Abstract
The Y-box binding protein 1 (YB-1) is a DNA/RNA-binding nucleocytoplasmic shuttling protein whose regulatory effect on many DNA and RNA-dependent events is determined by its localization in the cell. We have shown previously that YB-1 is cleaved by 20S proteasome between E219 and G220, and the truncated N-terminal YB-1 fragment accumulates in the nuclei of cells treated with DNA damaging drugs. We proposed that appearance of truncated YB-1 in the nucleus may predict multiple drug resistance. Here, we compared functional activities of the full-length and truncated YB-1 proteins and showed that the truncated form was more efficient in protecting cells against doxorubicin treatment. Both forms of YB-1 induced changes in expression of various genes without affecting those responsible for drug resistance. Interestingly, although YB-1 cleavage did not significantly affect its DNA binding properties, truncated YB-1 was detected in complexes with Mre11 and Rad50 under genotoxic stress conditions. We conclude that both full-length and truncated YB-1 are capable of protecting cells against DNA damaging agents, and the truncated form may have an additional function in DNA repair.
Keywords: YB-1, proteasome, cleavage, PEST, truncation, nuclear localization, DNA reparation, DNA damage, drug resistance
Introduction
Y-box binding protein 1 (YB-1) is а member of the family of DNA/RNA-binding proteins with an evolutionarily conserved cold-shock domain (CSD). Y-box binding proteins were first described as major protein components of cytoplasmic mRNPs.1 Independently, YB-1, also known as dbpB or NSEP-1, was cloned as а transcription factor that specifically recognizes the Y-box promoter element.2 The diverse biological functions of YB-1 appear to arise from its capability of binding to nucleic acids without apparent specificity and from its multiple protein–protein interactions. It is also known as a nucleocytoplasmic shuttling protein, capable of changing localization in response to growth and stress-induced signaling.3-5 In the majority of human tissues and cell lines, YB-1 is predominantly localized to the cytoplasm, where it acts as а growth suppressor inhibiting cap-dependent translation of many growth-related mRNAs.6,7 When in the cytoplasm,YB-1 additionally contributes to mRNA stabilization and storage.8
Subsequent studies revealed involvement of YB-1 in transcriptional regulation of numerous genes, including those lacking the Y-box element. In fact, recent paper has demonstrated that YB-1 does not recognize the Y-box promoter in vivo.9 YB-1 is believed to bind to promoter elements with purine–pyrimidine asymmetry and to stabilize the single-stranded DNA conformation.10,11 Nuclear levels of YB-1 are predictive of multidrug resistance and worse patient outcome in various human cancers, potentially due to its suggested ability to activate transcription of many growth- and stress-associated genes, such as thymidine kinase, proliferating cell nuclear antigen (PCNA), DNA topoisomerase II α, MDR1, epidermal growth factor receptor (EGFR), cyclin B1, etc.12 In addition to transcriptional regulation, YB-1 is likely to play а role in DNA repair, based on its ability to unwind DNA duplexes and to bind to drug-modified and apurinic DNA13-15 and DNA repair proteins.15,16
Nuclear translocation of YB-1 was observed in response to various stimuli, including UV irradiation or treatment with mitomycin С, cisplatin or doxorubicin, heat-shock, growth factors, and cytokines stimuli as well as during cell cycle progression.3-5,17-20 YB-1 has been shown to contain 2 types of signaling sequences, such as nuclear localization signal (NLS) and the cytoplasmic retention site (CRS).21 The CRS was suggested to dominate over NLS in normal cellular conditions, thereby promoting predominantly cytoplasmic localization of YB-1. Apparently, CRS dominance over NLS can be overpowered under certain conditions, as YB-1 could also be observed in cell nuclei. So far, only a few molecular mechanisms have been proposed to explain nuclear translocation of YB-1. One of them involves YB-1 phosphorylation by Akt or other kinases at S102 with subsequent relocation of full-length YB-1 to the nucleus.5,22 Another mechanism implicates proteasome-mediated cleavage of YB-1 between NLS and CRS and accumulation of truncated YB-1 lacking the CRS in nuclei of DNA damaged cells.23 In addition to cell lines, accumulation of truncated YB-1 was also observed in primary cancer cells taken from pleural fluids of patients with various types of carcinomas, including breast, lung, and ovarian cancers, and correlated with enhanced resistance of these cells to DNA damaging drugs, suggesting that generation of truncated YB-1 may be an important element of the cell defense system activated in response to genotoxic damage.23 In this study, we performed detailed analysis of the 20S proteasome-mediated cleavage mechanism and investigated the role of truncated YB-1 in DNA damage stress response. We established that protective effect of YB-1 against genotoxic stress mostly results from its more efficient nuclear import and involvement in DNA repair and not from activation of genes responsible for multiple drug resistance.
Results
Truncated YB-1 does not affect NIH3T3 cell proliferation but enhances survival of doxorubicin-treated cells
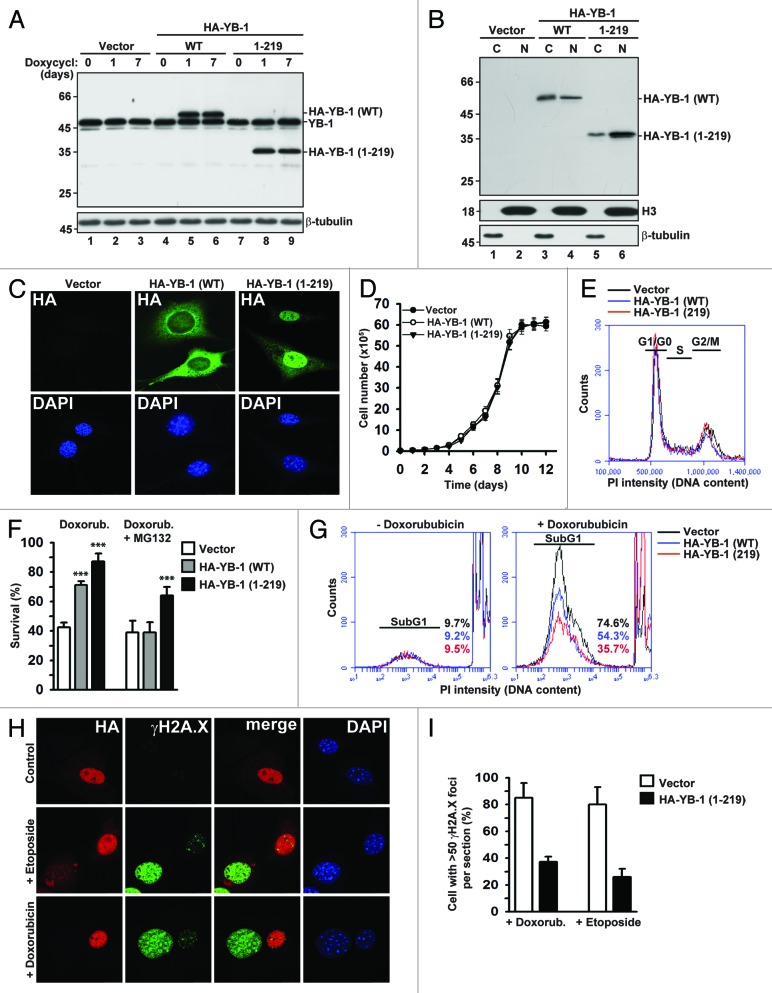
To compare effects of full-length and truncated YB-1 proteins on cell proliferation and survival during DNA damaging stress, we generated NIH3T3 fibroblasts stably expressing HA-YB-1 (WT) or HA-YB-1 (1–219) under the doxycycline-inducible promoter. In subsequent experiments, we used clones with equal protein expression levels. The maximal expression of full-length and truncated YB-1 was observed 24 h after induction with doxycycline and remained unchanged during subsequent cell passaging in the presence of doxycycline over a 7-d period (Fig. 1A). As expected, analysis of nuclear and cytoplasmic fractions showed that ~60% of full-length of YB-1 was localized to the cytoplasm (Fig. 1B, lanes 3 and 4), while truncated YB-1 was accumulated mainly in the nucleus (Fig. 1B, lanes 5 and 6). These results were supported by immunofluorescence microscopy (IF) (Fig. 1C). Since elevated levels of nuclear YB-1 were reported to be associated with increased or reduced proliferation, depending on the cell type, as well as with drug resistance,20,24-26 we tested whether full-length or truncated YB-1 proteins may exhibit similar effects in our experimental system. As seen in Figure 1D and E, neither full-length nor truncated YB-1 affected cell proliferation and cell cycle distribution, indicating that the effect of YB-1 on cell growth could be cell type-specific, and that in contrast to cancer cells, growth of immortalized NIH3T3 cells is not significantly affected by YB-1. The effects of YB-1 might also be influenced by its expression levels, and our experimental system did not produce artificially high YB-1 overexpression, which may be required to affect proliferation.
Figure 1. Effects of full-length and truncated YB-1 proteins on proliferation and survival during DNA damaging stress. (A) Expression of HA-tagged full-length and truncated YB-1 proteins in stably transfected NIH3T3 cells was induced by doxycycline. Whole-cell extracts were analyzed by WB using YB-1 antibodies. β-tubulin was used as loading control. (B) Nucleocytoplasmic distribution of HA-YB-1 (WT) and HA-YB-1 (1–219) was analyzed in cytoplasmic (C) and nuclear (N) fractions by WB using HA antibodies. Equal loading and fraction purity were controlled using Histone 3 and β-tubulin antibodies. (C) Subcellular localization of full-length and truncated YB-1 in NIH3T3 cells was examined by IF using HA antibodies. DAPI was used to visualize nuclei. (D) Growth curves of NIH3T3 cells expressing full-length or truncated YB-1. (E) Effect of YB-1 (WT) and YB-1 (1–219) expression in NIH3T3 cells on cell cycle progression. (F) Cells expressing full-length or truncated YB-1 were treated with 5 µM doxorubicin in the absence or presence of 10 µM MG132 for 14 h. Cell viability was determined by trypan blue staining (t test, P value [***] < 0.001). (G) Survival of YB-1 expressing NIH3T3 cells treated with 5 μM doxorubicin was monitored by FACS using PI staining. (H–I) NIH3T3 cells were transiently transfected with HA-YB-1 (1–219) and treated with 100 µM etoposide or 5 µM doxorubicin for 14 h and analyzed by IF using HA and γH2A.X antibodies, as indicated. DAPI was used to visualize nuclei (H). Percentage of cells containing more than 50 γH2A.X foci per section was determined by counting of >100 cells per field in 5 fields (I).
Despite the lack of pro-proliferative effects, both YB-1 proteins endowed cells with higher resistance to doxorubicin. Noteworthy, at lower doses of the drug both YB-1 proteins were almost equally effective (Fig. S1A and S1B), while YB-1 (1–219) was more efficient at higher doses (Fig. 1F and G; Fig. S1B). Moreover, pre-treatment with the proteasome inhibitor MG132 dramatically abolished the effect of full-length YB-1 while not affecting protective abilities of truncated YB-1 (Fig. 1F). Therefore, protective capacity of full-length YB-1 may depend on its proteasomal cleavage, and the truncated but not the full-length YB-1 could be required for enhanced survival during doxorubicin treatment.
An early marker of DNA damage is phosphorylated histone H2A.X (γH2A.X), as its phosphorylation occurs at double-stranded breaks a few minutes after the damage.27,28 Notably, cells expressing truncated YB-1 were less likely to have large amounts of γH2A.X foci in the nuclei after treatment with doxorubicin or etoposide compared with control cells, and those with higher YB-1 levels tended to have less γH2A.X foci in the nuclei (Fig. 1H and I), in agreement with its potential function in DNA repair. Taken together, these data indicate that YB-1, especially the truncated one, promotes cell survival upon DNA damaging stress.
Generation of 20S proteasome-resistant YB-1 mutants
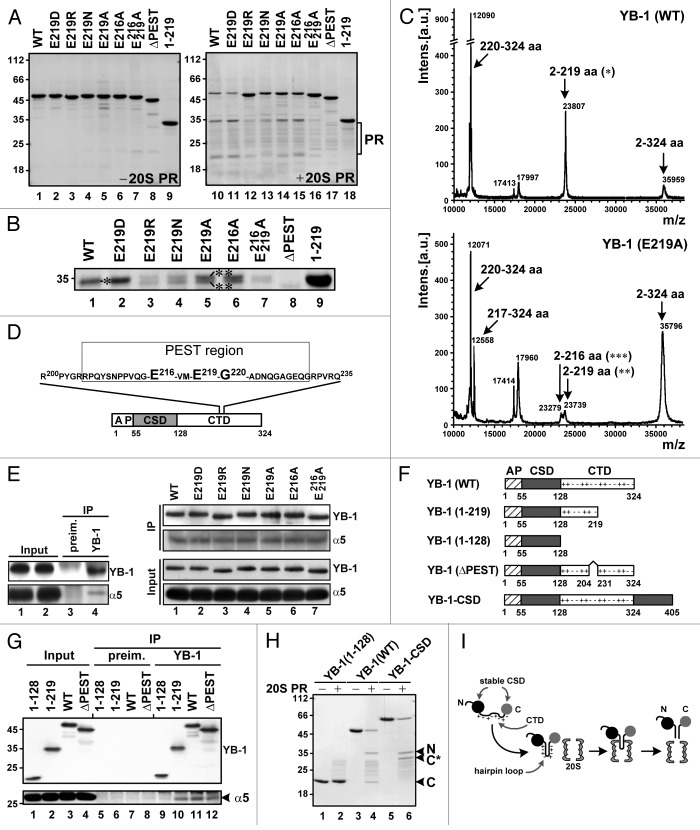
To further study the effect of the YB-1 processing on cellular responsiveness to DNA damage, we sought to generate YB-1 forms resistant to proteasomal cleavage. We have shown previously that YB-1 is cleaved by 20S proteasome after the negatively charged glutamic acid residue E219, indicating a possible involvement of caspase-like proteasomal activity in processing.29 As expected, substitution of E219 by similarly charged aspartic acid (E219D) produced no change in YB-1 sensitivity to 20S proteasome in vitro (Fig. 2A, lanes 10–11), while its replacement with positively charged arginine (E219R) made YB-1 almost completely resistant to 20S proteasome-mediated cleavage (Fig. 2A, lane 12). Neutral amino acid residues asparagine and alanine used to substitute for E219 (E219N and E219A) produced only partial resistance to YB-1 cleavage by 20S proteasome.
Figure 2. Mechanism of YB-1 cleavage by 20S proteasome. (A) YB-1 proteins were incubated in the absence (left panel) or presence (right panel) of 20S proteasome and analyzed by 12.5% SDS/PAGE and Coomassie Blue staining. (B) Magnified view of the large 35-kDa fragments from (A). (C) Mass spectrometry analysis of YB-1(WT) (upper panel) and E219A mutant (bottom panel) cleavage products. Arrows mark peaks corresponding to full-length protein and its major cleavage products. Asterisks in (B and C) indicate matching cleavage products. (D) Schematic representation of alternative cleavage sites and the PEST motif on YB-1. (E) YB-1 point mutants were incubated with 20S proteasome in the presence of MG132 and YB-1–20S proteasome complexes were precipitated using preimmune or YB-1 antibody. Co-IP was confirmed by WB using antibodies against YB-1 and α5 proteasomal subunit. (F) Schematic representation of YB-1 and its derivatives with indicated alanine/proline rich (AP), cold shock (CSD) and C-terminal (CTD) domains. (G) Co-IP of 20S proteasome and YB-1 mutants with YB-1 antibodies was performed as in (E). (H) Cleavage of YB-1 and its derivatives by 20S proteasome and Coomassie Blue staining was performed as in (A). N-terminal (N) and C-terminal (C) cleavage products of YB-1 (WT) are indicated. C* indicates the C-terminal cleavage product of YB-1-CSD. (I) The hairpin model for 20S proteasome-mediated YB-1 cleavage. The model depicts the YB-1-CSD fusion protein where the additional CSD (shown in gray) was attached to the C-terminal part of YB-1, thereby placing the supposedly unstructured proline-enriched CTD between 2 globular stable CSDs. As shown in (H), this did not prevent YB-1 cleavage, thereby excluding a possibility of processive digestion of CTD by 20S proteasome. This also indicates that the CTD may form a hairpin loop recognized by 20S proteasome, potentially through the interaction between positively and negatively charged amino acid clusters in this domain.
Importantly, the larger (N-terminal) cleavage products of YB-1 (WT) and (E219D) were represented by a single polypeptide (Fig. 2B, lanes 1–2), while cleavage products of YB-1 (E219R), (E219N) and (E219A) were heterogeneous (Fig. 2B, lanes 3–5), suggesting appearance of alternative cleavage sites, potentially due to changes in YB-1 secondary structure. Indeed, mass spectrometry analysis revealed that apart from the peptides 2–219 and 220–324, additional fragments 2–216 and 217–324 were detected among the cleavage products of YB-1 (E219A) (Fig. 2C), indicating that a peptide bond between the amino acid residues 216 and 217 serves as an alternative cleavage site for 20S proteasome. Of further note, both alternative cleavage sites, E216 and E219, are represented by glutamic acid, pointing to a possible involvement of the same caspase-like proteasome activity in YB-1 cleavage (Fig. 2D). Interestingly, both YB-1 (E216A) and (E219A), appear to be only partially resistant to 20S proteasome, and mutation of both sites YB-1 (E216/219A) was required for complete protection against 20S proteasomal cleavage (Fig. 2A, lanes 14–16 and Fig. 2B, lanes 5–7). Importantly, none of the introduced mutations affected YB-1 binding to 20S proteasome (Fig. 2E), indicating that there are 2 distinct sites on YB-1 that are required for 20S proteasome binding and cleavage, respectively. We thus identified E216 and E219 as 2 alternative cleavage sites on YB-1, and the corresponding mutant proteins resistant to 20S proteasome were utilized in the functional studies below.
Modeling YB-1 cleavage by 20S proteasome
We have shown previously that YB-1 is cleaved by 20S proteasome in an ubiquitin-independent manner.23 Short proteasome-recognition motifs targeting proteins for degradation in the absence of polyubiquitination are known to be enriched with proline (P), glutamic (E) or aspartic acid (D), serine (S), and threonine (T), and thus defined as PEST motifs.30 Many PEST-containing proteins are known to be polyubiquitinated and degraded by the 26S proteasome, and therefore must be recognized by the E2/E3 ligase system to become substrates for proteolysis.31 However, some PEST sequences appear to be constitutively active proteolytic signals, for example, the C terminus of mouse ornithine decarboxylase.32 Using the PESTFind algorithm (http://emboss.bioinformatics.nl/cgi-bin/emboss/epestfind), we found the highly conserved PEST regions within YB-1, including residues 1–26, 26–52, 118–137, 170–185, 205–231, 264–279, and 304–324. Since the YB-1 (1–219) mutant was resistant to proteasome, we concluded that the first 4 PEST-like regions on YB-1 were not involved in recognition by 20S proteasome (Fig. 2A, compare lanes 9 and 18). By contrast, deletion of the YB-1 PEST-like region 205–231 (ΔPEST) containing both alternative cleavage sites (E216 and E219; Fig. 2D) resulted in complete blockade of 20S proteasomal cleavage (Fig. 2A, compare lines 8 and 17).
Using a panel of YB-1 deletion mutants (Fig. 2F), we next performed co-immunoprecipitation (co-IP) experiments in the presence of the proteasome inhibitor MG132 to map a region on YB-1 required for binding to 20S proteasome. This identified YB-1 (WT), (1–219) and (ΔPEST) but not (1–128) as those interacting with 20S proteasome (Fig. 2G). Therefore, the C-terminal YB-1 domain appears to be responsible for binding to 20S proteasome.
In our previous study, we suggested a “hairpin loop” model for YB-1 cleavage by 20S proteasome, implying that the unstructured C-terminal YB-1 domain consisting of positively and negatively charged amino acids may form a hairpin loop that could be recognized and cleaved by 20S proteasome.23 To further verify this hypothesis, and to exclude an alternative possibility that the C-terminal part may spontaneously go through the catalytic cavity of 20S proteasome, thereby dragging the YB-1 cleavage site to a 20S proteasome active center, we generated a YB-1-CSD mutant protein containing an additional cold shock domain (CSD) proximal to the C-terminal domain (Fig. 2F). The CSD, which could not enter the 20S annulus due to its large size and highly globular structure was expected to prevent the C-terminal domain from freely entering the 20S proteasome cavity. Using this construct, we showed that cleavage of YB-1-CSD by 20S proteasome generated 2 fragments (Fig. 2H). Most importantly, identical sizes of the N-terminal fragments (~32 kDa) generated from both the WT and mutant proteins ascertained that the additional CSD was not able to block proteasomal cleavage. Therefore, although 20S proteasome binds to YB-1 via its C-terminal domain, YB-1 cleavage does not involve processive digestion of the C-terminal part and occurs endoproteolytically, in agreement with the proposed “hairpin loop” model (Fig. 2I).
20S proteasome-non-cleavable YB-1 mutants do not increase doxorubicin resistance
Next, we tested whether the YB-1 mutants that could not be cleaved by 20S proteasome may protect cells against DNA damaging agents, similar to WT YB-1. To this end, we generated the HA-tagged N-terminal YB-1 mutants using the pcDNA3 vector suitable for protein expression in eukaryotic cells (Fig. 3A). We first examined whether point mutations at the cleavage sites produced any effect on YB-1 stability in cells treated with doxorubicin. Similar to in vitro results, YB-1 (E219D) was degraded to the extent comparable to that of the WT protein, while levels of (E219R), (E219A) and (E216/219A) and (ΔPEST) mutant proteins were unchanged (Fig. 3B). The observed cleavage is likely to be unique for YB-1, since PABP, another major mRNA-binding protein, was not affected (Fig. 3B).
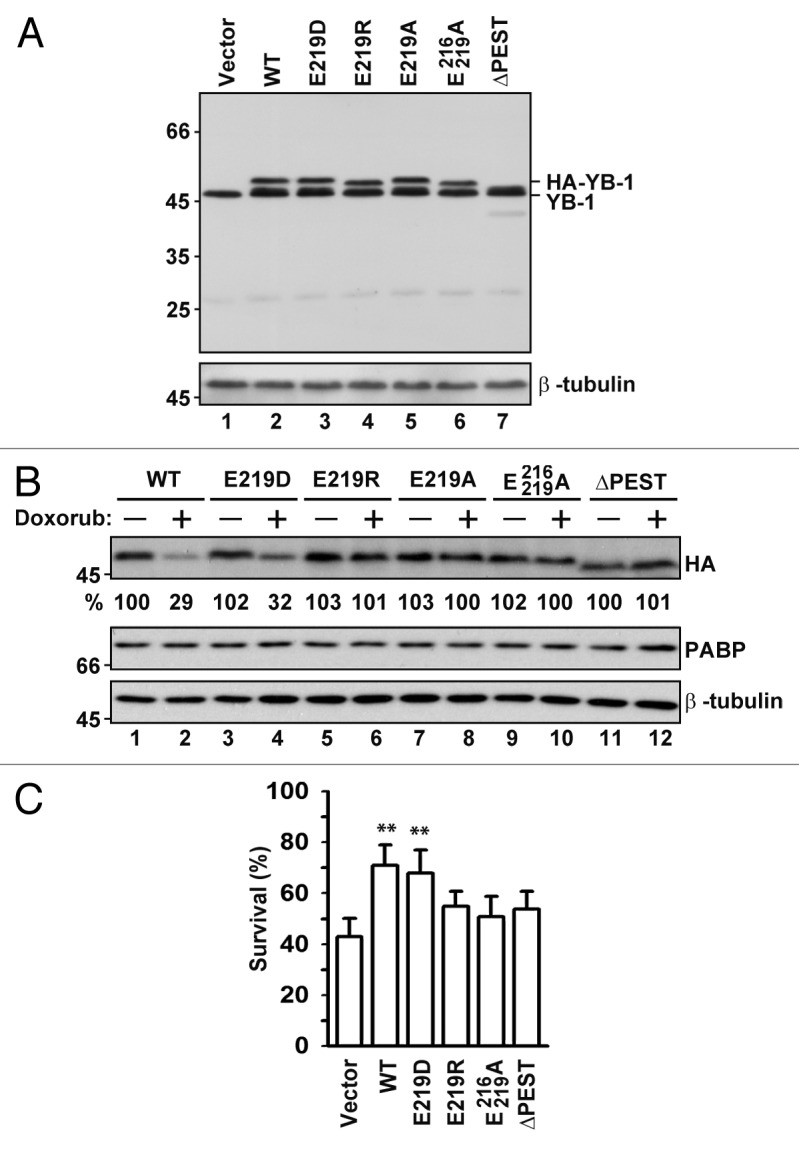
Figure 3. Expression, stability, and pro-survival effects of proteasome-resistant YB-1 mutants in cells treated with doxorubicin. (A) NIH3T3 cells were transfected with plasmids encoding YB-1 mutants, as indicated, and 24 h later analyzed by WB using YB-1 and β-tubulin antibodies. (B) Cells were transfected as in (A) and then left untreated or treated with 5 µM doxorubicin for additional 14 h. Whole-cell extracts were analyzed by WB. YB-1 was detected using HA antibodies. The numbers at the bottom indicate averaged percentage of the remaining protein as measured from 3 independent experiments by densitometry. β-tubulin was used as a loading control, and PABP was monitored as an additional control for protein stability. (C) Effect of YB-1 mutants on NIH3T3 cell survival after treatment with 5 µM doxorubicin for 14 h was monitored by trypan blue staining (t test, P value [**] <0.01).
To test whether expression of the 20S proteasome cleavage-resistant YB-1 mutants may protect cells against DNA damaging agents, NIH3T3 cells transiently transfected with the appropriate construct were treated with doxorubicin and analyzed for cell viability. Strikingly, only cleavable YB-1 proteins, (WT) and (E219D), exhibited significantly enhanced cell survival (Fig. 3C), confirming a notion that YB-1 cleavage is important for its protective activities during DNA damaging stress.
Proteasomal cleavage of YB-1 alleviates its nuclear import
The C-truncated YB-1 is known to be localized to the nucleus21,23 (see also Fig. 1C). Here, we investigated whether proteasomal processing might be sufficient to trigger nuclear import of YB-1 using an in vitro transport system prepared from nuclear and cytosolic fractions of HeLa cells grown under ambient conditions. To distinguish from endogenous YB-1 and to avoid potential implications of posttranslational modifications on nuclear import, we utilized the HA-tagged YB-1 proteins synthesized in E. coli. We first tested this system using model substrates, GST (glutathione S-transferase), which is normally localized to the cytoplasm, and its derivate GST-NLS (GST fusion with NLS from SV40 large T antigen). As expected, GST-NLS was readily detectable in nuclei, while GST remained in the cytosol and was washed away (Fig. 4A). We then tested the efficacy of nuclear import of WT and 1–219 YB-1 proteins in this system. As shown in Figure 4B, only truncated YB-1 was found in the nuclear fractions. Considering the lack of posttranslational modifications in these recombinant proteins produced in E. coli, these results demonstrate that the truncation itself represents a modification sufficient for nuclear import of YB-1, making it readily available during DNA damaging stress.
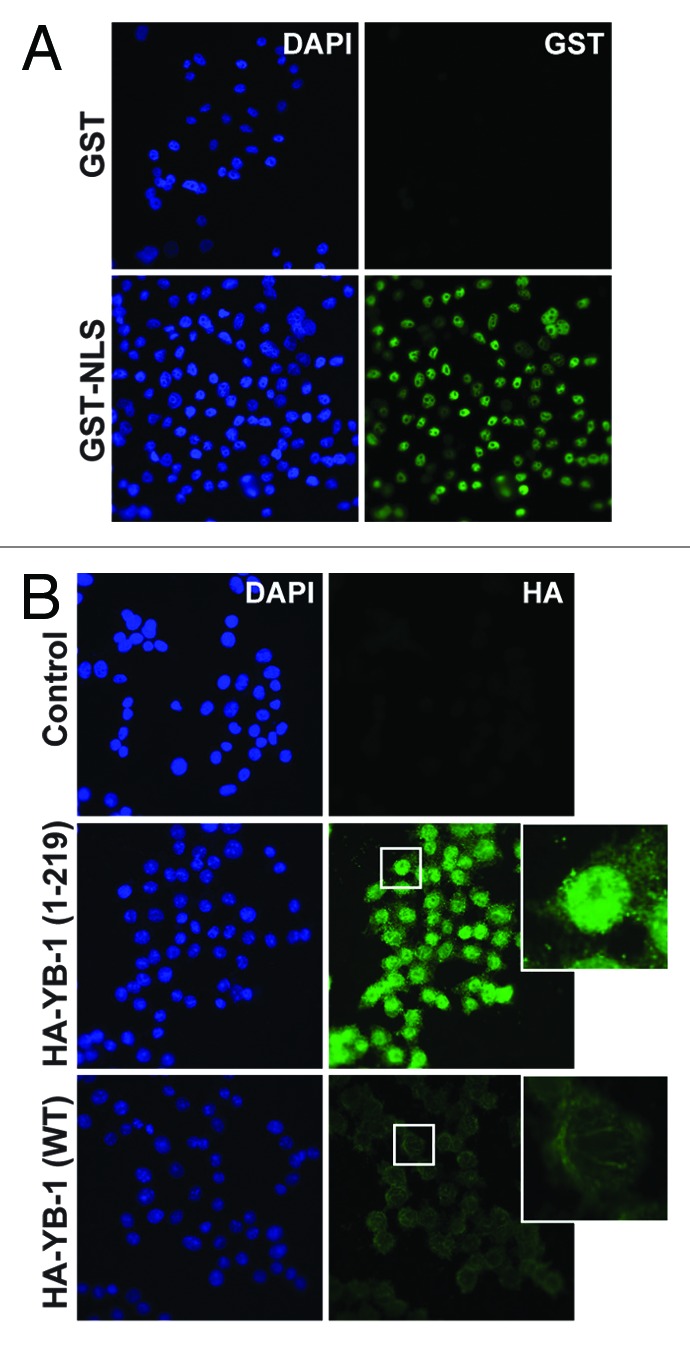
Figure 4. YB-1 cleavage activates its nuclear import. (A and B) GST and GST-NLS model substrates (A) or HA-YB-1 (WT) and HA-YB-1 (1–219) proteins (B) were used in the in vitro nuclear transport system, as described in “Materials and Methods”. IF of nuclear and cytosolic fractions using corresponding antibodies is shown. DAPI was used to visualize nuclei. Note that truncated YB-1 (1–219) was mostly localized to the nuclear fraction, in contrast to the WT YB-1 protein.
WT and truncated YB-1 proteins differentially affect gene expression
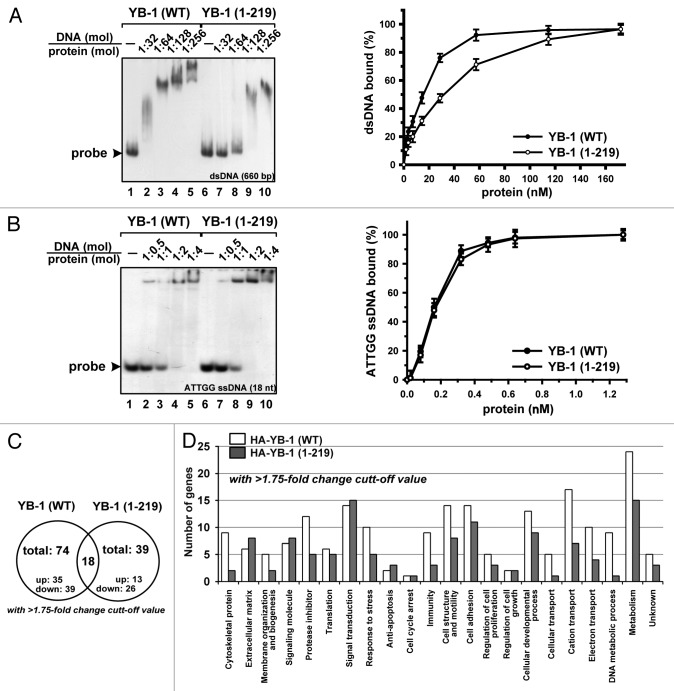
According to previous reports, YB-1 may protect cells indirectly, by inducing transcription of drug resistance-related genes, such as MDR1, or directly, by participating in the DNA repair process.12 We thus investigated whether cleavage may alter DNA binding properties of YB-1 or affect gene expression. Using EMSA and DNA binding assay we found that binding of truncated YB-1 to nonspecific 660-bp double-stranded (ds) DNA was reduced by ~2-fold (Fig. 5A), potentially due to the lack of a large portion of the C-terminal domain required for nonspecific nucleic acid binding. In contrast, binding of truncated YB-1 to a short Y-box-containing single-stranded (ss) DNA was comparable to that of the WT protein (Fig. 5B), suggesting that the truncated protein may retain similar sequence selectivity.
Figure 5. Effect of YB-1 cleavage on its DNA binding properties and gene expression regulation. (A and B) YB-1 binding to nonspecific [32P]-labeled 660-nt DNA duplex (A) or 18-nt Y-box-containing ssDNA (B) was analyzed by EMSA (left panels) and filter binding assay (right panels), as described in “Materials and Methods”. (C) Venn diagram showing common and differentially expressed RNA subsets identified by microarray analysis in NIH3T3 cells expressing WT or truncated YB-1 proteins. (D) Functional categories of genes whose expression was changed in NIH3T3 cells expressing WT or truncated YB-1 proteins, as compared with vector alone cells. Analysis was done using DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov).
Using mouse WG-6 gene expression array (Illumina), we compared mRNA profiles derived from the cells expressing WT or truncated YB-1 (1–219) proteins. For analysis, we selected RNA subsets based upon their statistical significance (P value < 0.01) and changes in their expression levels (>1.75-fold). Based upon these criteria, we selected 18 common genes whose expression was changed in both cell lines (Fig. 5C; Table S1). Interestingly, the expression changes were unidirectional for 14 of the selected genes, whereas 4 genes exhibited differential expression (Table S1). We also identified 56 and 21 genes in WT and truncated YB-1 expressing cell lines, respectively, whose expression was induced or reduced compared with control cells (Tables S2 and S3). Surprisingly, we have not identified MDR1 among the affected genes in our cell lines (Tables S4 and S5). Using DAVID Bioinformatics Database (http://david.abcc.ncifcrf.gov), we established that WT YB-1 mostly affects genes associated with cytoskeleton and DNA metabolism, whereas truncated YB-1 influenced genes responsible for extracellular matrix formation, signal transduction, and apoptosis inhibition (Fig. 5D). Together, these data indicate that YB-1 cleavage by 20S proteasome may produce protein with altered biological activity, which may differentially affect expression of specific subsets of genes.
Truncated YB-1 interacts and co-localizes with DNA repair complexes
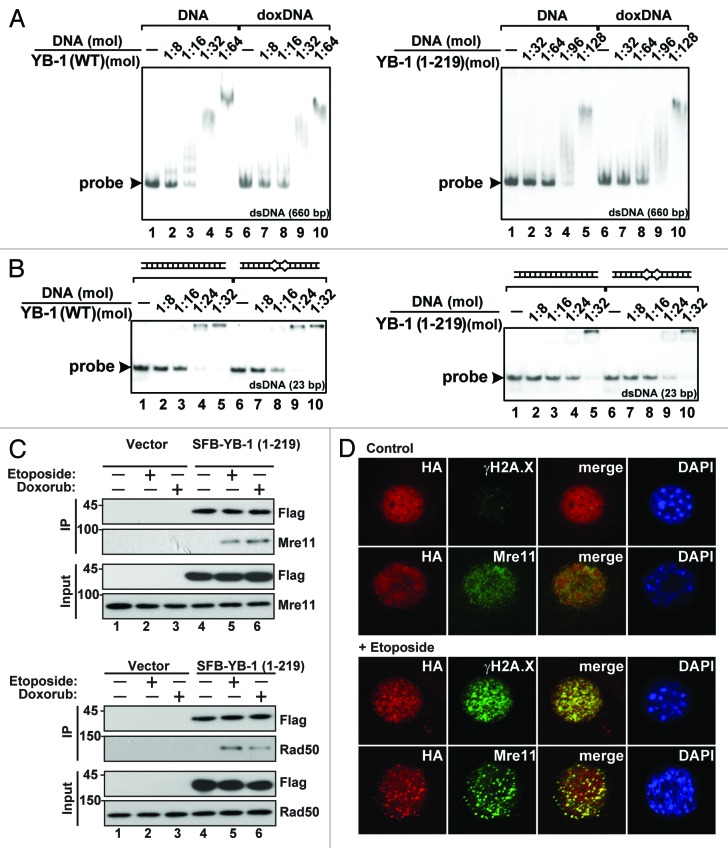
We next sought to determine whether the truncated YB-1 protein may be directly involved in DNA repair after DNA damage. To test if truncated YB-1 may bind to doxorubicin-modified dsDNA and mismatched DNA duplexes, we performed EMSA using the corresponding probes. We found that both WT and truncated YB-1 proteins exhibited similar affinity to doxorubicin-modified dsDNA and unmodified dsDNA (Fig. 6A). Notably, however, the truncated form was slightly more efficient in case of DNA with mismatches compared with perfect dsDNA (Fig. 6B, compare lanes 4 and 9), suggesting its potential involvement in recognition and binding to damaged DNA.
Figure 6. Both full-length and truncated YB-1 proteins interact with damaged DNA and DNA repair protein complexes. (A and B) Binding of YB-1 (WT) and YB-1 (1–219) with nonspecific [32P]-labeled 660-nt dsDNA, control or modified with doxorubicin, as indicated (A), or 23-nt perfect or mismatched DNA duplexes (B), was examined by EMSA. (C) NIH3T3 cells were transiently transfected with plasmids encoding SFB-tagged YB-1 (1–219) and 24 h later were treated with 5 µM doxorubicin or 100 µM etoposide for 14 h. Whole-cell extracts were used for IP with streptavidin conjugated beads, and immunoprecipitated proteins were examined by WB. (D) NIH3T3 cells transiently expressing HA-tagged YB-1 (1–219) for 24 h were treated with 100 µM etoposide for 14 h and analyzed by IF using the indicated antibodies. Note that in response to treatment, truncated YB-1 was localized to the nuclear speckles, together with other DNA repair proteins, such as γН2А.Х and Mre11.
We also tested a possibility that, similar to γH2A.X, YB-1 might be mostly involved in DNA repair as a component of DNA repair protein complexes. These complexes are known to comprise ATM, MDC1, 53BP1, BRCA1, and MRN complex (Mre11-Rad50-Nbs1) among others.33,34 Indeed, using Flag-tagged YB-1 (1–219), we were able to co-immunoprecipitate it with Mre11 and Rad50 from cells treated with doxorubicin or etoposide but not from those grown under ambient conditions (Fig. 6C). This was also confirmed by IF showing that truncated YB-1 was co-localized with γН2А.Х and Мrе11 in etoposide-treated cells (Fig. 6D). These results are consistent with a notion that truncated YB-1 functions in large DNA repair protein complexes, which are assembled in response to DNA damage.
Discussion
Involvement of YB-1 in tumor development and progression has long been a subject for debate. There are numerous reports stating that YB-1 mRNA and protein levels are significantly elevated in tumors of different origin.18,24,35-43 YB-1 was also shown to be involved in improved survival of cancer cells treated with various chemotherapeutic agents and development of multiple drug resistance.42,44-47 Of particular importance, YB-1 was reported to protect tumor stem cells during chemotherapy, which may contribute to tumor recurrence.44,48 We showed earlier that treatment of various cancer cells with DNA damaging agents results in 20S proteasome-mediated cleavage of YB-1, and its truncated form is accumulated in the nucleus. We proposed that the truncated and not the full-length YB-1 protein may play a major role in enhanced cancer cell survival and drug resistance.23 Here, we elucidated its role in cell defense mechanisms activated in response to DNA damaging stress.
In earlier studies, we and others employed shorter forms of YB-1, including (1–202), (1–204), and (1–205).21,23,49,50 Curiously, addition of only 2 amino acids to the YB-1 (1–205) fragment was shown to diminish its transcriptional activity, suggesting an importance of this region in regulating YB-1 functions.49 In our previous work,23 we identified E219 as a proteasomal cleavage site on YB-1. Here, using the full-length and truncated YB-1 (1–219) proteins, we further investigated mechanisms of 20S-mediated YB-1 processing and its functional significance.
We found that both full-length and truncated YB-1 forms were capable of increasing survival of cells treated with low doses of doxorubicin. However, the truncated form was more efficient in protecting cells treated with higher doses of doxorubicin. This is consistent with an earlier report showing that truncated YB-1 (1–205) increased cisplatin resistance above the levels conferred by wild-type YB-1.51 Given that in contrast to the full-length YB-1 its truncated CSD-containing derivatives are localized exclusively to the nucleus,21,23 we concluded that YB-1 cleavage and generation of its truncated (1–219) form in response to DNA damage could allow for its rapid nuclear translocation, where it fulfills its functions. Supporting this notion, 20S proteasome-resistant YB-1 mutants were not able to enhance cell survival, and pre-treatment with proteasomal inhibitor MG132 abolished protective activity of full-length YB-1, while not affecting pro-survival activity of the truncated form.
To gain further insights into potential mechanisms underlying YB-1 activities in cell defense, we performed genome-wide microarray analysis of gene expression in NIH3T3 cells expressing full-length or truncated YB-1 proteins. This revealed subsets of mRNAs whose expression was similarly or differentially affected by full-length and truncated YB-1, highlighting the fact that, directly or indirectly, YB-1 cleavage may alter expression of many genes. We believe that full-length YB-1 may exhibit rather indirect effects on gene expression, including at the levels of mRNA splicing, stability, and protein synthesis regulation, which may, in turn, affect transcription. Direct involvement of YB-1 in regulating expression of some genes, including MDR1, has recently been questioned,9,52 and we have not identified MDR1 among the affected genes in our cell lines (Tables S4 and S5).
The results of our study implicate truncated YB-1 in DNA repair pathways. This is based upon our findings that enhanced resistance to DNA damaging drugs in cells expressing truncated YB-1 was also associated with reduced abundance of γH2A.X, a marker for double-stranded DNA breaks. Moreover, in response to doxorubicin or etoposide treatment, truncated YB-1 appears to interact and to co-localize with key components of DNA repair machinery, such as histone γH2A.X, Mre11, and Rad50, characterizing it as a functional component of multiprotein DNA repair complexes. It should also be noted that full-length YB-1 is known to be involved in genotoxic stress response and to interact with many DNA repair proteins, including MSH2, DNA polymerase delta, Ku80, and Werner syndrome (WRN) proteins.15,16 It is therefore conceivable that truncated YB-1 preserves many activities of the full-length protein, and the main purpose of the 20S-mediated YB-1 cleavage is to allow for its rapid delivery into the nucleus. This could be especially important under severe stress conditions, such as higher doses of genotoxic agents. Blockade of YB-1 cleavage may thus be advantageous as an adjuvant treatment during chemotherapy to enhance action of genotoxic drugs. This possibility remains to be elucidated.
Materials and Methods
Expression constructs and recombinant proteins
YB-1 encoding plasmid pET-3-1-YB-1 (WT) was described previously.53 To generate constructs coding YB-1 mutant proteins, deletions or point mutations were introduced by PCR, and mutated YB-1 cDNAs were subcloned into the appropriate vectors. Resulting constructs were verified by sequensing. Recombinant YB-1 proteins were expressed in Escherichia coli and purified as described previously.8,53
Cell cultures, subcellular fractionation, and western blotting (WB)
NIH3T3 cells were purchased from American Type Culture Collection (ATCC) and maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum at 37 °C in 5% CO2 (v/v). Transient transfection of NIH3T3 cells was performed using Lipofectamin 2000 reagent (Invitrogen). Cell lines stably expressing HA-YB-1 (WT) or HA-YB-1 (1–219) were generated by sequential retroviral transduction of NIH3T3 cells with pRevTet-On vector (Clontech) and pRevTRE-HA-YB-1 (WT) or pRevTRE-HA-YB-1 (1–219), respectively. Overexpression of HA-YB-1 (WT) and HA-YB-1 (1–219) was induced by addition of 1 μg/ml of doxycycline (Sigma). Subcellular fractionation and WB were performed as described earlier.8 HA, GST, β-tubulin, and Rad50 antibodies were purchased from Sigma. Histone 3, γH2A.X, Mre 11 antibodies were from Abcam, α5 from Biomol; YB-1 antibodies were custom-made and described previously,54 and PABP antibodies were obtained from Dr N Sonenberg.
In vitro cleavage assay
Cleavage reaction was performed using 1.5 μg of YB-1 proteins and 0.1 μg of 20S proteasome (BioMol) in 20 μl reaction mixture containing 30 mM Tris-HCl (pH 7.6), 100 mM NaCl, 10 mM CaCl2, 2 mM MgCl2, 50 μM ATP, 1 mM DTT, 10% (v/v) glycerol. After 1 h incubation at 30°C the reaction was stopped by addition of 5× Laemmli sample buffer, reaction products were resolved by SDS/PAGE and stained with Coomassie brilliant blue R-250.
Mass spectrometry
YB-1 (WT) and YB-1 (E219A) cleaved with 20S proteasome, as described in the previous section, were precipitated with 10% TCA and dissolved in deionized water. Mass spectrometry was performed using a MALDI-TOF mass spectrometer (Reflex III model; Bruker Analytic GmbH) with 337 nm UV laser. YB-1 peptides were searched for among resulting peaks using MASCOT software (Matrix Sciences).
Co-immunoprecipitation (Co-IP)
YB-1 (WT) or YB-1 fragments (100 ng each) and 20S proteasome (100 ng) were incubated at 30°С for 30 min in а total volume of 100 µl of binding buffer containing 20 mM HEPES-КОН (рН 7.6), 100 mM КСl, 5 mM МgС12, 2 mM DTT, 20 µМ МG132, 0.25% (v/v) Nonidet Р-40. YB-1/20S proteasome complexes were precipitated with YB-1 antibody coupled to protein G Sepharose beads at 4 °С for 12 h. After extensive washes, samples were eluted with 2× Laemmli buffer, boiled, resolved by SDS/РАGЕ, and analyzed by WB using antibodies against YB-1 and the α5-subunit of 20S proteasome (BioMol). Co-IP of a C-terminal triple-epitope SFB (S protein, Flag, and streptavidin-binding peptide)-tagged YB-1 (1–219) with Mre11 and Rad50 was performed as previously described.55 Briefly, NIH3T3 cells were transiently transfected with a plasmid encoding SFB-YB-1 (1–219) and, 24 h later, were treated with 5 µM doxorubicin or 100 µM etoposide for 14 h. Whole-cell extracts were subjected to IP using streptavidin conjugated beads, and precipitated proteins were then subjected to WB.
Immunofluorescence microscopy (IF)
NIH3T3 cells overexpressing HA-YB-1 (WT) or HA-YB-1 (1–219) were grown on coverslips up to ~50% confluency, treated with 5 µM doxorubicin (Sigma) or 100 µM etoposide (Sigma), fixed in 3.7% paraformaldehyde-PBS for 20 min, and permeabilized in PBS-0.5% Triton X-100 for 20 min. For foci visualization, cells were permeabilized with PBS-0.5% Triton X-100 for 5 min before fixation in 3.7% paraformaldehyde-PBS and then permeabilized for the second time with PBS-0.5% Triton X-100 for 20 min. Coverslips were blocked with 5% goat serum in PBS for 30 min and incubated in 0.5% goat serum-PBS with primary antibodies for 1 h, followed by fluorophore-conjugated secondary antibodies (Invitrogen). Slides were assembled using Pro Long Gold antifade reagent with DAPI (Molecular Probes) and analyzed using Leica TCS SPE confocal microscope.
Cell counting and flow cytometry
Cells were stained with 0.4% trypan blue (Gibco) and counted using a Countess® Automated Cell Counter (Invitrogen). For flow cytometry (FACS) analysis, cells were collected, washed with 1.5 ml of cold PBS (4 °С), and fixed in 1.5 ml of cold 70% ethanol (−20 °С) at 4 °С for 14 h. Then cells were washed twice with PBS and stained in 1 ml of PBS containing 50 μg/ml propidium iodide (PI) (Sigma), 0.1% Triton X-100 (Sigma), and 1 mg/ml RNase A (Sigma) at 37 °С for 1 h and analized using a BD Accuri C6 flow cytometer (BD Biosciences).
Clonogenic assay
Cells (1000/10 cm plate) were seeded in triplicate and treated with increasing concentrations of doxorubicin (0–90 nM) for 14 h. Colonies were allowed to form for 8 d, then fixed in 3.7% paraformaldehyde-PBS for 30 min at room temperature, stained with crystal violet (0.05% w/v in water), and counted.
Electrophoretic mobility shift assay (EMSA) and filter binding assay
A 660-nt DNA duplex was generated by PCR amplification of the pET-3-1-YB-1(WT) fragment encoding the first 220 amino acids of YB-1 and 5′ end-labeled with [32P]-ATP using T4 polynucleotide kinase (Fermentas). Synthetic AAACTGATTG GCCAAAAA ssDNA and 23 nt oligonucleotides were [32P]-labeled at the 5′ end by T4 polynucleotide kinase. Twenty-three nt ssDNA molecules were annealed to generate a perfect duplex (5′-TGTCAGCACC TGCCATCACT TCT-3′ and 5′-AGAAGTGATG GCAGGTGCTG ACA-3′) or a duplex with mismatches (5′-AGAAGTGATG GCAGGTGCTG ACA-3′ and 5′-TGTCAGCACC CGGCATCACT TCT-3′). To generate modified DNA, DNA duplexes were incubated with 1 μM doxorubicin in 10 mM sodium phosphate buffer (pH 7.0) at 37 °C for 14 h. The 32P-labeled DNA duplexes modified or unmodified with doxorubicin were incubated with increasing amounts of YB-1 proteins in a final volume of 10 μl in a buffer A containing 150 mM NaCl, 10 mM Tris-HCl, pH 7.6 at 30 °C for 10 min. DNA protein complexes were analyzed by 3.8% PAGE in 44.5 mM Tris, 44.5 mM boric acid, 10 mM EDTA, followed by autoradiography. For filter binding assay, the same amounts of DNA and protein were incubated in a final volume of 50 μl of buffer A at 30 °C for 10 min. The mixture was filtrated through a nylon/nitrocellulose membrane sandwich (Hybond, GE Healthcare), followed by autoradiography.
Transcriptome microarray analysis
~70%-confluent cells stably expressing HA-YB-1 (WT), HA-YB-1 (1–219), or empty vector were grown in 10-cm plates, rinsed 3 times with PBS, and harvested in TRIzol (Life Technologies). RNA was quantified using Nanodrop and 200 ng of total RNA was amplified by Illumina® TotalPrep™ RNA Amplification Kit (Ambion). Amplified RNA was hybridized with Mouse WG-6 gene expression array (Illumina) in duplicates according to Illumina protocol. Data aquisition and analisys was done by BeadStudio software (Illumina) using gene expression module. Average signals with detection P values (based on Illumina replicate gene probes) >0.01 were removed from analyses. Functional categorization of affected genes was made using DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov).
In vitro transport assay
In vitro transport assay was assembled as described previously.56 Briefly, HeLa cells were seeded onto 18 × 18 mm coverslips to reach ~70% confluence the next day. The cells were permeabilized with 50 µg/ml digitonin (Sigma), and soluble cytoplasm proteins were washed off with import buffer containing 20 mM НЕРЕS-KOH рН 7.3, 0.11 М CH3COOК, 2 mM (CH3COO)2Mg. The coverslips were inverted over drops (10 μl) containing 5 mM creatine phosphate, 0.1 µg/µl creatine phosphokinase, 0.5 mM ATP and 0.5 mM GTP, 1 μg RNase A, 0.5× HeLa cytosol and 50 µg/ml of target protein. The reaction mixture was incubated at 30 °C for 30 min and washed with import buffer to remove all remaining cytosolic proteins. Cell were fixed with 4% formaldehyde and stained for GST or HA-tags. GST and GST-NLS (SV40 large T antigen NLS) synthesized in E. coli and purified using Glutathione Sepharose were used as controls.
Statistical analysis
Statistical significance of data was determined using 2-tailed t test.
Supplementary Material
Acknowledgments
The authors thank Prof AA Stavrovskaya for fruitful discussions and EV Serebrova for help in manuscript preparation. This study was supported by Programs on “Molecular and Cell Biology” and “Basic Sciences to Medicine” from the Presidium of the Russian Academy of Sciences.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/26670
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26670
References
- 1.Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry (Mosc) 2011;76:1402–33. doi: 10.1134/S0006297911130049. [DOI] [PubMed] [Google Scholar]
- 2.Didier DK, Schiffenbauer J, Woulfe SL, Zacheis M, Schwartz BD. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci U S A. 1988;85:7322–6. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koike K, Uchiumi T, Ohga T, Toh S, Wada M, Kohno K, Kuwano M. Nuclear translocation of the Y-box binding protein by ultraviolet irradiation. FEBS Lett. 1997;417:390–4. doi: 10.1016/S0014-5793(97)01296-9. [DOI] [PubMed] [Google Scholar]
- 4.Higashi K, Inagaki Y, Suzuki N, Mitsui S, Mauviel A, Kaneko H, Nakatsuka I. Y-box-binding protein YB-1 mediates transcriptional repression of human alpha 2(I) collagen gene expression by interferon-gamma. J Biol Chem. 2003;278:5156–62. doi: 10.1074/jbc.M208724200. [DOI] [PubMed] [Google Scholar]
- 5.Basaki Y, Hosoi F, Oda Y, Fotovati A, Maruyama Y, Oie S, Ono M, Izumi H, Kohno K, Sakai K, et al. Akt-dependent nuclear localization of Y-box-binding protein 1 in acquisition of malignant characteristics by human ovarian cancer cells. Oncogene. 2007;26:2736–46. doi: 10.1038/sj.onc.1210084. [DOI] [PubMed] [Google Scholar]
- 6.Evdokimova V, Sorokin A. YBX1 (Y box binding protein 1) Atlas Genet Cytogenet Oncol Haematol. 2011;15:598–604. [Google Scholar]
- 7.Evdokimova V, Ruzanov P, Anglesio MS, Sorokin AV, Ovchinnikov LP, Buckley J, Triche TJ, Sonenberg N, Sorensen PH. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol Cell Biol. 2006;26:277–92. doi: 10.1128/MCB.26.1.277-292.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evdokimova V, Ruzanov P, Imataka H, Raught B, Svitkin Y, Ovchinnikov LP, Sonenberg N. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J. 2001;20:5491–502. doi: 10.1093/emboj/20.19.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolfini D, Mantovani R. YB-1 (YBX1) does not bind to Y/CCAAT boxes in vivo. Oncogene. 2013;32:4189–90. doi: 10.1038/onc.2012.521. [DOI] [PubMed] [Google Scholar]
- 10.Tafuri SR, Wolffe AP. DNA binding, multimerization, and transcription stimulation by the Xenopus Y box proteins in vitro. New Biol. 1992;4:349–59. [PubMed] [Google Scholar]
- 11.Swamynathan SK, Nambiar A, Guntaka RV. Role of single-stranded DNA regions and Y-box proteins in transcriptional regulation of viral and cellular genes. FASEB J. 1998;12:515–22. doi: 10.1096/fasebj.12.7.515. [DOI] [PubMed] [Google Scholar]
- 12.Kuwano M, Oda Y, Izumi H, Yang SJ, Uchiumi T, Iwamoto Y, Toi M, Fujii T, Yamana H, Kinoshita H, et al. The role of nuclear Y-box binding protein 1 as a global marker in drug resistance. Mol Cancer Ther. 2004;3:1485–92. [PubMed] [Google Scholar]
- 13.Hasegawa SL, Doetsch PW, Hamilton KK, Martin AM, Okenquist SA, Lenz J, Boss JM. DNA binding properties of YB-1 and dbpA: binding to double-stranded, single-stranded, and abasic site containing DNAs. Nucleic Acids Res. 1991;19:4915–20. doi: 10.1093/nar/19.18.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ise T, Nagatani G, Imamura T, Kato K, Takano H, Nomoto M, Izumi H, Ohmori H, Okamoto T, Ohga T, et al. Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res. 1999;59:342–6. [PubMed] [Google Scholar]
- 15.Gaudreault I, Guay D, Lebel M. YB-1 promotes strand separation in vitro of duplex DNA containing either mispaired bases or cisplatin modifications, exhibits endonucleolytic activities and binds several DNA repair proteins. Nucleic Acids Res. 2004;32:316–27. doi: 10.1093/nar/gkh170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das S, Chattopadhyay R, Bhakat KK, Boldogh I, Kohno K, Prasad R, Wilson SH, Hazra TK. Stimulation of NEIL2-mediated oxidized base excision repair via YB-1 interaction during oxidative stress. J Biol Chem. 2007;282:28474–84. doi: 10.1074/jbc.M704672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohga T, Koike K, Ono M, Makino Y, Itagaki Y, Tanimoto M, Kuwano M, Kohno K. Role of the human Y box-binding protein YB-1 in cellular sensitivity to the DNA damaging agents cisplatin, mitomycin C, and ultraviolet light. Cancer Res. 1996;56:4224–8. [PubMed] [Google Scholar]
- 18.Bargou RC, Jürchott K, Wagener C, Bergmann S, Metzner S, Bommert K, Mapara MY, Winzer KJ, Dietel M, Dörken B, et al. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat Med. 1997;3:447–50. doi: 10.1038/nm0497-447. [DOI] [PubMed] [Google Scholar]
- 19.Stein U, Jürchott K, Walther W, Bergmann S, Schlag PM, Royer HD. Hyperthermia-induced nuclear translocation of transcription factor YB-1 leads to enhanced expression of multidrug resistance-related ABC transporters. J Biol Chem. 2001;276:28562–9. doi: 10.1074/jbc.M100311200. [DOI] [PubMed] [Google Scholar]
- 20.Jurchott K, Bergmann S, Stein U, Walther W, Janz M, Manni I, Piaggio G, Fietze E, Dietel M, Royer HD. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J Biol Chem. 2003;278:27988–96. doi: 10.1074/jbc.M212966200. [DOI] [PubMed] [Google Scholar]
- 21.Bader AG, Vogt PK. Inhibition of protein synthesis by Y box-binding protein 1 blocks oncogenic cell transformation. Mol Cell Biol. 2005;25:2095–106. doi: 10.1128/MCB.25.6.2095-2106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutherland BW, Kucab J, Wu J, Lee C, Cheang MC, Yorida E, Turbin D, Dedhar S, Nelson C, Pollak M, et al. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005;24:4281–92. doi: 10.1038/sj.onc.1208590. [DOI] [PubMed] [Google Scholar]
- 23.Sorokin AV, Selyutina AA, Skabkin MA, Guryanov SG, Nazimov IV, Richard C, Th’ng J, Yau J, Sorensen PH, Ovchinnikov LP, et al. Proteasome-mediated cleavage of the Y-box-binding protein 1 is linked to DNA damage stress response. EMBO J. 2005;24:3602–12. doi: 10.1038/sj.emboj.7600830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shibao K, Takano H, Nakayama Y, Okazaki K, Nagata N, Izumi H, Uchiumi T, Kuwano M, Kohno K, Itoh H. Enhanced coexpression of YB-1 and DNA topoisomerase II alpha genes in human colorectal carcinomas. Int J Cancer. 1999;83:732–7. doi: 10.1002/(SICI)1097-0215(19991210)83:6<732::AID-IJC6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 25.Bergmann S, Royer-Pokora B, Fietze E, Jürchott K, Hildebrandt B, Trost D, Leenders F, Claude JC, Theuring F, Bargou R, et al. YB-1 provokes breast cancer through the induction of chromosomal instability that emerges from mitotic failure and centrosome amplification. Cancer Res. 2005;65:4078–87. doi: 10.1158/0008-5472.CAN-04-4056. [DOI] [PubMed] [Google Scholar]
- 26.Evdokimova V, Tognon C, Ng T, Ruzanov P, Melnyk N, Fink D, Sorokin A, Ovchinnikov LP, Davicioni E, Triche TJ, et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15:402–15. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 28.Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12:162–9. doi: 10.1016/S0959-437X(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 29.Sorokin AV, Kim ER, Ovchinnikov LP. Proteasome system of protein degradation and processing. Biochemistry (Mosc) 2009;74:1411–42. doi: 10.1134/S000629790913001X. [DOI] [PubMed] [Google Scholar]
- 30.Rogers SW, Rechsteiner MC. Microinjection studies on selective protein degradation: relationships between stability, structure, and location. Biomed Biochim Acta. 1986;45:1611–8. [PubMed] [Google Scholar]
- 31.Rechsteiner M. Natural substrates of the ubiquitin proteolytic pathway. Cell. 1991;66:615–8. doi: 10.1016/0092-8674(91)90104-7. [DOI] [PubMed] [Google Scholar]
- 32.Ghoda L, van Daalen Wetters T, Macrae M, Ascherman D, Coffino P. Prevention of rapid intracellular degradation of ODC by a carboxyl-terminal truncation. Science. 1989;243:1493–5. doi: 10.1126/science.2928784. [DOI] [PubMed] [Google Scholar]
- 33.Kim JE, Minter-Dykhouse K, Chen J. Signaling networks controlled by the MRN complex and MDC1 during early DNA damage responses. Mol Carcinog. 2006;45:403–8. doi: 10.1002/mc.20221. [DOI] [PubMed] [Google Scholar]
- 34.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–95. doi: 10.1016/S0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 35.Shibahara K, Sugio K, Osaki T, Uchiumi T, Maehara Y, Kohno K, Yasumoto K, Sugimachi K, Kuwano M. Nuclear expression of the Y-box binding protein, YB-1, as a novel marker of disease progression in non-small cell lung cancer. Clin Cancer Res. 2001;7:3151–5. [PubMed] [Google Scholar]
- 36.Janz M, Harbeck N, Dettmar P, Berger U, Schmidt A, Jürchott K, Schmitt M, Royer HD. Y-box factor YB-1 predicts drug resistance and patient outcome in breast cancer independent of clinically relevant tumor biologic factors HER2, uPA and PAI-1. Int J Cancer. 2002;97:278–82. doi: 10.1002/ijc.1610. [DOI] [PubMed] [Google Scholar]
- 37.Rubinstein DB, Stortchevoi A, Boosalis M, Ashfaq R, Guillaume T. Overexpression of DNA binding protein B gene product in breast cancer as detected by in vitro-generated combinatorial human immunoglobulin libraries. Cancer Res. 2002;62:4985–91. [PubMed] [Google Scholar]
- 38.Yahata H, Kobayashi H, Kamura T, Amada S, Hirakawa T, Kohno K, Kuwano M, Nakano H. Increased nuclear localization of transcription factor YB-1 in acquired cisplatin-resistant ovarian cancer. J Cancer Res Clin Oncol. 2002;128:621–6. doi: 10.1007/s00432-002-0386-6. [DOI] [PubMed] [Google Scholar]
- 39.Saji H, Toi M, Saji S, Koike M, Kohno K, Kuwano M. Nuclear expression of YB-1 protein correlates with P-glycoprotein expression in human breast carcinoma. Cancer Lett. 2003;190:191–7. doi: 10.1016/S0304-3835(02)00590-6. [DOI] [PubMed] [Google Scholar]
- 40.Giménez-Bonafé P, Fedoruk MN, Whitmore TG, Akbari M, Ralph JL, Ettinger S, Gleave ME, Nelson CC. YB-1 is upregulated during prostate cancer tumor progression and increases P-glycoprotein activity. Prostate. 2004;59:337–49. doi: 10.1002/pros.20023. [DOI] [PubMed] [Google Scholar]
- 41.Yasen M, Kajino K, Kano S, Tobita H, Yamamoto J, Uchiumi T, Kon S, Maeda M, Obulhasim G, Arii S, et al. The up-regulation of Y-box binding proteins (DNA binding protein A and Y-box binding protein-1) as prognostic markers of hepatocellular carcinoma. Clin Cancer Res. 2005;11:7354–61. doi: 10.1158/1078-0432.CCR-05-1027. [DOI] [PubMed] [Google Scholar]
- 42.Schittek B, Psenner K, Sauer B, Meier F, Iftner T, Garbe C. The increased expression of Y box-binding protein 1 in melanoma stimulates proliferation and tumor invasion, antagonizes apoptosis and enhances chemoresistance. Int J Cancer. 2007;120:2110–8. doi: 10.1002/ijc.22512. [DOI] [PubMed] [Google Scholar]
- 43.Xu W, Zhou L, Qin R, Tang H, Shen H. Nuclear expression of YB-1 in diffuse large B-cell lymphoma: correlation with disease activity and patient outcome. Eur J Haematol. 2009;83:313–9. doi: 10.1111/j.1600-0609.2009.01285.x. [DOI] [PubMed] [Google Scholar]
- 44.Shibahara K, Uchiumi T, Fukuda T, Kura S, Tominaga Y, Maehara Y, Kohno K, Nakabeppu Y, Tsuzuki T, Kuwano M. Targeted disruption of one allele of the Y-box binding protein-1 (YB-1) gene in mouse embryonic stem cells and increased sensitivity to cisplatin and mitomycin C. Cancer Sci. 2004;95:348–53. doi: 10.1111/j.1349-7006.2004.tb03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chatterjee M, Rancso C, Stühmer T, Eckstein N, Andrulis M, Gerecke C, Lorentz H, Royer HD, Bargou RC. The Y-box binding protein YB-1 is associated with progressive disease and mediates survival and drug resistance in multiple myeloma. Blood. 2008;111:3714–22. doi: 10.1182/blood-2007-05-089151. [DOI] [PubMed] [Google Scholar]
- 46.Shiota M, Izumi H, Tanimoto A, Takahashi M, Miyamoto N, Kashiwagi E, Kidani A, Hirano G, Masubuchi D, Fukunaka Y, et al. Programmed cell death protein 4 down-regulates Y-box binding protein-1 expression via a direct interaction with Twist1 to suppress cancer cell growth. Cancer Res. 2009;69:3148–56. doi: 10.1158/0008-5472.CAN-08-2334. [DOI] [PubMed] [Google Scholar]
- 47.Gao Y, Fotovati A, Lee C, Wang M, Cote G, Guns E, Toyota B, Faury D, Jabado N, Dunn SE. Inhibition of Y-box binding protein-1 slows the growth of glioblastoma multiforme and sensitizes to temozolomide independent O6-methylguanine-DNA methyltransferase. Mol Cancer Ther. 2009;8:3276–84. doi: 10.1158/1535-7163.MCT-09-0478. [DOI] [PubMed] [Google Scholar]
- 48.To K, Fotovati A, Reipas KM, Law JH, Hu K, Wang J, Astanehe A, Davies AH, Lee L, Stratford AL, et al. Y-box binding protein-1 induces the expression of CD44 and CD49f leading to enhanced self-renewal, mammosphere growth, and drug resistance. Cancer Res. 2010;70:2840–51. doi: 10.1158/0008-5472.CAN-09-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stenina OI, Shaneyfelt KM, DiCorleto PE. Thrombin induces the release of the Y-box protein dbpB from mRNA: a mechanism of transcriptional activation. Proc Natl Acad Sci U S A. 2001;98:7277–82. doi: 10.1073/pnas.121592298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guay D, Evoy AA, Paquet E, Garand C, Bachvarova M, Bachvarov D, Lebel M. The strand separation and nuclease activities associated with YB-1 are dispensable for cisplatin resistance but overexpression of YB-1 in MCF7 and MDA-MB-231 breast tumor cells generates several chemoresistance signatures. Int J Biochem Cell Biol. 2008;40:2492–507. doi: 10.1016/j.biocel.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 51.Ohga T, Uchiumi T, Makino Y, Koike K, Wada M, Kuwano M, Kohno K. Direct involvement of the Y-box binding protein YB-1 in genotoxic stress-induced activation of the human multidrug resistance 1 gene. J Biol Chem. 1998;273:5997–6000. doi: 10.1074/jbc.273.11.5997. [DOI] [PubMed] [Google Scholar]
- 52.Kaszubiak A, Kupstat A, Müller U, Hausmann R, Holm PS, Lage H. Regulation of MDR1 gene expression in multidrug-resistant cancer cells is independent from YB-1. Biochem Biophys Res Commun. 2007;357:295–301. doi: 10.1016/j.bbrc.2007.03.145. [DOI] [PubMed] [Google Scholar]
- 53.Evdokimova VM, Wei CL, Sitikov AS, Simonenko PN, Lazarev OA, Vasilenko KS, Ustinov VA, Hershey JW, Ovchinnikov LP. The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J Biol Chem. 1995;270:3186–92. doi: 10.1074/jbc.270.7.3186. [DOI] [PubMed] [Google Scholar]
- 54.Davydova EK, Evdokimova VM, Ovchinnikov LP, Hershey JW. Overexpression in COS cells of p50, the major core protein associated with mRNA, results in translation inhibition. Nucleic Acids Res. 1997;25:2911–6. doi: 10.1093/nar/25.14.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sorokin AV, Chen J. MEMO1, a new IRS1-interacting protein, induces epithelial-mesenchymal transition in mammary epithelial cells. Oncogene. 2013;32:3130–8. doi: 10.1038/onc.2012.327. [DOI] [PubMed] [Google Scholar]
- 56.Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–16. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.