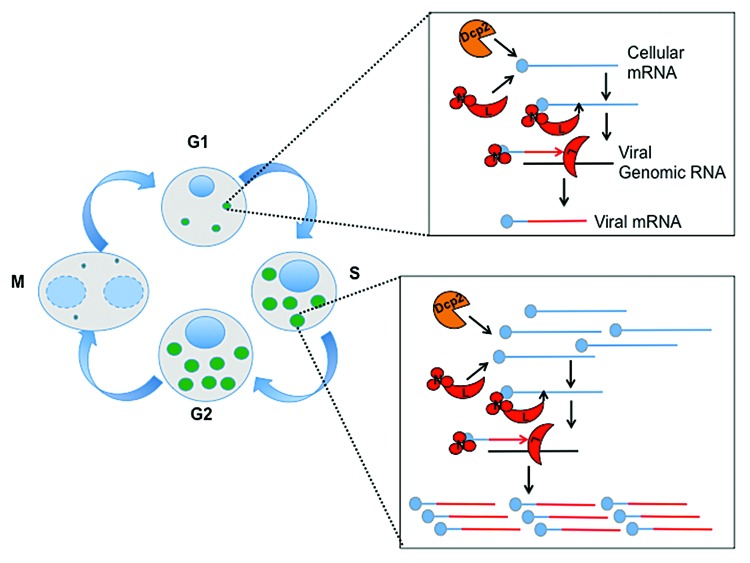
As obligate intracellular parasites with limited coding capacity, viruses utilize cellular pathways for replication. Arthropod-borne viruses in particular replicate in disparate, evolutionarily distant hosts (arthropods and vertebrates), suggesting that interactions between these viruses and cellular processes are deeply conserved. To identify factors affecting the replication of the mosquito-transmitted bunyavirus Rift Valley fever virus (RVFV), we used Drosophila as a model insect and performed a genome-wide RNAi screen in Drosophila cells. We validated 131 genes, and found that the canonical mRNA decapping enzyme, Dcp2, along with 2 decapping activators, restricted viral infection at the level of mRNA transcription.1 Furthermore, ectopic expression of Dcp2 restricted infection, suggesting that levels of Dcp2 define viral susceptibility. Interestingly, bunyaviruses cap their mRNAs at the 5′ end by “cap-snatching,” wherein the 5′ cap and 10–18 nucleotides of cellular mRNAs are recognized by the virally encoded nucleocapsid (N) and cleaved by the viral RNA-dependent RNA polymerase (RdRp), L, which uses this fragment as a primer for viral mRNA synthesis (Fig. 1, upper inset). The bunyaviral cap-snatching machinery localizes to processing (P) bodies in insects1 and mammals.2 P bodies are dynamically regulated cytoplasmic RNP granules that contain much of the RNA decay machinery, including Dcp2.3 Therefore, we hypothesized Dcp2 affects RVFV either by directly decapping viral mRNAs or by decapping cellular mRNAs targeted by RVFV for cap-snatching, thus creating a bottleneck for replication. We found that both viral mRNA stability and the levels of capped viral mRNA were unaffected by Dcp2 depletion, suggesting that Dcp2 limits the cellular substrate targeted by RVFV for cap-snatching (Fig. 1, upper inset).
Figure 1. Decapping and cap-snatching machinery compete for cell cycle regulated mRNAs. Left: Schematic of P body dynamics during cell cycle progression. During G1, P bodies (green) are present at low levels in cells. As cells exit S phase and progress into G2, P bodies increase in number and size.1,4 As cells enter mitosis, P bodies are lost.4 Upper inset: RVFV cap-snatching and Dcp2 mRNA decapping are competing processes. Viral mRNA transcription initiates upon the binding of RVFV N (red circles) to 5′ caps of cellular mRNAs (blue). Next, RVFV L (red moons) is recruited, and its endonuclease activity cleaves 10–18bp downstream of the cap and uses this primer to initiate viral transcription from the genomic RNA (black line), producing cellular-virus conjugate mRNAs (blue and red line). Dcp2 (orange) targets and degrades the same pool of cellular mRNAs that RVFV uses for transcription, creating a bottleneck. Lower inset: During the S/G2 phase of the cell cycle, as mRNAs required for DNA replication are targeted for degradation (blue), this increased level of substrates alleviates the bottleneck, allowing the viral cap-snatching machinery to increase viral transcription (blue and red lines). Thus, arresting cells in S/G2 increases bunyaviral replication.
This indicates that a common pool of mRNAs is targeted by Dcp2 and RVFV. To test this, we performed 5′ RACE, and found that half of the mRNAs snatched by the virus had gene ontology (GO) terms associated with the cell cycle. Furthermore, Dcp2 depletion increased basal levels of these transcripts, as well as their incorporation into viral mRNAs, without causing cell cycle arrest. Interestingly, previous reports in human cells found that P body dynamics are intricately linked to cell cycle progression.4 We show that this connection is deeply conserved, with P body size and number in Drosophila peaking as cells transition from S to G2, a time where mRNAs encoding genes necessary for DNA replication are targeted for degradation (Fig. 1, left). Since P bodies are an aggregate of RNAs destined for degradation, it is likely that this accumulation of targets accounts for the increases in RVFV replication. Indeed, our screen also identified 39 antiviral genes with the GO term “cell cycle.” Experimentally, we also identified 28 genes that, when depleted, led to both an arrest in S/G2, as measured by increased nuclear area, and increased infection of bunyaviruses. This effect on infection was specific to S/G2, as arrest in other phases of the cell cycle did not impact RVFV replication. Altogether, these data suggest that the increased accumulation of mRNAs destined for degradation in P bodies at S/G2 promotes cap-snatching and, thus, RVFV replication (Fig. 1, lower inset). Studies by others have also shown that RVFV causes cell cycle arrest in late S phase in human cells through activation of the DNA damage response, and that this arrest is advantageous for viral output.5 These data further suggest that there is an intricate interplay between RVFV and the cell cycle, and that S/G2 is favored due to increased mRNA targets in P bodies, alleviating the bottleneck from insects to humans.
We set out to identify genes affecting RVFV replication, which led to the discovery that cell cycle mRNAs are targets of both viral cap-snatching and Dcp2-dependent decapping. This suggests pools of cell cycle RNAs are under precise control by Dcp2; how these particular mRNAs are targeted to Dcp2 remains unknown. Furthermore, increasing evidence suggests that P bodies are not compartments of uniform composition; rather, they may possess potential specializations, since P body components only partially overlap and can be in separate structures.6,7 Indeed, our own studies found that while Dcp2 tightly co-localizes with RVFV, the canonical binding partner of Dcp2, Dcp1, only partially co-localizes with either Dcp2 or RVFV N.1 It is likely that in addition to specialization in their protein composition, these granules vary in their specificity for RNA targets. Therefore, RVFV and other bunyaviruses may provide a useful tool for probing the RNA composition of subsets of granules.
To add further complexity, while Drosophila encode only one known decapper, in mammals there are at least two decappers (DCP2 and NUDT16) possessing both specific and redundant functions in mRNA decay pathways.8 This suggests a tight degree of regulation in both the specificity of target selection to decapping enzymes, which likely reside in distinct compartments, and decapping activation itself. Further work will elucidate the regulation of RNA targeting to decapping enzymes, as these are potentially druggable targets. The possibility of inducing decapping to restrict bunyavirus infection is exciting, as no therapeutic interventions currently exist. Additionally, our finding that the stability of cell cycle mRNAs is specifically regulated by decapping suggests the possibility that decapping could be induced to restrict the expression of cell cycle regulated genes during cell cycle dysregulation, including cancer.
Hopkins KC, et al. Genes Dev. 2013;27:1511–25. doi: 10.1101/gad.215384.113.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26878
References
- 1.Hopkins KC, et al. Genes Dev. 2013;27:1511–25. doi: 10.1101/gad.215384.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mir MA, et al. Proc Natl Acad Sci U S A. 2008;105:19294–9. doi: 10.1073/pnas.0807211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ingelfinger DD, et al. RNA. 2002;8:1489–501. [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z, et al. J Cell Sci. 2004;117:5567–78. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- 5.Baer A, et al. J Biol Chem. 2012;287:7399–410. doi: 10.1074/jbc.M111.296608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weil TT, et al. Nat Cell Biol. 2012;14:1305–13. doi: 10.1038/ncb2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sen GL, et al. Nat Cell Biol. 2005;7:633–6. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, et al. RNA. 2011;17:419–28. doi: 10.1261/rna.2439811. [DOI] [PMC free article] [PubMed] [Google Scholar]