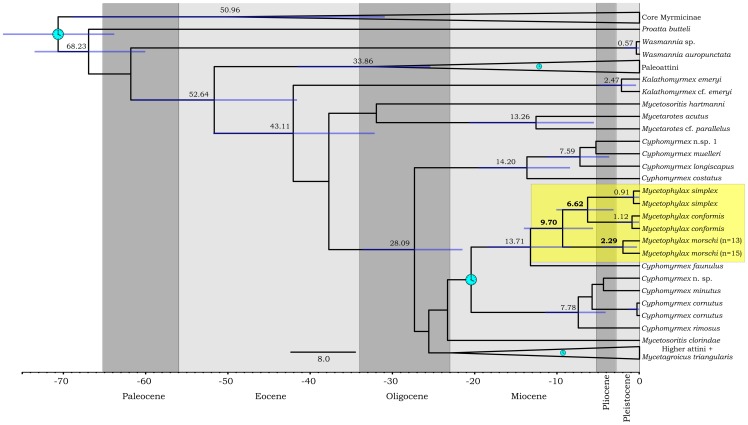
The Bayesian time–calibrated tree allowed us to infer that the Mycetophylax species diverged from Cyphomyrmex during the Miocene, around ∼13 Ma (95% CI = 8.49–18.91, Figure 5). This divergence was probably followed by chromosome changes. Considering the three species of Mycetophylax, M. morschi split early, around 9.1 Ma (95% CI = 5.75–14.34), whereas M. conformis and M. simplex split around ∼6.62 Ma (95% CI = 3.22–10.32). The initial emergence of M. morschi karyotypes was estimated to have occurred during the Pleistocene, around ∼2.29 Ma (95% CI = 0.31–4.72).
Figure 5. Bayesian time–calibrated maximum clade–credibility tree using a relaxed clock.
Two calibration points are indicated with blue clocks and the third and fourth are suppressed within the Paleoattini clade and Higher attini clade (for calibration point details see Schultz and Brady, 2008). The numbers on the upper branches are the inferred age of the nodes (shown in red for Mycetophylax), while the 95% credibility intervals are indicated as blue bars on the nodes.
Discussion
We detected wide chromosomal variability among the species of the genus Mycetophylax. The three species analyzed showed different karyotypes and, in fact, we verified two different diploid chromosome numbers for M. morschi. However, the karyotypes did not show significant geographic structuring, since they were found in both the northern and southern occurrence areas of this species. In three sampled localities, Angra dos Reis beach in the State of Rio de Janeiro and Mostardas and Chuí beaches in the State of Rio Grande do Sul (see Figure 1, Table S1), we detected the karyotypes n = 13 and n = 15, though not living sympatrically on the same beach.
Despite the large number of colonies analyzed, we did not find hybrid karyotypes. Intermediate karyotypes (hybrid karyotypes or heterokaryotypes) would be expected if there was still gene flow among the M. morschi citotypes. Besides, due the differences with respect to chromossome number among karyotypes would be expected heterokaryotypes that show chromosomes that do not exactly match in pairs. Since this unmatched chromosome were not observed in the colonies analyzed we conclude that the two M. morschi citotypes are in fixed populations and should be treated as separate species (to be further taxonomically described). The role of chromosome changes in the speciation has been extensively reported in literature [1], [28], [29], and it has been proposed to accelerate evolution of the species [30]. One hypothetical scenario that could explain the observed differences in karyotype number suggests that changes in chromosome number evolve gradually overtime by multiple events of rearrangements that culminate in different races due to fixation of a single or few chromosomal changes, followed by extinction of intermediate karyotypes [31]. The observed citotypes of M. morschi may be a result of this step-by-step mechanism of chromosome evolution. Moreover, this is in agreement with the general rule that changes in the karyotype occurs throughout species diversification in ants. Usually when species from one genus are cytogenetically analyzed they show polymorphic karyotypes regarding both number and morphology [2].
The most commonly invoked mechanism for chromosome evolution in ants is “the minimum-interaction theory” proposed by Imai et al. [5]. According to this theory, karyotype changes tend toward increasing the number of chromosomes in order to minimize the threats of deleterious rearrangements due to the interaction of chromosomes within the nucleus. In general, this model predicts an increase in chromosome number due to centric fission, followed by chromatin addition (mainly heterochromatin) or pericentrometric inversions (for details see [5], [32]). Therefore, in the course of evolution the number of chromosomes will increase in number and reduce in size. Although the minimum interaction theory does not disregard fusions, this chromosome rearrangement is considerate rare and fixed or positively selected when it bring about short-term advantages [33].
Considering the minimum interaction theory, the ancestral karyotype of M. morschi would be n = 13, reaching n = 15 by mean of fission rearrangements. However, based on our analysis of chromosome evolution, the recovered ancestral haploid chromosome number between karyotypes of M. morschi was n = 15, suggesting that the karyotype n = 13 probably arose due to tandem fusion from the karyotype with n = 15 chromosomes. The karyotypes do not show any absence of the medium size chromosomes, which would be expected in the case of centric fission from 13 to 15 haploid chromosomes, or acrocentric chromosomes in the karyotypes n = 15, which could have occurred in the case of centric fusion from n = 15 chromosomes to n = 13. Likewise, the contemporary haploid chromosome number of M. conformis seems to be produced by fusion, decreasing from n = 17 to n = 15. The estimated ancestral haploid chromosome number between M. confomis and M. simplex was n = 17, which is also the ancestral state estimated for the genus Mycetophylax. Thus, the karyotype number verified for M. simplex may have evolved due to centric fission instead of fusion, since it shows a haploid number of 18 chromosomes.
Several studies that evaluate karyotype evolution within an ant genera advocate in favor of centric fission as the main chromosomal rearrangement determining karyotypes, e.g. regarding Ponerinae ants, the suggestion is that chromosome changes occurred in the evolution of the genera Odontomachus and Anochetus [34]. The authors explained that centric fission is the principal evolutionary force acting on the karyotypes of Odontomachus, resulting in a larger, more stable karyotype mainly composed of subtelocentric chromosomes, compared with Anochetus, which is characterized by extreme karyotype diversification ranging from n = 12 to n = 23 and mainly composed of metacentric chromosomes. However, tandem fusion has been proposed to drive karyotype differentiation in a few cases (reviewed by [2]). In Myrmecia pilosula, this chromosome rearrangement was used to explain the origin of a long metacentric chromosome through the fusion of a subtelocentric and an acrocentric chromosome [35]. Chromosome fusion was also suggested to be involved in the genus Acromyrmex, due the decrease in the chromosome number, from n = 19 to n = 18, in Acromyrmex ameliae [36]. Notwithstanding, tandem fusion has also been reported to be involved in chromosome rearrangements for great number of other animals, including grasshoppers [37], [38]; wasps [39] and bats [40]. Our findings suggest that both fusion and fission may interplay during karyotype evolution in ants, promoting speciation by reducing or impairing gene flow. Chromosome changes are known to limit gene flow in parapatric and sympatric populations by means of gene isolations (unable to rearrange during meiosis) that can accumulate in consequence of rearrangements [31], [41], hence promoting population differentiation and speciation due to deficient recombination.
According to molecular phylogenetic analysis [17], Mycetophylax is divided into two major linages: one is composed only by the species M. morschi and the other comprises M. conformis and M. simplex. Since the last two species are more closely related to each other, the majority of metacentric and submetacentric chromosomes may be a characteristic shared by M. conformis and M. simplex. Furthermore, the pair of acrocentric chromosomes common to the karyotypes of M. morschi may be a symplesiomorphic chromosomal character retained from the ancestor that was lost in the lineage that diversified into M. simplex and M. conformis. On the other hand, this chromosome rearrangement could be one of the karyotypical characters that differentiate M. morschi from the others.
The C–banding technique and fluorochrome staining confirmed the cytotaxonomic groups distinguished by chromosome morphology analysis. Both karyotypes of M. morschi showed minimal amounts of heterochromatin and uniform fluorochrome staining. In contrast, M. conformis and M. simplex comprise a distinct group with intermediate to large amounts of AT-rich heterochromatin and a pair of chromosomes bearing a CG–rich region. The banding patterns shared by M. conformis and M. simplex suggest that their chromosomes underwent rearrangements following the split from a common ancestor related to M. morschi. Moreover, the amount of heterochromatin found in M. simplex is in agreement with the higher DNA content of this species. The 1C DNA amount estimated to M. simplex was 381.42 Mbp, whereas both karyotype of M. morschi and M. conformis have identical genome sizes with 312.96 Mbp [14].
The interspecific karyotype variability found among the species of the genus Mycetophylax could be associated with the biological environment where these species are restricted. As mentioned above, these species are confined to sand dune habitats along the Atlantic coast [12], [15]. This area is known to have been strongly influence during the Quaternary, having been remodeled due to periods of transgression and regression of the sea level [42]. Our results suggested that the genus Mycetophylax diverged from Cyphomyrmex during the middle Miocene (∼13 Ma) and diversified into the current species between the end of the Miocene and the beginning of the Pliocene. These periods are marked by deep modifications in the landscape that could facilitate the isolation of populations, culminating in the accumulation of chromosome mutations that could favor speciation and the evolution of new taxa. We suggest that an ancestor of these species was distributed along the Atlantic coast and later, due to transgressive movements of the sea, the geographic distribution was split by rising sea levels, producing barriers and sandy islands where the speciation process took place. The distinct karyotypes of M. morschi arose during the Pleistocene, a period extensively reported to have influenced the diversification of species in the Brazilian Atlantic Forest [43], which includes its coastline. Highly intra and inter–specific karyotype variability is also reported for the genus Ctenomys [44], a subterranean rodent that has habitat requirements and a distribution pattern similar to Mycetophylax. Thus, the sand dune environments on the Atlantic coast of Brazil and their geological history could have acted as a trigger for chromosomal rearrangements and the subsequent speciation in these areas.
This is the first comprehensive cytogenetic description and evolutionary analysis of an Attini genus based on molecular data and provides a baseline for future comparative and integrative studies. Based on our chromosome evolution approach and cytogenetic banding techniques, we hypothesized that fusions instead of fission could be involved in the chromosome evolution of the Mycetophylax. These chromosome rearrangements likely took place by involving complete genetic isolation of the two major lineages within Mycetophylax that therefore established their own evolutionary strategies. One of these lineages diversified into the M. morschi group complex and the other diversified into M. simplex and M. conformis. Overall, the results presented in this study confirm that tandem fusion could very well participate in chromosome ant evolution. Our karyological analysis based on phylogenetic framework suggests that some chromosome rearrangements may be more recurrent than previously thought. Besides, this integrative approach can be helpful to avoid misinterpretations on chromosome changes during species diversification. It is important that studies involving cytogenetic data within genus and between related genera are continued and that these studies take into account molecular phylogenic methods in the evaluation of the cytogenetic data of ants, as well in other taxa.
Supporting Information
Chromosome number evolution and inferred ancestral chromosome state in the genus Mycetophylax inferred under Bayesian and Maximum likelihood optimization with inferred frequency of fusion and fission events estimated throughout the phylogenetic tree. Green numbers at the branches and tips represent the inferred frequency of gain events (fission) and purple loss events (fusion) that had a probability >0.5. The analysis was carried out including other Attini ants and Wasmannia auropunctata as outgroup (Myrmicinae subfamily). Boxes at the nodes present the inferred ancestral haploid chromosome number for each node by Bayesian and ML analysis, respectively. Numbers at the tips are the known haploid chromosome numbers of species.
(TIF)
Sampled localities and number of the colonies analyzed cytogenetically per collected site and species. RS – Rio Grande do Sul state, SC – Santa Catarina state, PR – Paraná state, SP – São Paulo state, RJ – Rio de Janeiro state.
(XLS)
GenBank accession numbers of specimens used for phylogenetic inference in molecular clock analysis and to reconstruct the Bayesian tree used in the haploid ancestral state reconstruction and chromosome evolution analysis (in bold).
(XLS)
Acknowledgments
We are grateful to the many people that made this work possible. We thank all colleagues for their help with field sampling. We also thank Tathiana G. Sobrinho, Jorge A. Dergam and Antonio J. Mayhé–Nunes for suggestion on previous version of the manuscript. This work forms part of the D.Sc. thesis of the first author. The samples were collected with authorization of ICMBio (Instituto Chico Mendes de Conservação da Biodiversidade) by permission number 24869–2.
Funding Statement
This work was supported by a Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), process numbers: CRA-APQ-00540-11 and CBB-22004-11. Financial support was also provided by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.King M (1993) Species Evolution: The role of chromosome change. Cambridge University Press.
- 2. Lorite P, Palomeque T (2010) Karyotype evolution in ants (Hymenoptera: Formicidae), with a review of the known ant chromosome numbers. Myrmecol News 13: 89–102. [Google Scholar]
- 3. Imai HT, Crozier RH, Taylor RW (1977) Karyotype evolution in Australian ants. Chromosoma 59: 341–393. [Google Scholar]
- 4. Imai HT, Marayama T, Gojobori YI, Crozier RH (1986) Theoretical bases for karyotype evolution. 1. The minimum–interaction hypothesis. Am Nat 128: 900–920. [Google Scholar]
- 5. Imai HT, Taylor RW, Crozier RH (1994) Experimental bases for the minimum interaction theory. I. Chromosome evolution in ants of the Myrmecia pilosula species complex (Hymenoptera: Formicidae: Myrmeciinae). Jpn J Genet 69: 137–182. [Google Scholar]
- 6. Guerra M (2012) Cytotaxonomy: the end of childhood. Plant Biosyst 146: 703–710. [Google Scholar]
- 7. Mehdiabadi NJ, Mueller UG, Brady SG, Himler AG, Schultz TR (2012) Symbiont fidelity and the origin of species in fungus–growing ants. Nat Commun 3: 840. [DOI] [PubMed] [Google Scholar]
- 8. Schultz TR, Brady SG (2008) Major evolutionary transitions in ant agriculture. Proc Nat Acad Sci USA 105: 5435–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klingenberg C, Brandao CRF (2009) Revision of the fungus–growing ant genera Mycetophylax Emery and Paramycetophylax Kusnezov rev. stat., and description of Kalathomyrmex n. gen. (Formicidae: Myrmicinae: Attini). Zootaxa (2052): 1–31. [Google Scholar]
- 10. Schultz TR, Solomons SA, Mueller UG, Villesen P, Boomsma JJ, et al. (2002) Cryptic speciation in the fungus–growing ants Cyphomyrmex longiscapus Weber and Cyphomyrmex muelleri Schultz and Solomon, new species (Formicidae, Attini). Insectes Soc 49: 331–343. [Google Scholar]
- 11. Sosa–Calvo J, Schultz TR (2010) Three remarkable new fungus–growing ant species of the genus Myrmicocrypta (Hymenoptera: Formicidae), with a reassessment of the characters that define the genus and its position within the Attini. Ann Entomol Soc Am 103: 181–195. [Google Scholar]
- 12. Klingenberg C, Brandão CRF, Engels W (2007) Primitive nest architecture and small monogynous colonies in basal Attini inhabiting sandy beaches of southern Brazil. Stud Neotrop Fauna E 42: 121–126. [Google Scholar]
- 13. Cardoso DC, Cristiano MP, Tavares MG (2011) Methodological remarks on rearing basal Attini ants in the laboratory for biological and evolutionary studies: overview of the genus Mycetophylax . Insectes Soc 58: 427–430. [Google Scholar]
- 14. Cardoso DC, Carvalho CR, Cristiano MP, Soares FA, Tavares MG (2012) Estimation of nuclear genome size of the genus Mycetophylax Emery, 1913: evidence of no whole–genome duplication in Neoattini. C R Biol 335: 619–624. [DOI] [PubMed] [Google Scholar]
- 15. Cardoso DC, Cristiano MP, Tavares MG, Schoereder JH (2012) Co-occurrence of putatively allopatric species of the genus Mycetophylax: first record of Mycetophylax simplex (Emery, 1888) (Hymenoptera: Formicidae) from Rio de Janeiro State, Brazil. Myrmecol News 16: 57–59. [Google Scholar]
- 16. Kempf WW (1972) Catálogo abreviado das formigas da Região Neotropical. Studia Entomologica 15: 3–344. [Google Scholar]
- 17. Cardoso DC, Cristiano MP, Heinze J, Tavares MG (2014) A nuclear DNA based phylogeny of endemic sand dune ants of the genus Mycetophylax (Emery, 1913): how morphology is reflected in molecular data. Mol Phylogenet Evol 70: 378–382. [DOI] [PubMed] [Google Scholar]
- 18. Mayrose I, Barker MS, Otto SP (2010) Probabilistic models of chromosome number evolution and the inference of polyploidy. Syst Biol 59: 132–144. [DOI] [PubMed] [Google Scholar]
- 19. Imai HT, Taylor RW, Crosland MWJ, Crozier RH (1988) Modes of spontaneous chromosomal mutation and karyotype evolution in ants with reference to the minimum interaction hypothesis. Jpn J Genet 63: 159–185. [DOI] [PubMed] [Google Scholar]
- 20. Levan A, Fredga K, Sandberg AA (1964) Nomeclature for centromeric position on chromosomes. Hereditas 52: 201–220. [Google Scholar]
- 21. Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75: 304–306. [DOI] [PubMed] [Google Scholar]
- 22. Pompolo SG, Takahashi CS (1990) Chromosome numbers and C–banding in two wasp species of the genus Polistes (Hymenoptera Polistine, Polistini). Insectes Soc 37: 251–257. [Google Scholar]
- 23. Schweizer D (1980) Simultaneous fluorescent staining of R bands and specific heterochromatic regions (DA–DAPI bands) in human chromosomes. Cytogenetics and Cell Genetics 27: 190–193. [DOI] [PubMed] [Google Scholar]
- 24. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 26. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rabeling C, Gonzales O, Schultz TR, Bacci M, Garcia MV, et al. (2011) Cryptic sexual populations account for genetic diversity and ecological success in a widely distributed, asexual fungus–growing ant. Proc Nat Acad Sci USA 108: 12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White MJD (1973) Animal cytology and evolution. Cambridge University Press.
- 29. Livingstone K, Rieseberg L (2004) Chromosomal evolution and speciation: a recombination-based approach. New Phytol 161: 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lukhtanov VA, Dincă V, Talavera G, Vila R (2011) Unprecedented within-species chromosome number cline in the wood white butterfly Leptidea sinapis and its significance for karyotype evolution and speciation. BMC Evol Biol 11: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Imai HT, Satta Y, Takahata N (2001) Integrative study on chromosome evolution of mammals, ants and wasps based on the Minimum Interaction Theory. J Theor Biol 210: 475–497. [DOI] [PubMed] [Google Scholar]
- 33. Schubert I (2007) Chromosome evolution. Curr Opin Plant Biol 10: 109–115. [DOI] [PubMed] [Google Scholar]
- 34. Santos IS, Costa MA, Mariano CSF, Delabie JHC, Andrade–Souza V, et al. (2010) A cytogenetic approach to the study of neotropical Odontomachus and Anochetus ants (Hymenoptera: Formicidae). Ann Entomol Soc Am 103: 424–429. [Google Scholar]
- 35. Imai HT, Taylor RW (1989) Chromosomal polymorphisms involving telomere fusion, centromeric inactivation and centromere shift in the ant Myrmecia (pilosula) n = 1 . Chromosoma 98: 456–460. [Google Scholar]
- 36. Barros LAC, Aguiar HJAC, Mariano CSF, Delabie JHC, Pompolo SG (2010) Cytogenetic Characterization of the Lower–Attine Mycocepurus goeldii (Formicidae: Myrmicinae: Attini). Sociobiology 56: 57–66. [Google Scholar]
- 37. Warchałowska–Śliwa E, Grzywacz B, Maryańska–Nadachowska A, Karamysheva TV, Chobanov DP, et al. (2013) Cytogenetic variability among Bradyporinae species (Orthoptera: Tettigoniidae). Eur J Entomol 110: 1–12. [Google Scholar]
- 38. Hemp C, Heller KG, Warchalowska–Sliwa E, Hemp A (2013) The genus Aerotegmina (Orthoptera, Tettigoniidae, Hexacentrinae): chromosomes, morphological relations, phylogeographical patterns and description of a new species. Org Divers Evol 13: 521:530. [Google Scholar]
- 39.Gokhman VE (2009) Karyotypes of Parasitic Hymenoptera. Springer London.
- 40. Rodrigues LRR, Barros RMS, Marques–Aguiar S, Assis MF, Pieczarka JC, et al. (2003) Comparative cytogenetics of two phyllostomids bats. A new hypothesis to the origin of the rearranged X chromosome from Artibeus lituratus (Chiroptera, Phyllostomidae). Caryologia 56: 413–419. [Google Scholar]
- 41. Faria R, Navarro A (2010) Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol Evol. 25: 660–669. [DOI] [PubMed] [Google Scholar]
- 42.Dillenburg SR, Hesp PA (2009) Geology and geomorphology of Holocene coastal barriers of Brazil. Springer Berlin Heidelberg.
- 44. Freitas TR (2006) Cytogenetics status of four Ctenomys species in the south of Brazil. Genetica 126: 227–335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chromosome number evolution and inferred ancestral chromosome state in the genus Mycetophylax inferred under Bayesian and Maximum likelihood optimization with inferred frequency of fusion and fission events estimated throughout the phylogenetic tree. Green numbers at the branches and tips represent the inferred frequency of gain events (fission) and purple loss events (fusion) that had a probability >0.5. The analysis was carried out including other Attini ants and Wasmannia auropunctata as outgroup (Myrmicinae subfamily). Boxes at the nodes present the inferred ancestral haploid chromosome number for each node by Bayesian and ML analysis, respectively. Numbers at the tips are the known haploid chromosome numbers of species.
(TIF)
Sampled localities and number of the colonies analyzed cytogenetically per collected site and species. RS – Rio Grande do Sul state, SC – Santa Catarina state, PR – Paraná state, SP – São Paulo state, RJ – Rio de Janeiro state.
(XLS)
GenBank accession numbers of specimens used for phylogenetic inference in molecular clock analysis and to reconstruct the Bayesian tree used in the haploid ancestral state reconstruction and chromosome evolution analysis (in bold).
(XLS)