Abstract
Centrosomes are comprised of 2 orthogonally arranged centrioles surrounded by the pericentriolar material (PCM), which serves as the main microtubule organizing center of the animal cell. More importantly, centrosomes also control spindle polarity and orientation during mitosis. Recently, we and other investigators discovered that several nucleoporins play critical roles during cell division. Here, we show that nucleoporin Nup62 plays a novel role in centrosome integrity. Knockdown of Nup62 induced mitotic arrest in G2/M phases and mitotic cell death. Depletion of Nup62 using RNA interference results in defective centrosome segregation and centriole maturation during the G2 phase. Moreover, Nup62 depletion in human cells leads to the appearance of multinucleated cells and induces the formation of multipolar centrosomes, centriole synthesis defects, dramatic spindle orientation defects, and centrosome component rearrangements that impair cell bi-polarity. Our results also point to a potential role of Nup62 in targeting gamma-tubulin and SAS-6 to the centrioles.
Keywords: nucleoporin, centrosome, centriole, Nup62, gamma-tubulin, SAS-6
Introduction
In eukaryotes, the nuclear envelope (NE) forms a membrane barrier around the chromosomes. Most of the molecular transportation in and out of the nucleus is mediated by nuclear pore complexes (NPC), which are large multiprotein channels composed of ~30 different nuclear pore proteins (Nups) that span the NE.1-3 Multiples of 8 units of each subcomplex roughly assemble into the segmental structures that span the nuclear envelope, creating pores that allow for the regulated transport of macromolecules into and out of the nucleus.4-8 In addition to mediating transport, nuclear pore complexes have also been implicated in cell division.
The mammalian protein Nup62 and its yeast homolog Nps1p are found in 2 distinct NPC sub-complexes, the Nsp1p-Nup57p-Nup49p-Nic96p and the Nsp1p-Nup82p-Nup159p complex.9 Notably, Nsp1 was used to form an in vitro phenylalanine-glycine (FG)-repeat hydrogel consisting of this single nucleoporin only that was able to mimic fundamental aspects of selective protein trafficking.10 This suggests that core biophysical features of the nuclear pore central channel are contained in the amino acid sequence of Nup62/Nsp1. Recently, Arabidopsis Nup62 was found through its sequence similarity to mammalian Nup62 and yeast Nsp1p. Overexpression-based co-suppression of AtNup62 leads to severely dwarfed, early flowering plants, suggesting an important function for Nup62 in plants.11
The mammalian Nup62 subcomplex assembles from O-glycosylated proteins of molecular masses 62, 58, 54, and 45 kDa.12,13 The 62-kDa component of the complex, Nup62, contains 3 domains: N-terminal FG-repeat, central threonine/alanine-rich linker, and C-terminal α-helical coiled-coil. The N-terminal FG-rich region of Nup62 serves as a docking site for NTF2 (nuclear transport factor 2),14 while the C terminus of Nup62 is predicted to adopt a coiled-coil structure and to facilitate the anchoring of Nup62 to the NPC.1,15 The C-terminus of Nup62 has been shown to interact with the transport receptor importin-β in vitro16 and to mediate interactions with other members of the Nup62 complex, including the NPC proteins Nup58, Nup54, and Nup45.17-19 The mucin 1 C-terminal subunit (MUC1-C) was reported to interact directly with the Nup62 central domain and indirectly with the Nup62 C-terminal α-helical coiled-coil domain.20 Similarly, Nup62 was reported to bind heat shock proteins, hsp90, hsp70, p23, and the TPR domain proteins FKBP52 and PP5 during nuclear importation.21 Nup62 is also reported to bind the N-terminal domain of the exocyst complex component Exo70 through its coiled-coil domain but not through its FG-repeat domain.22 Clinically, Nup62 was also suggested to play a role in human immunodeficiency virus type 1 (HIV-1) nucleocytoplasmic shuttling23 and in the degeneration of the basal ganglia. In humans, Nup62 mutations cause autosomal recessive infantile bilateral striatal necrosis.24
Our recent findings revealed that several NPC proteins, such as RNA export factor 1 (Rae1),25-28 Nup98,29 Tpr,30 Nup88,31 and Nup35832 do not simply disperse into the mitotic cytoplasm, but instead preferentially associate with kinetochores, mitotic spindles, and centrosomes, where they are crucial in maintaining spindle bipolarity and thus prevent aneuploidy and carcinogenesis.5 Despite these advances, the role of Nup62 during mitosis has not been investigated. Therefore, we investigated the mitotic role of Nup62.
The centrosome is a small cytoplasmic non-membranous organelle capable of duplicating itself once per cell cycle under normal conditions. This process is initiated by the splitting of mother and daughter centrioles, most likely through the regulation of centriole components (e.g., Ninein, SAS-6, and C-Nap1) and kinases (e.g., Plk4).33 Centrioles are also essential for the formation of cilia and flagella.34 Thus, centrosome duplication is initiated in mammalian cells during late G1 phase, as daughter centrioles begin to grow semi-conservatively from their parents. During S and G2 phases, centrioles continue to elongate, and during this time, centrosomes are situated near the nucleus and lie in proximity to one another. However, as cells enter the prophase, the centrosomes begin to separate, migrating to opposite poles and establishing the mitotic spindle.35
Here, we show that Nup62 is critical for centrosome and centriole homeostasis in mammalian cells.
Results
Nup62 down-modulation induces G2/M phase arrest, mitotic cell death, and aberrant centrosome/centriole formation
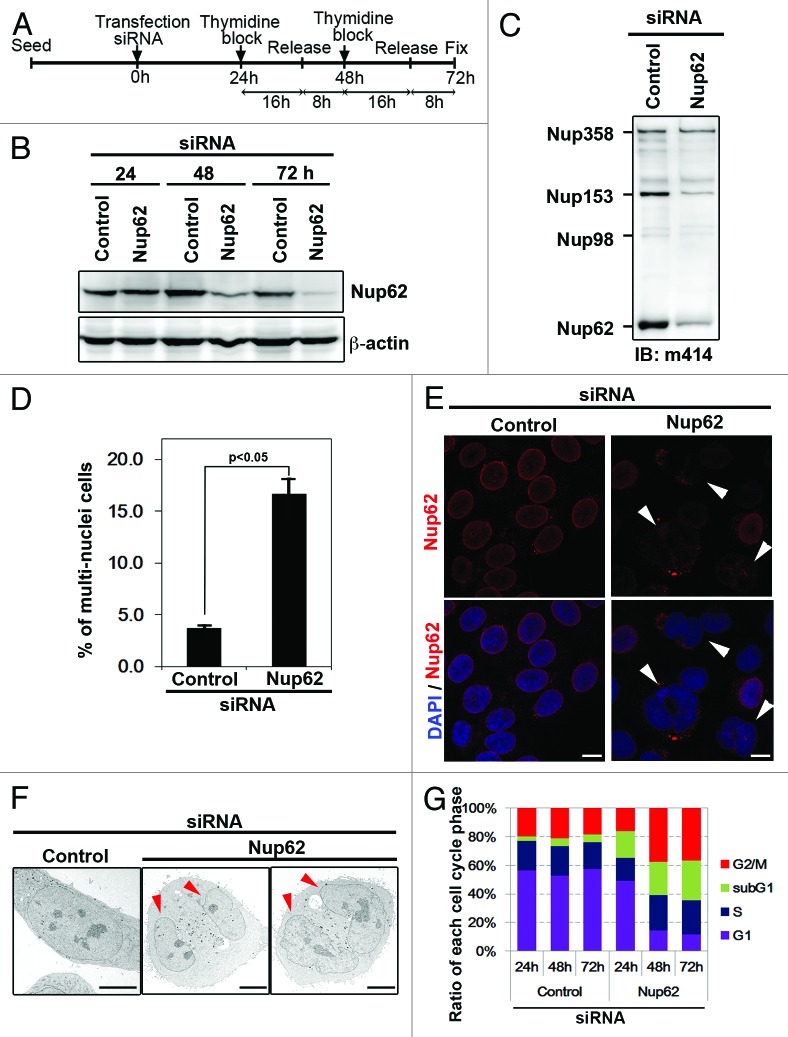
To understand the mitotic role of Nup62 in cell division, we used siRNAs to inhibit Nup62 expression in HeLa cells. Immunoblot analysis revealed that the Nup62 siRNA could reduce its expression in a time-dependent manner (Fig. 1A). After 72 h, Nup62 expression in siRNA-transfected HeLa cells was 85% lower than in controls (Fig. 1B). The reduction of Nup62 was most obvious 3 d post-transfection. Therefore, 3 d post-transfection was chosen as the analysis time point for further experiments throughout this study. The same immunoblot membrane was reprobed with β-actin to ensure equivalent loading. We also checked other FG nucleoporins with the m414 antibody, and we found that only Nup153 showed reduced expression (Fig. 1C). To identify potential defects after Nup62 down-modulation, cells were fixed, stained with fluorescent markers for Nup62 (red) and DNA (blue), and examined by confocal microscopy. We found that the number of multinucleated cells was dramatically higher in Nup62-depleted cells compared with control siRNA cells (P < 0.05) (Fig. 1D and E). Many cells had aberrant nuclei that appeared to be composed of fragmented or interconnected micronuclei. Consistently, multinucleated cells were also detectable by electron microscopy (EM). As shown in Figure 1F, Nup62-siRNA transfection led to significantly more multi-nucleated cells. Next, we performed cell cycle analysis. As shown in Figure 1G, compared with control siRNA-transfected cells, the Nup62-siRNA-treated HeLa cells exhibited a marked increase in the percentage of cells in the G2/M phase. To rule out the possibility that Nup62’s effect on the cell cycle might arise from mitotic delay rather than mitotic arrest, we monitored cell cycle distribution at 24, 48, and 72 h by flow cytometry (Fig. S1). Cell cycle arrest should result in a time-dependent increase in the percentage of cells in G2, while a delay will probably not affect the distribution beyond the difference observed. We found that Nup62 depletion led to a larger dead cell fraction (subG1), G2/M arrest, and a concomitant decrease in the percentage of cells in the G1 phase 48 and 72 h after transfection (Fig. 1G; Fig. S1). Together, these data suggest that Nup62 depletion induces multi-nuclei formation and leads to cell cycle arrest during S and G2/M phases. These data also indicate that in addition to its role in nuclear transport and assembly, Nup62 plays a role in cell cycle regulation.
Figure 1.
Nup62 regulates proper chromosome segregation and cell cycle progression. (A) Schedule of collecting cells after Nup62 depletion via siRNA transfection. (B) HeLa cells transfected with control or Nup62 siRNA, collected at 24, 48, and 72 h after transfection, were analyzed by immunoblot (IB) for Nup62 and β-actin expression. (C) HeLa cells transfected with control or Nup62 siRNA, collected at 72 h after transfection, were analyzed by IB for anti-m414 antibody. (D) Quantification (relative %) of multi-nuclei phenotypes in control or Nup62 siRNA-transfected cells. Values are based on 3 independent experiments, counting 100 cells in each experiment. Mean values ± SD (error bars) are shown. (E) Representative images of asynchronous HeLa cells, transfected with control or Nup62 siRNA. Seventy-two hours after transfection, cells were stained with an anti-Nup62 antibody (red) and analyzed by confocal laser microscopy. Chromatin was stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar, 5 μm. White arrows indicate cells with multi-nuclei when Nup62 was completely depleted. (F) HeLa cells transfected with control or Nup62 siRNA, fixed at 72 h after transfection, were visualized by electron microscopy. Red arrowheads indicated each of 2 nuclei in the cell shown. (G) Representative cell cycle distributions after transfection with control or Nup62 siRNA for 24, 48, and 72 h. Percentage of G1, G2/M, S, and sub-G1 cells were calculated.
Nup62’s role in centrosome and centriole formation
To explore the potential role of Nup62 in the regulation of the cell cycle and cell fate, we next asked whether Nup62 depletion disrupts normal NPC components and eventually affects the nuclear trafficking process. To further characterize the effect of Nup62 knockdown on nano-scale structural characteristics; we decided to investigate the effect of Nup62 depletion on NPC numbers and morphology using high-resolution EM during interphase. We performed thin-section transmission electron microscopy to gain more detailed physiological observations of the structural components of the HeLa cell NPC. However, unlike our recent findings for Tpr,36 we did not observe significant visible differences between Nup62 siRNA and control siRNA cells (each n = 20 cells) in terms of NPC gross morphology (data not shown). Consistent with recent immunolabeling data,37 the absence of changes in the EM observations suggests that removal of Nup62 does not disrupt nuclear pores or overtly alter their distribution.
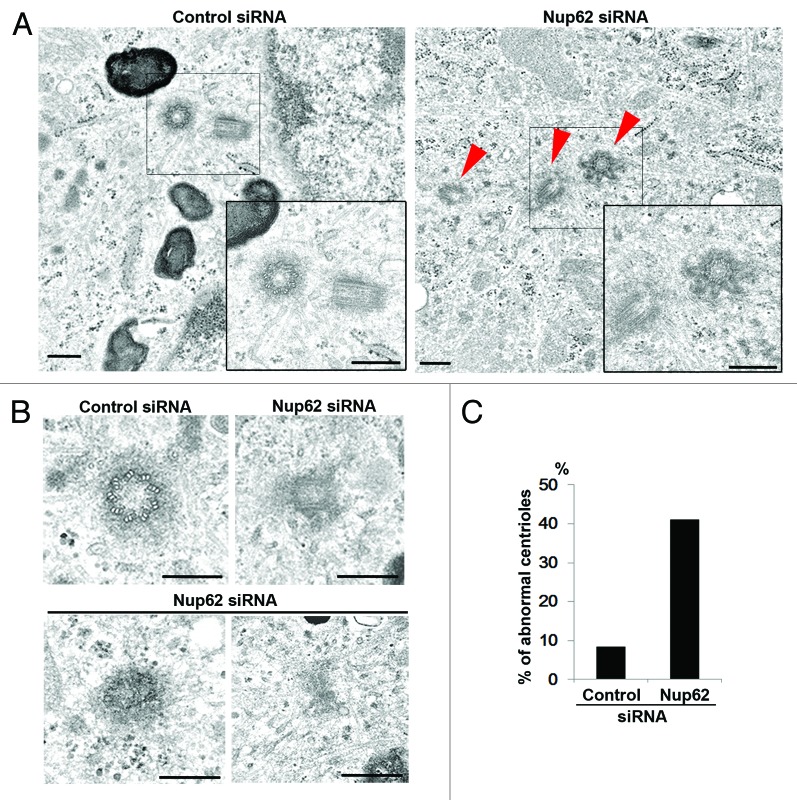
As shown in Figure 2A (left panel), we found normal orthogonally arranged centrioles of 9 triplet microtubules surrounded by electron-dense pericentriolar material (PCM) in control siRNA-depleted cells. In the Nup62-depleted cells shown in Figure 2A (right panel), the centrosome contained 3 centrioles compared with the 2 centrioles typically found in control cells during late G1 to early S phase. A more severe abnormality was found in the shape of the third centriole at high magnification, where it formed a protruding “swelling star” structure from the centriolar cylinder (right panel). Figure 2B showed various degrees of morphological abnormality in centrioles detected in Nup62 siRNA-depleted cells. The classical orthogonal configuration and accurate centriole duplication that characterize control cells were severely disrupted in Nup62 siRNA-transfected cells. We randomly selected a dozen EM images with centrioles from control siRNA or Nup62 siRNA. To our surprise, we noted that over 40% of Nup62 siRNA-depleted cells (n = 12) contained aberrant centrioles and only 8% in control siRNA cells (n = 12) (Fig. 2C). These data indicate that Nup62 function might contribute to centriole formation and integrity.
Figure 2.
Nup62 regulates proper centriole configuration. (A and B) Electron microscopic images of control or Nup62 siRNA-transfected HeLa cells. (A). Orthogonal configuration of 2 centrioles (control siRNA); red arrowhead indicates abnormal configuration of 3 centrioles (Nup62 siRNA). Scale bars, 250 nm. Insets are magnifications of the centriole areas in the observed cells (B). A typical centriole with 9 triplet microtubules (control siRNA) and abnormal centriole configuration (Nup62 siRNA). Scale bars, 250 nm (C). Percentage of abnormal centrioles of control or Nup62 siRNA-transfected HeLa cells from EM.
Depleting Nup62 induces the formation of supernumerary centrosomes and leads to aberrant spindle orientation
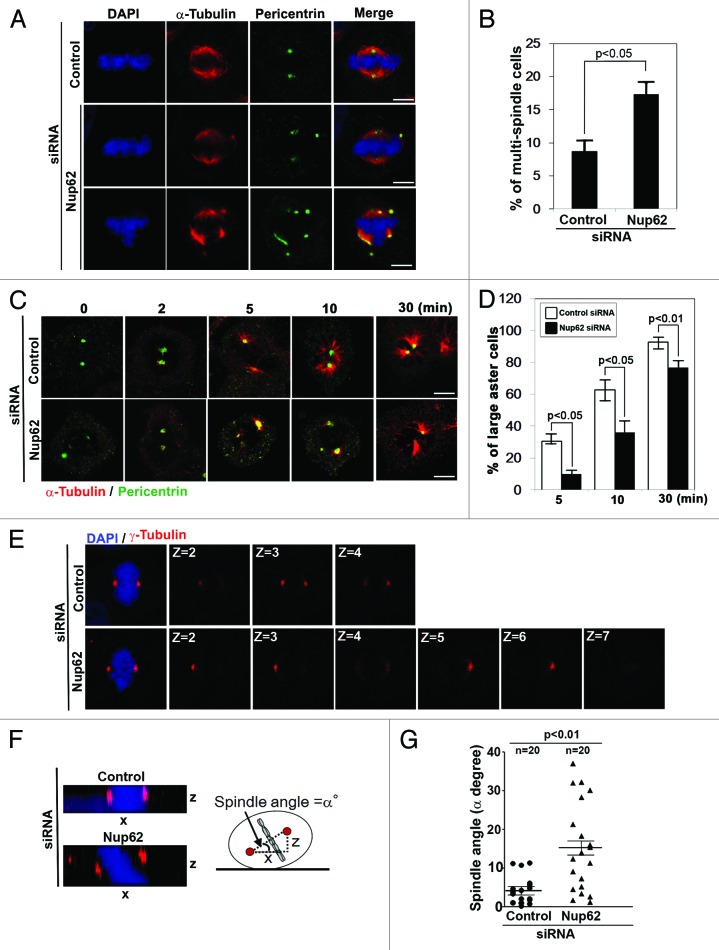
The above-described results prompted us to examine the consequence of Nup62 depletion on mitotic centrosomes. To determine a possible role for Nup62 in regulating centrosomal formation, we transiently transfected HeLa cells with control siRNA or Nup62 siRNA and examined the status of the centrosomes 72 h after transfection. Nup62 depletion produced a much greater number of cells with centrosome amplification and multiple spindles as compared with siRNA control cells (Fig. 3A and B). In control cells, multipolar spindles were found in 8.7% of total cells, whereas in Nup62-depleted cells, the percentage dramatically increased to 17.3% (Fig. 3A and B, n = 300 cells). In addition, we also observed that down-modulation of Nup62 in HeLa cells results in a less prominent MT aster when compared with control siRNA cells during re-polymerization of the MT network (Fig. 3C and D, n = 100).
Figure 3.
Nup62 is required for metaphase spindle maintenance, microtubule nucleation, and centrosome positioning. (A, C, E, and F) Confocal microscopic images of control or Nup62 siRNA-transfected HeLa cells. Scale bars, 5 μm. (A) Cells were stained for anti-α-tubulin (red) and anti-Pericentrin (green) antibodies. (B) The proportion of metaphase cells with multiple spindles characterized by displaced centrosomes was quantified. Values are based on 3 independent experiments counting 100 mitotic cells in each experiment mean values ± SD (error bars) are shown with the Student t test P value. (C) Following thymidine release for 4 h, cells were treated with nocodazole for 4 h to depolymerize microtubules. Nocodazole was then washed out and cells incubated on ice for 30 min. To allow microtubule regrowth, cells were then incubated in medium at 37 °C and stopped at the indicated time points by fixation and stained for anti-α-tubulin (red) and anti-Pericentrin (green) antibodies. (D) The ratio of mitotic cells harboring large asters (>5 µm) at indicated time points. Values are based on 3 independent experiments counting 100 mitotic cells in each experiment. Mean values ± SD (error bars) are shown with the Student t test P values. (E) Individual z-stacks of anti-γ-tubulin staining (red) in control or Nup62 siRNA-transfected HeLa cells. Overlay images (left panel) show maximum intensity projections of z-stacks. (F) Orthogonal slice along spindle polarity axis viewed from a side-on perspective of γ-tubulin immunostained z-sections reveals spindle rotation in z-axis. “α” is the angle of rotation from planar orientation. (G) The angle of spindle rotation was measured in control or Nup62 siRNA-treated HeLa cells and plotted on a vertical scatter plot. Horizontal lines depict means ± s.e.m. and n values are the total number of cells counted from two independent experiments with the Student t test P values.
In addition, we found that in the control group, the majority of mitotic cells exhibited normal bipolar spindles, with both spindle poles on the same focal plane. However, in the Nup62 siRNA groups, approximately 25% of mitotic cells showed disorganized spindles (without clear spindle poles) or disoriented spindles (exhibiting two spindle poles but at different focal planes) (Fig. 3A and B). These data suggest that Nup62 might be involved in spindle orientation.
During mitosis, centrosomes participate in both the organization and orientation of the mitotic spindle and, thus, the arrangement and segregation of chromosomes. Control of spindle orientation is essential for defining the plane of cell division during the symmetric and asymmetric divisions of stem cells and epithelial cells.38 Indeed, Nup62 depletion resulted in abnormal metaphase spindles characterized by a displacement of centrosomes from the spindle (Fig. 3E). The metaphase spindle polarity axis in Nup62-depleted cells displayed substantial rotation in the z-axis (Fig. 3G). The rotation of the spindle in the z-axis was more clearly seen by orthogonal sectioning through the z-stacks (Fig. 3F). We measured the angle between the mitotic spindle axis and the growth plane. As shown in Figure 3F, the majority of control cells assembled bipolar spindles parallel to the growth plane with a spindle angle of 0–10°. In contrast, Nup62 siRNA resulted in severe defects in spindle orientation, as evidenced by the much wider distribution of spindle angles 10–40° (Fig. 3G). Thus, these data suggest that centrosomal Nup62 is also required for the organization and orientation of the mitotic spindle.
Nup62 localization to centrosomes during cell division
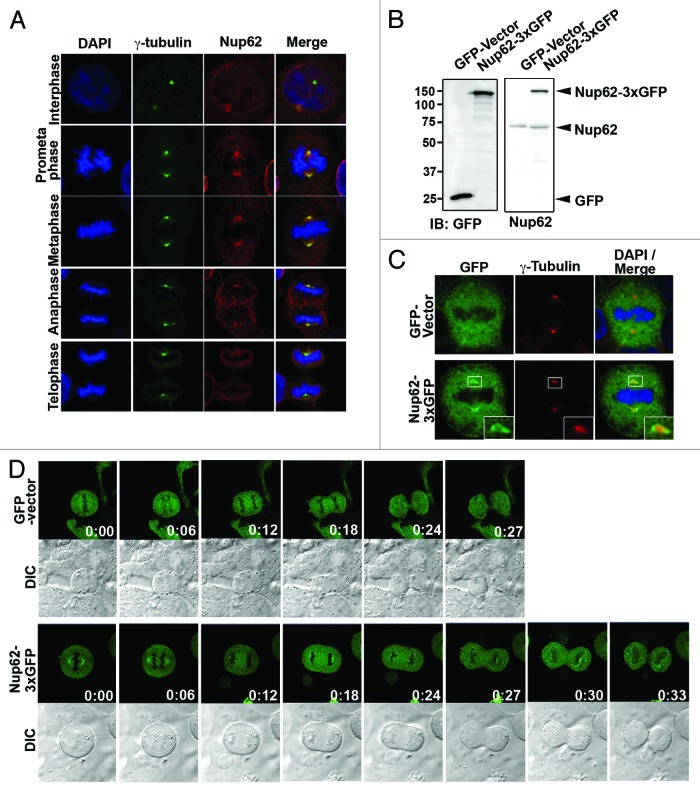
To confirm the Nup62 mitotic topography with respect to the centrosomal apparatus, we used specific antibodies against Nup62 and γ-tubulin (centrosome marker) to examine their localizations at different stages of the cell cycle. Confocal microscopy of HeLa cells during interphase revealed that Nup62 was predominantly distributed on the nuclear envelope, with typical nuclear rim staining, whereas γ-tubulin was localized as prominent spots close to the nucleus (Fig. 4A). From late prophase through anaphase, Nup62 staining overlapped with that of γ-tubulin at the spindle poles/centrosomes (Fig. 4A, metaphase, anaphase). A faint Nup62 signal was also detected on the mitotic spindle. At telophase, Nup62 was detected in the cytoplasm as well as in the newly developed nuclear envelope membrane (Fig. 4A). Identical results were obtained with SW480 human cell lines (data not shown). To further ensure that this centrosomal staining was as a result of Nup62 protein and not an unexpected antibody cross-reaction, we examined the localization of Nup62 by transfecting GFP-tagged Nup62, and similar results were obtained (Fig. 4B and C). Next, we studied GFP-Nup62 in HeLa cells by live cell imaging. A representative series from time-lapse images of GFP-Nup62 during metaphase–anaphase transitions is shown in Figure 4D. We observed a GFP-Nup62 staining pattern at the centrosome during metaphase that dynamically re-localized back to the newly developed chromatin boundary in late anaphase (Fig. 4D; Videos S1 and 2). Taken together, these data plainly pinpoint Nup62 localization to the centrosome during mitosis.
Figure 4.
Nup62 is localized on centrosome during mitosis. (A) Confocal microscopy images of HeLa cells at different cell cycle stages, stained with anti-γ-tubulin (green) and anti-Nup62 (red) antibodies. Chromatin was stained with DAPI (blue). Scale bars, 5 μm. (B) HeLa cells transfected with GFP-vector or Nup62–3xGFP expression plasmid were analyzed by IB for GFP or Nup62–3xGFP and Nup62 expression. (C) Confocal microscopy images of GFP-vector or Nup62–3xGFP expressed HeLa cells, stained with GFP (green) and γ-tubulin (red). Insets are magnifications of the centrosome areas in the observed cells. (D) Time-lapse images of GFP-vector or Nup62–3xGFP-expressed HeLa cells from metaphase to cytokinesis. Images were taken every 3 min.
C-terminal regions of Nup62 involved in its localization to centrosomes
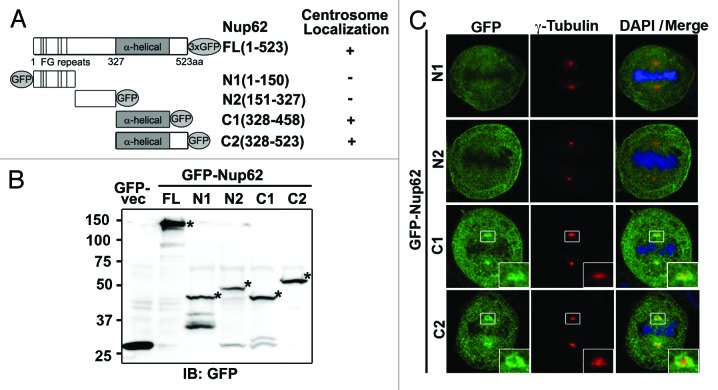
We next investigated which region of Nup62 was required for its targeting to centrosomes. Based on its structural character, we constructed some Nup62 truncated mutants containing residues 1–523 (full-length, FL), 1–150 (N1), 151–327 (N2), 328–458 (C1), or 328–523 (C2) (Fig. 5A and B). We transfected HeLa cells with GFP-tagged full-length or truncated mutants of Nup62 and observed the distributions of these truncated proteins by confocal microscopy. The results showed that ~85% of the cells transfected with GFP-Nup62328–458(C1) or GFP-Nup62328–523(C2) had bright spots co-localized with γ-tubulin. In contrast, we found that a portion of GFP-Nup621–150(N1) and GFP-Nup62151–327(N2) localized around the mitotic spindle. More importantly, GFP-Nup621–150(N1) and GFP-Nup62151–327(N2) did not have obvious centrosomal localization (Fig. 5C). These data suggest that C-terminal (α-helical regions) of Nup62 contain signals promoting its association with centrosomes.
Figure 5.
C-terminal of Nup62 required for centrosome localization. (A) Scheme of full-length Nup62 and 4 fragments with GFP tags. Numbers on the right refer to amino acids. (B) HeLa cells transfected with GFP-vector or an expression plasmid carrying cDNA for GFP tagged full-length Nup62 or Nup62 fragments were analyzed by IB for GFP expression. Numbers indicate molecular mass markers in kilodaltons. Asterisks indicate the GFP fusion proteins. (C) Confocal microscopy images of GFP tagged Nup62 fragments expressing HeLa cells, stained with anti-GFP (green) and anti-γ-tubulin (red) antibodies.
Nup62 associates with γ-tubulin and centriole components
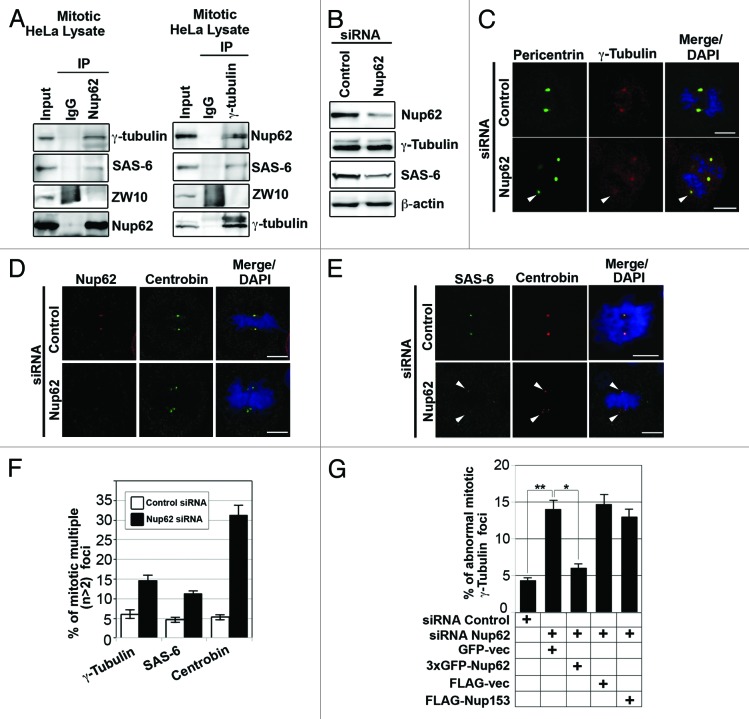
Given the apparent colocalization of Nup62 with γ-tubulin at the centrosome, we sought to investigate whether or not Nup62 physically interacts with γ-tubulin. As predicted, we found that Nup62 colocalized with the centrosomal markers γ-tubulin and SAS-6 in HeLa cells (Fig. 6A). We next examined whether Nup62 was associated with γ-tubulin in HeLa cells that were synchronized by a double thymidine block. Using immunoblotting anti-Nup62 immunoprecipitates, we detected coprecipitating γ-tubulin, SAS-6, but not the kinetochore protein ZW10 (Fig. 6A, left). Conversely, using anti-γ-tubulin antibodies, we immunoprecipitated SAS-6, Nup62, but not ZW10 (Fig. 6A, right). These data suggested that Nup62, along with several other Nups, interacts with γ-tubulin at the spindle pole/centrosome during mitosis. Next, we employed a Nup62 down-modulation approach. Immunoblot analysis of HeLa cells subjected to Nup62 siRNA treatment for 3 d revealed an 85% reduction of Nup62 compared with controls (Fig. 6B). The same immunoblot membrane was re-probed with β-actin to ensure equivalent loading. Interestingly, we did not find any significant changes in γ-tubulin, but we noted reduction of SAS-6 protein expression (Fig. 6B). We observed that when Nup62 was almost completely knocked down, multiple centrosome defects were found (Fig. 6C). Notably, mitotic Nup62 siRNA treatment was consistently associated with abnormal localization of the centriole markers centrobin and SAS-6 (Fig. 6D and E, white arrow). These data also suggest that Nup62 might recruit some centrosomal components (e.g., SAS-6) to the centrosome after nuclear envelope breakdown (NEBD).
Figure 6.
Nup62 is associated with centrosome proteins and knockdown of Nup62 alters centriole component expression and localization. (A) Immunoprecipitation from mitotic HeLa cell extracts with anti-Nup62, anti-γ-tubulin or nonspecific rabbit antibodies (IgG) were analyzed by SDS-PAGE, followed by IB with anti-γ-tubulin, anti- SAS-6, anti-ZW10, or anti-Nup62 antibodies. (B) HeLa cells transfected with control or Nup62 siRNA, collected at 72 h after transfection, were analyzed by IB with anti-Nup62, anti-γ-tubulin, anti- SAS-6, and β-actin antibodies. (C–E) Representative images of mitotic HeLa cells, transfected with control or Nup62 siRNA. Seventy-two hours after transfection, cells were stained with centrosome marker (C) anti-γ-tubulin (red), anti-pericentrin (green), or counter stained with centriole markers (D) anti-centrobin (green) with Nup62 (red); antibodies (E) anti- SAS-6 (red), and centrobin antibody (green), and analyzed by confocal laser microscopy. Chromatin was stained with DAPI (blue). Scale bars, 5 μm. White arrowheads indicate cells with γ-tubulin (red) or SAS-6 (red) mis-localized/mis-targeting to the centrosomes, whereas pericentrin (green) or centrobin (green) as centrosome/centriole marker. Abnormal centriole and multi-centriole phenotypes were often found in Nup62-depleted cells. (F) Quantification (relative %) of multiple foci in the control or Nup62 siRNA-transfected cells. Values are based on three independent experiments counting 100 mitotic cells in each experiment Mean values ± SD (error bars) are shown. All cells were treated with double thymidine block, stained with DAPI, and visualized by confocal microscopy. (G) Quantification (relative percentage) of centrosomal defect phenotypes for the indicated siRNA and/or plasmid. Values are based on 3 independent experiments, counting 100 mitotic cells in each experiment. Mean values ± SD (error bars) are shown. All cells were treated with double thymidine block, stained with anti-γ-tubulin, DAPI and visualized by confocal microscopy.
Moreover, more than 32% of metaphase cells had multiple foci (>2) when counterstained with the daughter centriole maker centrobin, whereas these defects were not found in control siRNA cells (Fig. 6F) in 3 independent experiments (n = 100 mitotic cells). Together, these data indicate that knockdown of Nup62 causes SAS-6 reduction and aberrant defects in centrosome and centriole formation. In addition, we also revealed that a portion of γ-tubulin, centrobin, and SAS-6 was diffuse and mis-localized to the cytoplasm (Fig. 6C–E). In addition, since we found that Nup62 depletion also caused Nup153 reduction, to address whether the observed Nup62 phenotype was an indirect consequence of depleting Nup153 or not, we co-transfected GFP (vector alone), GFP-Nup62, Flag (vector alone), or Flag-Nup153 with siRNA Nup62 into HeLa cells. We observed that only Nup62 but not Nup153 or vector alone could partially rescue the defects (Fig. 6G; Fig. S2). These data confirm the importance of Nup62 in centrosome homeostasis.
Discussion
Nucleoporins, once thought to be solely structural blocks of the nuclear pore with roles only in nuclear transportation, are emerging as regulators of diverse cellular functions.2,39,40 We and other investigators demonstrated that a number of nuclear pore proteins (Nups) play active roles during mitosis.4-8
In the present study, we discovered a novel role for human Nup62 in regulating centrosome homeostasis (Fig. 7). We found that Nup62 localized to centrosomes, and knockdown of Nup62 was able to induce uncontrolled centrosome amplification and abnormal centriole separation. More importantly, Nup62-depleted cells arrested in the G2/M phase owing to the formation of severe chromosome defects. We also showed that Nup62 regulates microtubule aster formation and spindle orientation during mitosis. In addition, we demonstrated that the C-terminal domain of Nup62 localized to the centrosome. Our data converge into a novel role in which Nup62 targets centriole components during NEBD and functions as an assembly and maturation factor for mitotic centrosomes during mitosis.
Figure 7.
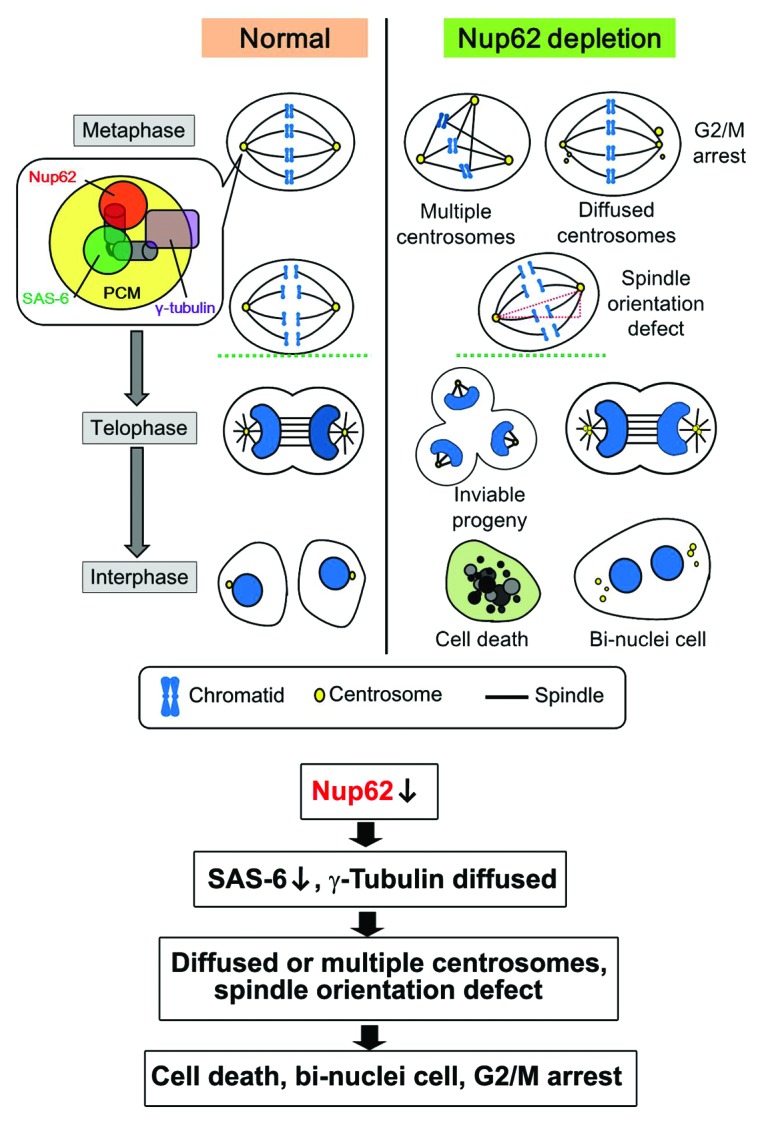
Speculative working model for Nup62 in mitotic progression and centrosomes/centrioles homeostasis. Absence of Nup62 during mitosis caused abnormal mitotic spindles, supernumerary centrosomes, impaired spindle orientation, mitotic arrest, and cell death.
Consistent with the observation in TOV112D-9 cell culture,41 we also observed that Nup62 plays a role in cell cycle entry/exit (cellular dormancy) in HeLa cells. The mechanisms of cancer cell relegation and tumor dormancy are still not well understood. It is worth noting that the centrosome is both structurally and functionally linked to the basal body and the cilium, and some nucleoporins also localize at the basal body and the ciliary axoneme.42,43 Therefore, an interesting question for future study is whether Nup62 is involved in the pathogenesis of ciliary diseases and other disorders.
Because Nup62 siRNA only partially knocks down Nup62 accumulation, the block to cell cycle progression is not absolute and most likely behaves similarly to a weir. The defects in cell cycle progression we detected in the Nup62 siRNA-treated cells (Fig. 1) may be a consequence of the alteration of Nup62 or related nuclear protein expression in these cells. For example, siRNA knockdown of Nup15344 and lamin A/C leads to cell cycle G1 arrest.45 Alternatively, the defects in cell cycle progression could also be explained by a role for Nup62 in regulating cell cycle gene expression, as Nup62 has been recently demonstrated to control gene expression through a Nup62-chromatin interaction at the NPC or within the nucleoplasm.46 Further work is necessary to address whether or not Nup62 subcomplex proteins (Nup54, Nup58) or Nup62-chromatin play a role in regulating cell cycle gene expression.
Although our in vitro studies did not observe significant differences with inhibitors (Fig. S3), further studies will be required to determine the finer detailed post-translational modifications (O-GluNac glycosylation and phosphorylation) of Nup62 during mitosis and their functional consequences.
Our analysis also reveals that Nup62 is essential for the organization and orientation of the mitotic spindle (Fig. 3), which is consistent with the function of the centrosome as the driving force for bipolar spindle assembly in mammalian cells.47,48 At this moment, the precise mechanism of how Nup62 contributes to spindle orientation is unclear. The dynein–dynactin (p150) complex has been shown to exert pulling forces on astral microtubules, ensuring correct orientation of the spindle,49 and to interact with nuclear pore proteins.30 We did not find a significant difference in p150 and NuMA when Nup62 was depleted (data not shown). However, it is also conceivable that Nup62 may participate with other centrosomal components to modulate the pulling forces on astral microtubules that are critical for proper spindle orientation. Indeed, down-modulation of Nup62 reduced the MT astral formation (Fig. 3).
One straightforward observation is that depleting Nup62 altered centrosome/centriole synthesis and segregation. We also found extra mini-centrioles, as if they had been synthesized prematurely and split from their mother centrioles prior to completion of synthesis (Fig. 2). Therefore, it could be suggested that Nup62 may play a role in mechanisms for recruiting those newly duplicated centrosomes/centriole components during nuclear envelope break down. We do not completely exclude the possibility that the mitotic defects (cell cycle arrest, multiple nuclei, spindle mis-orientation, and supernumerary centrosomes) were also related to the role of Nup62 during interphase. We imagine that the concentration of Nup62 could influence sensitivity to centrosome synthesis and segregation during interphase; however, reduction of nucleocytoplasmic transport mediated by Nup62 does not appear to be part of this mechanism.37,41 It is very difficult to imagine that Nup62 is solely responsible for trafficking the majority of centrosome components (>100 proteins) or those proteins responsible for cell cycle arrest.
Our Nup62 centrosomal findings are novel and consistent with proteomic analysis of the human centrosome. Nup62 peptides were identified from in-solution-digested centrosome preparations.50 In recent years, much progress has been made toward assembling a list of human centrosome components and key proteins important for centriole biogenesis, duplication, pericentriolar material (PCM) recruitment, and basal body functions.51 The molecular details of each novel centrosomal component still need to be characterized individually. However, it will surely not be so surprising to find other nucleoporins in the centrosome. We hope to use highly sensitive mass spectrometry and proteomic analysis together with super-resolution fluorescence microscopy techniques in the near future to investigate this area. Furthermore, we found interactions and co-localization with γ-tubulin and SAS-6. Understanding the mechanistic details of Nup62’s function at the centrosome clearly represents a major challenge for future investigation. Moreover, Nup62 is known to be in a complex with Nup54. Whether Nup54 forms a complex with Nup62 during mitosis and plays a role in the centrosome/centriole is another key question to be addressed.
Accumulating evidence suggests that nucleoporins play a role in mitosis. In mitosis, Rae1 binds to microtubules and is required for spindle formation. We found that Rae1 interacts and colocalizes with NuMA (nuclear mitotic apparatus protein)28 and the Cohesin complex,27 which may promote microtubule bundling at spindle poles. In addition to Rae1, the Nup107–160 nucleoporin subcomplex has been implicated in spindle formation using X. laevis egg extracts and in mammalian cells.52 On the other hand, there are contradictory reports that at least a prominent fraction of the NuP107–160 complex localizes to kinetochores in both C. elegans embryos53 and mammalian cells.7,54-56 The Nup107–160 complex also helps to attract other nucleoporins to kinetochores, namely the Ran binding protein 2 (RanBP2; also known as NuP358)–RanGAP1 complex.7 The depletion of RanBP2–RanGAP1 leads to defects in bipolar spindle formation or the accumulation of unaligned chromosomes.57,58 Consistent with recent proteomics data, we found at least a portion of RanBP2/Nup358 localized to the centrosomes.32 In fact, Nup358 could be co-immunoprecipitated with Nup62.1 From our depletion EM data, we think Nup62 and Nup358 may play a role in centriole synthesis and maturation during mitosis. Besides, we and others also demonstrated that Tpr regulates spindle checkpoints via the dynein complex.4,5 Furthermore, mitotic roles for Nup15359,60 and Nup18861 were also reported recently. We suggest that different nucleoporin subcomplexes have individual scaffolding roles in the kinetochore, mitotic spindle, and centrosomes governing proper chromosome segregation.
Collectively, our data suggest that nucleoporin Nup62 localizes to the centrosome and plays a role in proper centrosome/centriole maturation and synthesis during mitosis.
Materials and Methods
Mammalian cell culture
HeLa and SW480 cells were obtained from the American Type Culture Collection (ATCC). All cell lines were cultured in DMEM (Invitrogen) with 10% fetal bovine serum (FBS) and penicillin/streptomycin. All cell lines were maintained at 37 °C in an air/5% CO2 incubator.
DNA constructs and RNA interference
HeLa cell cDNA was synthesized using a SuperScript™ III CellsDirect cDNA Synthesis System Kit (Life Technologies). The full-length Nup62 coding region was PCR amplified from cDNA and subcloned into pJET 1.2 (Thermo Scientific). Four Nup62 fragments (N1, N2, C1, and C2) were subcloned by PCR from pJET-Nup62FL into pEGFP-C3, pEGFP-N1 vectors. The plasmid encoding full-length mouse Nup62, tagged with 3xGFP was from Euroscarf. The plasmid encoding full-length human Nup153 was from Origene and subcloned into p3XFLAG-CMV (Sigma), and GFP-H2B plasmid was obtained from Addgene. The details of expression constructs and cloning primers are listed in Tables S1 and S2. All constructs were confirmed by DNA sequencing. siRNA duplexes targeting Nup62 (siRNA ID: SASI_Hs01_00038069;) and control siRNA were purchased from Sigma-Aldrich, Japan. siRNA transfections were performed using Lipofectamine 2000, following the manufacturer's protocol (Life Technologies), except for flow cytometry assays. For flow cytometry, Fugene HD (Roche) was used, following the manufacturer’s protocol. HeLa cells were usually imaged 72 h after transfection. If necessary, transfection efficiency was monitored with Block-iT (Life Technologies).
Immunoprecipitation
For immunoprecipitation, 107 cells were seeded and synchronized as described previously.27,28 Briefly, mitotic HeLa cells were collected, washed with PBS, spun at 400 × g for 10 min, and lysed in 1 ml of cold NP-40 lysis buffer (50 mM Tris-HCl [pH 7.2], 250 mM NaCl, 0.1% Nonidet P-40, 2 mM EDTA, 10% glycerol) containing 1× protease inhibitor mixture (Roche Applied Science) and 1 mM phenylmethylsulfonyl fluoride. Lysates were centrifuged for 30 min at 4 °C at 14 000 × g. The resulting lysate supernatants were pre-cleared with 50 μl of protein A/G bead slurry (Santa Cruz Biotechnology), mixed with 10 μl of various antibodies as specified, and incubated for 1 h at 4°C with rocking. The beads were then washed 5 times with 500 μl of lysis buffer. After the last wash, 50 μl of 1× SDS-PAGE blue loading buffer (New England Biolabs) was added to the bead pellet before loading.
Electron microscopy
HeLa cells adhered to culture dishes were washed with phosphate-buffered saline (PBS) and fixed in 2% paraformaldehyde and 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for 20 min and then 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) overnight. They were then scraped off and sedimented at 200 g for 10 min. After rinsing in 0.1 M phosphate buffer (pH 7.4), the cells were post-fixed in 0.1 M phosphate buffer (pH 7.4) with 2% osmium tetraoxide, dehydrated in ethanol (50, 70, 90 and 100% × 4; each for 1 h) at room temperature. The cells were consecutively incubated in a 2:1 and then a 1:2 (v/v) mixture of ethanol and epoxy resin at 20 °C for 1 h each, followed by infiltration with pure epoxy resin at 20 °C and polymerization at 70 °C for 15 h. Sections of approximately 70-nm thickness were transferred onto 200 mesh copper grids without supporting film and stained with 2% uranyl acetate solution and then stained with lead stain solution before carbon vacuum deposition. Micrographs were recorded with a JEOL JEM-1200EX at 70–100 kV.
Antibodies, immunocytochemistry, and confocal microscopy
Anti-Nup62 (sc-166870) and anti-β-actin (sc-47778) mouse monoclonal antibodies were from Santa Cruz Biotechnology. Anti-mAb414 (MMS-120R) antibody was from COVANCE. Anti-GFP (A6455) rabbit polyclonal antibody was from Life Technologies. Anti-pericentrin rabbit polyclonal antibody (ab4448) was from Abcam. Anti-α-tubulin (DM1A), anti-γ-tubulin (GTU-88) mouse monoclonal antibodies, anti-Nup62 rat monoclonal antibody, and anti-Centrobin rabbit polyclonal antibody were from Sigma-Aldrich. Anti-FLAG antibody was from MBL. Secondary antibodies were from Molecular Probes (Life Technologies). For immunofluorescence, synchronized HeLa cells were washed in phosphate-buffered saline (PBS) and fixed for 10 min in ice-cold methanol. Cells were then permeabilized with 0.3% Triton X-100 in PBS for 10 min at room temperature. For Nup62 staining, cells were washed with PBS and permeabilized with 0.3% Triton X-100 in PBS for 4 min before fixation, and then fixed with 4% paraformaldehyde for 10 min. Coverslips were blocked with 4% bovine serum albumin in PBS for 30 min and incubated with primary antibodies for 2 h at room temperature and then secondary antibodies for 2 h at room temperature. Coverslips were mounted onto slide glasses using Pro-Long Gold Antifade reagent (Life Technologies) and were examined on a Zeiss LSM5 EXCITER confocal microscope with 4 laser beams, and all images were acquired using a plan-Apochromat 63× with a 1.4-N.A. objective or at 100× with a 1.4-N.A. objective.
Flow cytometry
HeLa cells transfected with siRNA were trypsinized, washed twice with PBS, and fixed in 70% ethanol at −20 °C overnight. The fixed cells were resuspended in PBS containing 50 μg RNase A/ml and 50 μg PI (propidium iodide)/ml. Cellular DNA content was analyzed using a FACSCanto II (BD Biosciences) with FACS Diva software (BD Biosciences).32,62
Spindle orientation analysis
Images were obtained with a Zeiss LSM5 confocal microscope and the angle between the spindle axis and the growth plane was measured with the ZEN2008 software.38
Time-lapse microscopy
Time-lapse analysis of Nup62 dynamics during metaphase-to-anaphase transition in live cells was recorded from Nup62–3xEGFP stably expressing HeLa cells. Cells were placed in a microincubation chamber (7136; Corning) on the stage of a Zeiss LSM5 confocal microscope, which was heated to 37 °C and equipped with CO2 supply (Electric CO2 Microscope Stage Incubator; OKO Lab). Time-lapse series were generated by collecting photographs every 3 min; the photographs were then converted to 8-bit images and processed by Adobe Photoshop CS5 and Quick Time software.
In vitro microtubule regrowth assay
HeLa cells were seeded on coverslips, transfected with siRNAs, and grown for 48 h. For synchronization, cells were treated with 2 mM of thymidine for 16 h and then released for 4 h and treated with 100 ng/ml of nocodazole. After 4 h, most cells were arrested in mitosis. Nocodazole was washed out, and microtubules were completely depolymerized for 30 min on ice. To allow microtubule regrowth, cells were then incubated in medium at 37 °C, and stopped at time points by fixation.
Statistical analysis
Data are expressed as means ± SD. Comparisons between groups were determined using the unpaired t test. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Tatsuyoshi Funasaka for technical support in Fig. S3 and helpful discussions. This work was supported by Grants-in-Aid for Scientific Research on Innovative Areas, Grants-in-Aid for Challenging Exploratory Research and Grants-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) Japan, and by grants from the Asahi Glass Foundation, the Suzuken Memorial Foundation, the Sumitomo Foundation, the Kowa Life Science Foundation, the Mochida Memorial Foundation, the Sagawa Foundation, the Inamori Foundation, the Uehara Memorial Foundation, and the Takeda Science Foundation (to RW). CH was supported by the Hayashi Memorial Foundation for Women Natural Scientists.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/26671
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26671
References
- 1.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 2.Adams RL, Wente SR. Uncovering nuclear pore complexity with innovation. Cell. 2013;152:1218–21. doi: 10.1016/j.cell.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–66. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakano H, Wang W, Hashizume C, Funasaka T, Sato H, Wong RW. Unexpected role of nucleoporins in coordination of cell cycle progression. Cell Cycle. 2011;10:425–33. doi: 10.4161/cc.10.3.14721. [DOI] [PubMed] [Google Scholar]
- 5.Bukata L, Parker SL, D’Angelo MA. Nuclear pore complexes in the maintenance of genome integrity. Curr Opin Cell Biol. 2013;25:378–86. doi: 10.1016/j.ceb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Chatel G, Fahrenkrog B. Nucleoporins: leaving the nuclear pore complex for a successful mitosis. Cell Signal. 2011;23:1555–62. doi: 10.1016/j.cellsig.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 7.Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–91. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 8.Wozniak R, Burke B, Doye V. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell Mol Life Sci. 2010;67:2215–30. doi: 10.1007/s00018-010-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailer SM, Balduf C, Hurt E. The Nsp1p carboxy-terminal domain is organized into functionally distinct coiled-coil regions required for assembly of nucleoporin subcomplexes and nucleocytoplasmic transport. Mol Cell Biol. 2001;21:7944–55. doi: 10.1128/MCB.21.23.7944-7955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jovanovic-Talisman T, Tetenbaum-Novatt J, McKenney AS, Zilman A, Peters R, Rout MP, Chait BT. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature. 2009;457:1023–7. doi: 10.1038/nature07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Q, Meier I. Identification and characterization of the Arabidopsis FG-repeat nucleoporin Nup62. Plant Signal Behav. 2011;6:330–4. doi: 10.4161/psb.6.3.13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan T, Müller S, Klier G, Panté N, Blevitt JM, Haner M, Paschal B, Aebi U, Gerace L. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol Biol Cell. 1995;6:1591–603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheffield LG, Miskiewicz HB, Tannenbaum LB, Mirra SS. Nuclear pore complex proteins in Alzheimer disease. J Neuropathol Exp Neurol. 2006;65:45–54. doi: 10.1097/01.jnen.0000195939.40410.08. [DOI] [PubMed] [Google Scholar]
- 15.Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci U S A. 2006;103:2172–7. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Percipalle P, Clarkson WD, Kent HM, Rhodes D, Stewart M. Molecular interactions between the importin alpha/beta heterodimer and proteins involved in vertebrate nuclear protein import. J Mol Biol. 1997;266:722–32. doi: 10.1006/jmbi.1996.0801. [DOI] [PubMed] [Google Scholar]
- 17.Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J Cell Biol. 1991;114:169–83. doi: 10.1083/jcb.114.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solmaz SR, Chauhan R, Blobel G, Melčák I. Molecular architecture of the transport channel of the nuclear pore complex. Cell. 2011;147:590–602. doi: 10.1016/j.cell.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park N, Skern T, Gustin KE. Specific cleavage of the nuclear pore complex protein Nup62 by a viral protease. J Biol Chem. 2010;285:28796–805. doi: 10.1074/jbc.M110.143404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leng Y, Cao C, Ren J, Huang L, Chen D, Ito M, Kufe D. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. J Biol Chem. 2007;282:19321–30. doi: 10.1074/jbc.M703222200. [DOI] [PubMed] [Google Scholar]
- 21.Echeverría PC, Mazaira G, Erlejman A, Gomez-Sanchez C, Piwien Pilipuk G, Galigniana MD. Nuclear import of the glucocorticoid receptor-hsp90 complex through the nuclear pore complex is mediated by its interaction with Nup62 and importin beta. Mol Cell Biol. 2009;29:4788–97. doi: 10.1128/MCB.00649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubert T, Vandekerckhove J, Gettemans J. Exo70-mediated recruitment of nucleoporin Nup62 at the leading edge of migrating cells is required for cell migration. Traffic. 2009;10:1257–71. doi: 10.1111/j.1600-0854.2009.00940.x. [DOI] [PubMed] [Google Scholar]
- 23.Monette A, Panté N, Mouland AJ. HIV-1 remodels the nuclear pore complex. J Cell Biol. 2011;193:619–31. doi: 10.1083/jcb.201008064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basel-Vanagaite L, Muncher L, Straussberg R, Pasmanik-Chor M, Yahav M, Rainshtein L, Walsh CA, Magal N, Taub E, Drasinover V, et al. Mutated nup62 causes autosomal recessive infantile bilateral striatal necrosis. Ann Neurol. 2006;60:214–22. doi: 10.1002/ana.20902. [DOI] [PubMed] [Google Scholar]
- 25.Wong RW. Interaction between Rae1 and cohesin subunit SMC1 is required for proper spindle formation. Cell Cycle. 2010;9:198–200. doi: 10.4161/cc.9.1.10431. [DOI] [PubMed] [Google Scholar]
- 26.Wong RW. An update on cohesin function as a ‘molecular glue’ on chromosomes and spindles. Cell Cycle. 2010;9:1754–8. doi: 10.4161/cc.9.9.11806. [DOI] [PubMed] [Google Scholar]
- 27.Wong RW, Blobel G. Cohesin subunit SMC1 associates with mitotic microtubules at the spindle pole. Proc Natl Acad Sci U S A. 2008;105:15441–5. doi: 10.1073/pnas.0807660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong RW, Blobel G, Coutavas E. Rae1 interaction with NuMA is required for bipolar spindle formation. Proc Natl Acad Sci U S A. 2006;103:19783–7. doi: 10.1073/pnas.0609582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Funasaka T, Nakano H, Wu Y, Hashizume C, Gu L, Nakamura T, Wang W, Zhou P, Moore MA, Sato H, et al. RNA export factor RAE1 contributes to NUP98-HOXA9-mediated leukemogenesis. Cell Cycle. 2011;10:1456–67. doi: 10.4161/cc.10.9.15494. [DOI] [PubMed] [Google Scholar]
- 30.Nakano H, Funasaka T, Hashizume C, Wong RW. Nucleoporin translocated promoter region (Tpr) associates with dynein complex, preventing chromosome lagging formation during mitosis. J Biol Chem. 2010;285:10841–9. doi: 10.1074/jbc.M110.105890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hashizume C, Nakano H, Yoshida K, Wong RW. Characterization of the role of the tumor marker Nup88 in mitosis. Mol Cancer. 2010;9:119. doi: 10.1186/1476-4598-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashizume C, Kobayashi A, Wong RW. Down-modulation of nucleoporin RanBP2/Nup358 impaired chromosomal alignment and induced mitotic catastrophe. Cell Death Dis. 2013;4:e854. doi: 10.1038/cddis.2013.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bornens M. The centrosome in cells and organisms. Science. 2012;335:422–6. doi: 10.1126/science.1209037. [DOI] [PubMed] [Google Scholar]
- 34.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–78. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 35.Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell. 2002;3:339–50. doi: 10.1016/S1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- 36.Funasaka T, Tsuka E, Wong RW. Regulation of autophagy by nucleoporin Tpr. Sci Rep. 2012;2:878. doi: 10.1038/srep00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinoshita Y, Kalir T, Dottino P, Kohtz DS. Nuclear distributions of NUP62 and NUP214 suggest architectural diversity and spatial patterning among nuclear pore complexes. PLoS One. 2012;7:e36137. doi: 10.1371/journal.pone.0036137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogoyevitch MA, Yeap YY, Qu Z, Ngoei KR, Yip YY, Zhao TT, Heng JI, Ng DC. WD40-repeat protein 62 is a JNK-phosphorylated spindle pole protein required for spindle maintenance and timely mitotic progression. J Cell Sci. 2012;125:5096–109. doi: 10.1242/jcs.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raices M, D’Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol. 2012;13:687–99. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- 40.Funasaka T, Wong RW. The role of nuclear pore complex in tumor microenvironment and metastasis. Cancer Metastasis Rev. 2011;30:239–51. doi: 10.1007/s10555-011-9287-y. [DOI] [PubMed] [Google Scholar]
- 41.Kinoshita Y, Kalir T, Rahaman J, Dottino P, Kohtz DS. Alterations in nuclear pore architecture allow cancer cell entry into or exit from drug-resistant dormancy. Am J Pathol. 2012;180:375–89. doi: 10.1016/j.ajpath.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol. 2012;14:431–7. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obado SO, Rout MP. Ciliary and nuclear transport: different places, similar routes? Dev Cell. 2012;22:693–4. doi: 10.1016/j.devcel.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Zhou L, Panté N. The nucleoporin Nup153 maintains nuclear envelope architecture and is required for cell migration in tumor cells. FEBS Lett. 2010;584:3013–20. doi: 10.1016/j.febslet.2010.05.038. [DOI] [PubMed] [Google Scholar]
- 45.Pekovic V, Harborth J, Broers JL, Ramaekers FC, van Engelen B, Lammens M, von Zglinicki T, Foisner R, Hutchison C, Markiewicz E. Nucleoplasmic LAP2alpha-lamin A complexes are required to maintain a proliferative state in human fibroblasts. J Cell Biol. 2007;176:163–72. doi: 10.1083/jcb.200606139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell cycle genes inside the nucleoplasm. Cell. 2010;140:360–71. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Tanenbaum ME, Medema RH. Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell. 2010;19:797–806. doi: 10.1016/j.devcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 48.O’Connell CB, Khodjakov AL. Cooperative mechanisms of mitotic spindle formation. J Cell Sci. 2007;120:1717–22. doi: 10.1242/jcs.03442. [DOI] [PubMed] [Google Scholar]
- 49.Pease JC, Tirnauer JS. Mitotic spindle misorientation in cancer--out of alignment and into the fire. J Cell Sci. 2011;124:1007–16. doi: 10.1242/jcs.081406. [DOI] [PubMed] [Google Scholar]
- 50.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–4. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 51.Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1:965–76. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, Briggs S, Dasso M, Forbes DJ. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell. 2006;17:3806–18. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galy V, Askjaer P, Franz C, López-Iglesias C, Mattaj IW. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr Biol. 2006;16:1748–56. doi: 10.1016/j.cub.2006.06.067. [DOI] [PubMed] [Google Scholar]
- 54.Belgareh N, Rabut G, Baï SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–60. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–64. doi: 10.1016/S1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- 56.Loïodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–44. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salina D, Enarson P, Rattner JB, Burke B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol. 2003;162:991–1001. doi: 10.1083/jcb.200304080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol. 2004;14:611–7. doi: 10.1016/j.cub.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 59.Lussi YC, Shumaker DK, Shimi T, Fahrenkrog B. The nucleoporin Nup153 affects spindle checkpoint activity due to an association with Mad1. Nucleus. 2010;1:71–84. doi: 10.4161/nucl.1.1.10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mackay DR, Elgort SW, Ullman KS. The nucleoporin Nup153 has separable roles in both early mitotic progression and the resolution of mitosis. Mol Biol Cell. 2009;20:1652–60. doi: 10.1091/mbc.E08-08-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Itoh G, Sugino S, Ikeda M, Mizuguchi M, Kanno S, Amin MA, Iemura K, Yasui A, Hirota T, Tanaka K. Nucleoporin Nup188 is required for chromosome alignment in mitosis. Cancer Sci. 2013;104:871–9. doi: 10.1111/cas.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Endo A, Moyori A, Kobayashi A, Wong RW. Nuclear mitotic apparatus protein, NuMA, modulates p53-mediated transcription in cancer cells. Cell Death Dis. 2013;4:e713. doi: 10.1038/cddis.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.