Abstract
Gibberella fujikuroi species complex (GFSC) was isolated from rice (Oryza sativa L.) seed samples from ten Asian countries and investigated for incidence of GFSC, molecular characteristics, and pathogenicity. Regardless of geographic origin, GFSC was detected with incidences ranging from 3% to 80%. Four species, Fusarium fujikuroi, F. concentricum, F. proliferatum, and F. verticillioides, were found to show an association with rice seeds, with F. fujikuroi being the predominant species. In phylogenetic analyses of DNA sequences, no relationship was found between species, isolates, and geographic sources of samples. Unidentified fragments of the β-tubulin gene were observed in ten isolates of F. fujikuroi and F. verticillioides. With the exception of three isolates of F. fujikuroi, F. fujikuroi, F. proliferatum, and F. verticillioides were found to have FUM1 (the fumonisin biosynthetic gene); however, FUM1 was not found in isolates of F. concentricum. Results of pathogenicity testing showed that all isolates caused reduced germination of rice seed. In addition, F. fujikuroi and F. concentricum caused typical symptoms of bakanae, leaf elongation and chlorosis, whereas F. proliferatum and F. verticillioides only caused stunting of seedlings. These findings provide insight into the characteristics of GFSC associated with rice seeds and might be helpful in development of strategies for management of bakanae.
Keywords: Gibberella fujikuroi species complex, Incidence, Molecular characteristics, Pathogenicity, Rice seed
Rice (Oryza sativa L.), a staple crop for many people worldwide, is subject to a number of fungal diseases. Among the major diseases affecting rice, 'bakanae' is widely distributed in all rice-growing countries and causes significant yield losses of up to 50% [1]. Occurrence of bakanae has shown a recent increase as environmentally-friendly cultivation of rice increases. Despite continuous development of new chemical and sterilization methods, control of this disease remains difficult. The causal organism of bakanae was first identified as Fusarium heterosporum Nees by Hori in 1898, and its teleomorph was described as Lisea fujikuroi by Sawada [2]. This organism was later transferred to Gibberella as G. fujikuroi (Sawada) Ito [3]. G. fujikuroi (Sawada) Ito is a polytypic species complex with anamorphs in Fusarium species from section Liseola [4, 5]. Some members of G. fujikuroi species complex (GFSC) (F. fujikuroi Nirenberg [6], F. proliferatum (Mats.) Nirenberg [6, 7] and F. verticillioides (Sacc.) Nirenberg [6, 7]), have been found to show an association with bakanae [6, 7]. However, it is unclear whether all three species are associated with symptoms of bakanae or if F. proliferatum and F. verticillioides are present only as saprophytes [8].
Studies of the incidence, genetic diversity, and pathogenicity of GFSC in rice plants and seed materials have been conducted in several countries. The incidence and severity of bakanae were investigated in Asian countries, including Malaysia [9], Indonesia [9], Pakistan [10], and India [11]. Genetic diversity, pathogenicity, and toxigenicity of GFSC strains isolated from rice seeds from nine Asian and African countries have been reported [8]. A recent study also investigated the species composition and genetic diversity of putative fumonisin-producing GFSC associated with rice seeds collected in Korea [12].
Species identification of Fusarium based on morphological characteristics is complicated and time-consuming [4]. In addition, species of GFSC have similar morphologies and differentiation of F. fujikuroi from F. proliferatum morphologically is almost impossible. Therefore, molecular identification of fungal species by DNA sequence has been used to support morphological identification of Fusarium species. In addition, phylogenetic analysis of DNA sequences has been used for differentiation and evaluation of the genetic relationship among closely related Fusarium species. The internal transcribed spacer (ITS) regions of nuclear ribosomal RNA gene repeats have commonly been applied; however, many fusaria within the Gibberella clade possess non-orthologous copies of ITS2, which can lead to incorrect phylogenetic inference [13, 14]. The translation elongation factor-1α gene (TEF) is regarded as a good tool for use in species identification for Fusarium because it is highly informative at the species level and non-orthologous copies of the gene have not been detected in the genus [15]. The histone H3 (H3) and β-tubulin (BT) genes were also used for species identification of Fusarium.
The most evident symptom of bakanae is chlorotic and abnormally elongated leaves induced by gibberellic acid (GA) produced by the pathogens [16]. The angle of the leaf with the stem was found to be wider in diseased seedlings than in healthy seedlings [17]. However, not all infected seedlings have symptoms of bakanae, and they are sometimes only stunted or normal in appearance. Currently, it is still not clear which species are associated with the various symptoms. Each species of GFSC has a specific profile of mycotoxins, including fumonisin. A gene cluster of fumonisin biosynthesis (FUM) was identified and characterized in F. verticillioides [18], revealing that fumonisin non-producing species lack the FUM genes [19]. Nevertheless, little information is available regarding the incidence, pathogenicity to rice, and mycotoxin profiles of F. concentricum, a member of GFSC.
The current study was conducted using rice seed samples from Asian countries in order to: 1) determine infection status with GFSC, 2) identify species of GFSC isolates, 3) characterize molecular traits of the isolates, 4) determine the presence of the FUM1 gene in the isolates, and 5) evaluate the pathogenicity of the isolates on rice.
MATERIALS AND METHODS
Seed samples and isolation of GFSC
A total of 110 seed samples of rice (Oryza sativa L.) from ten Asian countries, including 45 samples from Korea were used for isolation of the GFSC. Seed samples had been stored at 4℃ for 3~28 years. A total of 100 seeds of each sample were placed on two layers of filter paper (Advantec No. 2; Advantec, Dublin, CA, USA) moistened with sterile distilled water in a 9-cm Petri dish and then incubated at 25℃ under near ultra violet light for seven days. White and fluffy mycelia were removed from the seed surface under a stereomicroscope and transferred to potato dextrose agar (PDA) medium supplemented with 100mg/L of streptomycin sulfate. The samples were then incubated at 25℃ under the same light conditions for seven days; single spores were then isolated from suspected colonies and incubated at 25℃ in the dark for seven days. Isolates grown on slant PDA media were stored at 4℃ for further investigation.
DNA extraction, PCR amplification, and sequencing
Mycelia grown in potato dextrose broth medium were removed and transferred to a 1.5-mL Eppendorf tube. After lyophilization, the dry mycelia were ground to fine powder using TissueLyser II (Qiagen, Hilden, Germany) with 7-mm stainless beads. Genomic DNA was extracted from the mycelia powder using a DNeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions. The DNA concentration was adjusted to 20 ng/µL and 1 µL of DNA template was used for PCR amplification. The incidence of GFSC was determined by PCR using the Gib2-F (5'-GAGGCGCGGTGTCGGTGTGCTTG-3') and Fgc-R (5'-CTCTCATATACCCTCC G-3') primer set [20], which specifically detects strains of GFSC. Amplification reactions of three genes, TEF, H3, and BT, were performed for subsequent sequence analyses. Briefly, reactions were performed in 20 µL of AccuPower PCR premix (Bioneer, Daejeon, Korea), composed of 250 µM dNTP, 10 mM Tris-HCl (pH 9.0), 30 mM KCl, 1.5 mM MgCl2, and 1 unit of Top DNA polymerase. Each gene was partially amplified using the specific primer sets as described previously: EF-1 (5'-ATGGGTAAGGAGGACGACAAGAC-3') and EF-2 (5'-GGAAGTACCAGTGATCATGTT-3') for TEF [12], H3-1a (5'-ACTAAGCAGACCGCCCGCAGG-3') and H3-1b (5'-GCGGGCGAGCTGGATGTCCTT-3') for H3 [21], and Bt2a (5'-GGTAACCAAATCGGTGCTGCTTTC-3') and Bt2b (5'-ACCCTCAGTGTAGTGACCCTTGGC-3') for BT [21]. The PCR products were then purified using a HiGene PCR Purification Kit (Solgent, Daejeon, Korea) according to the manufacturer's instructions; then, they were directly sequenced in both directions using the same primers. DNA sequences were edited using SeqMan Pro (DNASTAR Inc., Madison, WI, USA) and the consensus sequences were used for species identification. The FUM1 gene was amplified using the rp32 (5'-ACAAGTGTCCTTGGGGTCCAGG-3') and rp33 (5'-GATGCTCTTGGAAGTGG CCTACG-3') primers as described by Procter et al. [18].
Molecular characterization and phylogenetic analysis
The DNA sequences of the TEF, H3, and BT genes were analyzed for species identification of GFSC isolates. Sequences were then compared with other sequences found in NCBI database (http://blast.ncbi.nih.gov) and Fusarium-ID (http://isolate.fusariumdb.org) [15] using Clustal W with a gap opening penalty of 3.0 and a gap extension penalty of 1.8. The neighbor-joining (NJ) method was used for phylogenetic analysis. For tree construction, the combined sequences were analyzed using the Tamura-Nei model as a nucleotide substitution model, including transitions and transversions with MEGA 5.1. The reliability of the NJ tree was estimated using the bootstrap method with 1,000 replications. Sequences of 12 strains of Asian and African clades of GFSC established by O'Donnell et al. [14] were used as references: NRRL13566 and KACC44004 (F. fujikuroi), NRRL25181 and NRRL26794 (F. concentricum), NRRL22944 and NRRL53578 (F. proliferatum), NRRL25226 (F. mangiferae Britz, M. J. Wingf. & Marasas), NRRL13999 [F. sacchari (E. J. Butler & Hafiz Khan) W. Gams], NRRL22172 (F. verticillioides), NRRL13308 (F. acutatum Nirenberg & O'Donnell), NRRL22045 (F. thapsinum Klittich, J. F. Leslie, P. E. Nelson & Marasas), and NRRL25446 (F. brevicatenulatum Nirenberg, O'Donnell, Kroschel & Andrianaivo). The tree was rooted with NRRL34079 (F. graminearum Schwabe). The partial BT sequence of Ophiocordyceps sinensis (Berk.) G. H. Sung, J. M. Sung, Hywel-Jones & Spatafora (accession No. JX968023) from the NCBI GenBank was used for analysis of the 360-bp fragments of BT.
Pathogenicity tests.
Pathogenicity tests were performed using seed inoculation assays. Seeds of the Chucheongbyeo rice cultivar were tested for the presence of any seed-borne Fusarium spp. by incubation on Komada's medium. Briefly, seeds were sterilized by soaking in 62℃ water for 10 min and then air-drying on sterile filter paper in a flow cabinet; then, seeds found to be Fusarium-free were used for inoculation of strains. Fungal inoculum was prepared for each strain from single spore cultures. The concentration of the inoculum suspension was determined using a hemocytometer and adjusted to 1 × 106 spores/mL with sterile water. Thirty seeds were soaked in 10 mL of the inoculum suspension in a 50-mL conical tube with gentle shaking for 20 hr at room temperature. Control seeds were soaked in sterile water instead of the inoculum suspension. Inoculated and control seeds were then placed on solidified MS basal medium in a glass test tube with a diameter of 30-mm and height of 200-mm (four tubes per strain/five seeds per tube) and incubated at 28℃ under alternating 12-hr fluorescent light/12-hr darkness for 14 days. Seven days after incubation, the number of germinated seeds was counted. At seven and 14 days after incubation, the seedlings were observed for symptoms of bakanae, slender and chlorotic leaves, elongated seedlings, leaves with wide angles between stems, and withered seedlings. Analysis of variance and Duncan's multiple range tests (p < 0.05) in each species were performed using R version 2.15.2 [22].
RESULTS
Infection of rice seeds with GFSC and molecular identification of isolates
The infection status of rice seeds with GFSC was examined in 110 seed samples from ten Asian countries using the blotter method (Table 1). At least 33% of samples from each country were found to be infected with GFSC, regardless of the geographic sources of the seed samples. All samples from Myanmar and Uzbekistan and more than 50% of samples from China, Japan, the Philippines, Russia, and Korea were found to be infected with GFSC, and the incidence (% infected seeds in a sample) of GFSC varied from 3% to 80%. After molecular identification with DNA sequences of TEF and histone H3 (H3), 48 isolates of four species were finally selected (39 F. fujikuroi, 2 F. concentricum Nirenberg & O'Donnell, 4 F. proliferatum and 3 F. verticillioides) (Table 2). F. fujikuroi was the species found most frequently, regardless of geographic sources. However, seed samples from Myanmar were only infected with F. proliferatum.
Table 1.
Geographic sources of rice seed samples, number of samples tested, occurrence and incidence of Gibberella fujikuroi species complex (GFSC), and storage period of seed samples
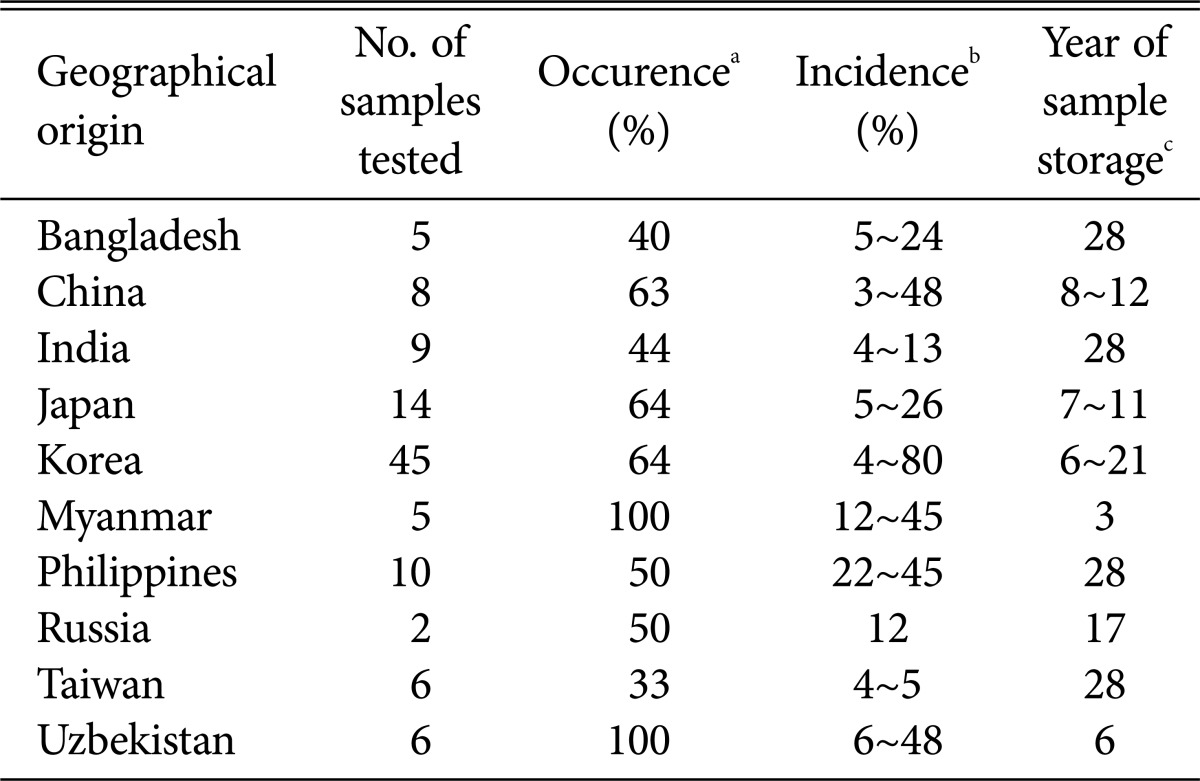
a% of samples infected with GFSC.
bLowest and highest % of seed infected with GFSC in a sample.
cLongest and shortest period of the infected sample storage at 4℃ and with 45% relative humidity.
Table 2.
Isolates of Gibberella fujikuroi species complex recovered from rice seed samples
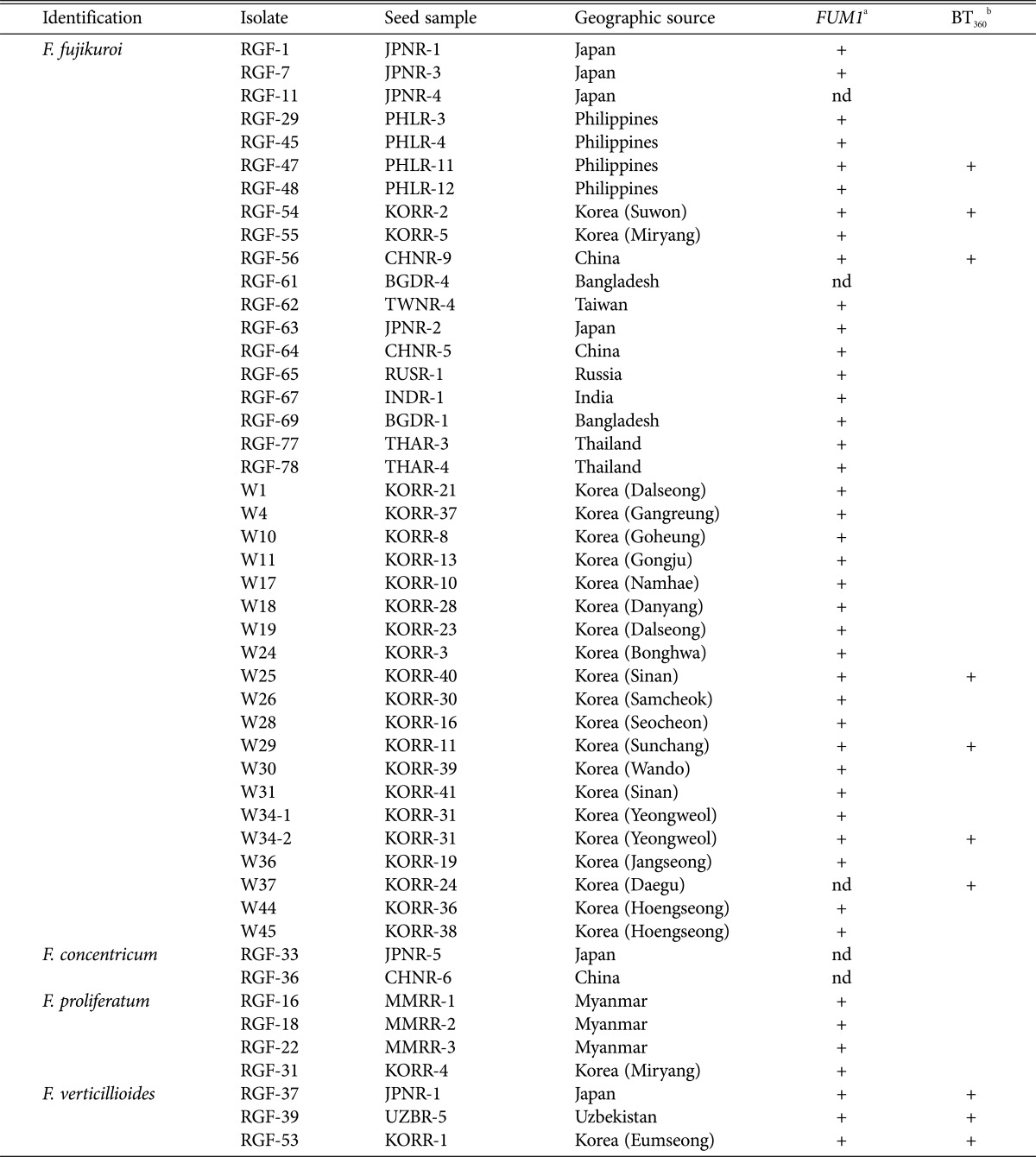
Species identification was based on DNA sequences of translation elongation factor-1α, histone H3, and β-tubulin genes.
a '+' indicates that FUM1 was detected, 'nd' indicates that the gene was not detected.
b '+' indicates that 360-bp fragments were detected.
Phylogenetic analysis
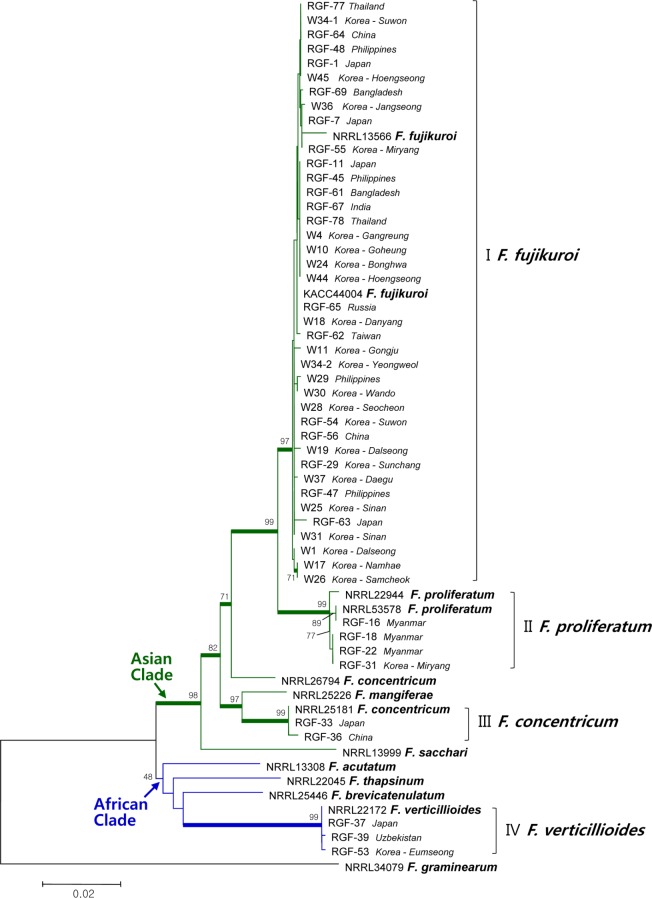
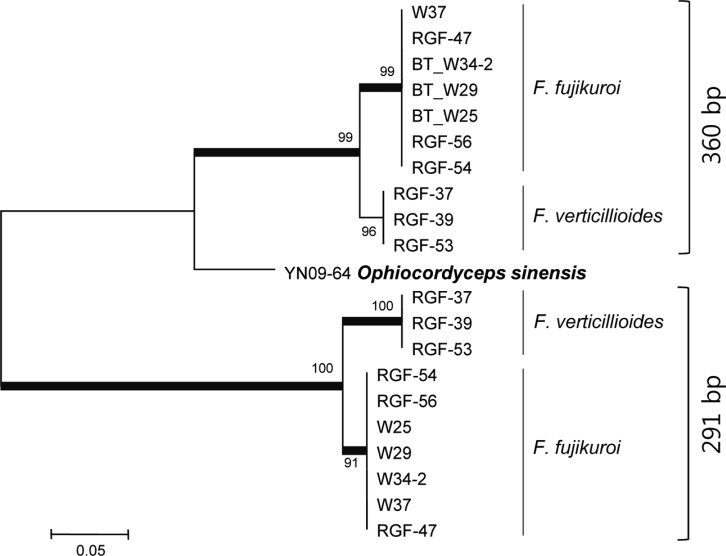
The phylogenetic tree constructed from combined DNA sequences of TEF, H3, and BT genes (1,283 sites with gaps) consisted of four main clades (Fig. 1). Clade I was composed of 39 F. fujikuroi isolates from samples from nine countries with 97% of the bootstrap value, clade II consisted of four isolates of F. proliferatum from samples from Myanmar and Korea with 99% of the bootstrap value, and clade III consisted of two isolates of F. concentricum from samples from China and Japan with 99% unity. Isolates of Clades I, II, III, NRRL25226 (F. mangiferae), and NRRL13999 (F. sacchari) were grouped into a large clade with bootstrap support of 98%. Three isolates of F. verticillioides were grouped into clade IV with 99% of the bootstrap values and this clade was included in a single clade with NRRL13308 (F. acutatum), NRRL22045 (F. thapsinum), and NRRL25446 (F. brevicatenulatum) with 48% of the bootstrap value. No relationship was observed between geographic sources of seed samples and phylogenetic position of the isolates. Of particular interest, PCR for amplification of the BT gene revealed that seven isolates of F. fujikuroi and three isolates of F. verticillioides had an additional fragment with a size of 360 bp. Sequence analysis of the fragments (302 sites with gaps) showed that those of F. verticillioides and F. fujikuroi were highly homologous (Fig. 2). The sequence identities of the additional fragments to the original BT fragments (291 bp) were 74~76% in F. fujikuroi and 68~69% in F. verticillioides. The greatest similarity was observed in the partial BT gene of Ophicordyceps sinensis, with identity of 83%.
Fig. 1.
Neighbor-Joining tree based on combined DNA sequences of translation elongation factor-1α, histone H3, and β-tubulin of Gibberella fujikuroi species complex isolated from rice seed samples. The bootstrap values based on 1,000 replications are indicated as percentages in the internodes. Thick branches are supported by bootstrap values > 70%. The tree is rooted with Fusarium graminearum (NRRL 34079).
Fig. 2.
Neighbor-Joining tree based on DNA sequences of β-tubulin in ten isolates of Fusarium fujikuroi and F. verticillioides. β-Tubulin sequences with sizes of 360 bp and 291 bp from the same isolates were compared. The bootstrap values based on 1,000 replications are indicated as percentages in the internodes. Thick branches are supported by bootstrap values > 70%. YN09-64 had the β-tubulin sequence with maximum homology to the 360-bp fragment.
Distribution of the FUM1 gene.
Most isolates were found to contain the FUM1 gene (Table 1), and the size of the amplified PCR fragments was approximately 680 bp. All isolates of F. proliferatum and F. verticillioides were found to contain the gene. In addition, 36 isolates of F. fujikuroi had specific fragments that encompassed all isolates of this organism except RGF-61, RGF-11, and W37. The size of isolates of F. concentricum (RGF-33 and RGF-36) was approximately 430-bp fragments instead of 680-bp fragments.
Pathogenicity
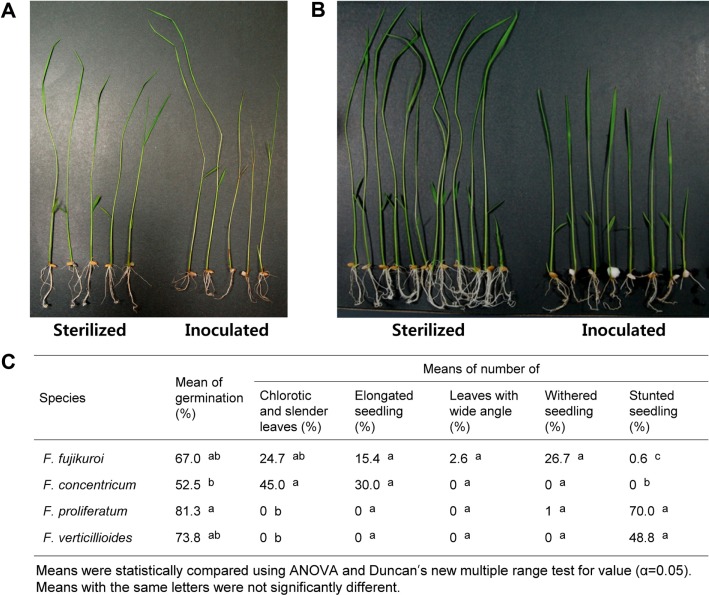
Compared with the control treatment, all isolates examined inhibited seed germination (Fig. 3), with germination rates of 67.0% for F. fujikuroi, 52.5% for F. concentricum, 81.3% for F. proliferatum, and 73.8% for F. verticillioides. However, results of ANOVA and Duncan's multiple range tests showed no significant differences. Isolates of F. fujikuroi and F. concentricum induced leaf chlorosis, slender leaves and/or elongation of seedlings (Fig. 3A and 3C). Overall, a wide angle between leaves and stems was observed in 2.6% of rice seedlings inoculated with isolates of F. fujikuroi (Fig. 3C), while isolates of F. proliferatum and F. verticillioides showed no symptoms of leaf chlorosis, slender leaves, or elongation, however, they did show stunted seedlings with a mean of 70.0% and 48.8%, respectively (Fig. 3B and 3C).
Fig. 3.
Pathogenicity of Gibberella fujikuroi species complex isolates to rice. Leaf elongation, chlorosis, and wide angle between leaf and stem in rice seedlings inoculated with isolates of Fusarium fujikuroi or F. concentricum (A), stunted seedlings inoculated with isolates of F. proliferatum or F. verticillioides (B), pathogenicity of each species to rice (C).
DISCUSSION
The results of this study demonstrate the importance of GFSC associated with rice seeds in Asian countries, including Korea. Bakanae is primarily seed-borne, and high levels of its incidence in rice seeds have been reported in recent studies. In the current study, the occurrence and incidence of GFSC in rice seeds was found to vary independently of sample sources and storage period of seeds. Seed samples dried to 15% seed moisture content were stored at 4℃ in air-tight containers; therefore, they had little chance of being contaminated during storage. It is notable that GFSC survived in dry seeds for up to 28 years because F. moniliforme (synonym of G. fujikuroi) was reported to be viable in seeds of Zea mays for eight years [23].
Three species of GFSC, F. fujikuroi, F. proliferatum, and F. verticillioides, which are known to be the predominant Fusarium species [6], were recovered from rice seed samples. In addition, F. concentricum, which was first described in banana (Musa sapientum) from Gatemala and Costa Rica and Asian brown leaf hopper (Nilaparvata lugens) from Korea [5], was also identified from rice seed samples from China and Japan. Among GFSC isolates, F. fujikuroi was the predominant species, and 92% of samples collected from Korea were infected with these species. The most abundant contamination of rice seeds with F. fujikuroi was previously reported in another study [12]. In addition, in the current study, seed samples from Myanmar were found to be infected with F. proliferatum, making this the first report of their occurrence in rice seeds from Myanmar.
Through multigene phylogenetic analysis, O'Donnell et al. [14] established three large clades in species of GFSC, which were designated as the "African", "American", and "Asian" based on the geographical origins of the plant hosts. The "Asian" clade is the smallest of the three GFSC clades. All isolates of F. fujikuroi, F. proliferatum, and F. concentricum recovered in the current study were grouped into this clade together with strains of F. mangiferae and F. sacchari. The "African" clade, the largest of the three clades, includes the agriculturally important pathogens, F. verticillioides, F. thapsinum, F. acutatum, and F. brevicatenulatum. Three isolates of F. verticillioides found in the current study were grouped into this clade, although the bootstrap value was low. Phylogenetic analysis with DNA sequences of isolates from rice seed samples found no relationship between species, isolates, and geographic sources of seed samples. Phylogenetic species recognition appeared to be the most effective method for differentiation of F. fujikuroi and F. proliferatum because they were closely related in terms of morphology and biology. Topologies of the trees from the TEF and H3 sequences were the same as that from combined sequences, whereas the tree from the original BT sequences had slightly different topology (data not shown); F. fujikuroi, F. concentricum, and F. verticillioides formed a large clade, however, F. proliferatum was separated into a distinct group in the BT tree. Grouping of F. fujikuroi and F. verticillioides was supported by 93% of bootstrap value. Additional unidentified fragments (approximately 360bp) of the BT gene observed in ten isolates of F. fujikuroi and F. verticillioides showed sequence identities to the original BT below 76%. These findings are interesting because the BT gene is known to be non-orthologous and there is no possibility of contamination by other fungal species at any experimental stage. More detailed investigations are needed in order to determine the identity of these additional BT fragments and why F. fujikuroi (Asian clade) and F. verticillioides (African clade) are closely located in the tree of BT, different from those from TEF and H3. Investigation of any relationship between these unidentified BT and resistance to chemicals such as benomyl is worthwhile because the BT gene was shown to confer cold-sensitive benomyl resistance to F. moniliforme [24]. The intraspecific variability of F. fujikuroi isolates was found to be very low, which was confirmed by Universally Primed-PCR analysis in the current study (data not shown).
Inhibition of seed germination is a primary symptom induced by spore inoculation of GFSC isolates in rice. Most isolates inhibited seed germination compared to the control treatment independent of species. Typical symptoms of bakanae, such as chlorotic/slender leaves and elongated seedlings are caused by most F. fujikuroi and F. concentricum isolates. In addition, F. fujikuroi was found to cause an increase in the angles between the rice leaf and stem, as reported by Imura [17], as well as withering of shoots. However, the pathogenicity of F. fujikuroi and F. concentricum to rice varied among isolates. These differences do not originate from the environment and nutritional status of the rice seedlings because the pathogenicity tests were performed under well-controlled conditions with MS agar media in a growth chamber. Although F. concentricum was determined not to present a threat to agricultural crops [25], in the current study, it induced disease symptoms in rice. Abnormal leaf elongation is known to be induced by production of GA by the pathogen. Although there have been no reports of GA production in F. concentricum to date, revaluation of the effects and role of F. concentricum on agricultural crops, including rice, is needed. The widespread occurrence of F. proliferatum suggests that its role in the complex symptoms of bakanae of rice may be significant [6]. In a recent study reported by Wulff et al. [8], F. verticillioides and F. proliferatum isolates from rice seeds showed pathogenicity to rice; therefore, they were considered potential pathogens of rice and not merely saprophytes. However, in the current study, rice seedlings inoculated with F. proliferatum or F. verticillioides isolates only showed stunted growth and did not show typical bakanae symptoms. Conduct of a more detailed study using a large number of isolates will provide a better understanding of the relationship between symptoms of bakanae and F. proliferatum and F. verticillioides.
The FUM1 gene is a member of the fumonisin biosynthetic gene cluster (FUM), which has been identified and characterized in F. verticillioides [18]. Fumonisin, nonproducing species of Fusarium, lack this gene [19]. FUM1 was detected in all isolates of F. proliferatum and F. verticillioides examined in the current study, which is reasonable because these are well known as fumonisin-producers in maize. Approximately 90% of isolates of F. fujikuroi examined contained FUM1; however, production of fumonisin in each isolate should be investigated further because FUM1 was also detected in isolates that did not produce fumonisins [19], and most F. fujikuroi isolates obtained from rice seeds in Korea produced only a small amount of fumonisins [12]. Two isolates of F. concentricum were found to lack FUM1, which was consistent with the results of a previous study showing that F. concentricum was a non-producer [23]. Although fumonisin production does not appear to be directly related to disease occurrence in rice, production of fumonisins might facilitate colonization of plant tissue and competition against other fungal species during infection [26]. Glenn et al. [27] reported that complementation of a FUM gene cluster deletion in F. verticillioides resulted in restoration of both fumonisin production and pathogenicity in maize seedlings; however, in the current study, the pathogenicity of the isolates examined was not related to FUM1, indicating that fumonisin production does not play a role in bakanae occurrence in rice and might depend on the host plant.
The current study provides insight into understanding the incidence, molecular characteristics, and pathogenicity of GFSC in rice seeds from Asian countries. A more detailed analysis of DNA sequences, pathogenicity, toxin production, and longevity of the pathogens in rice seeds might be helpful in development of more accurate tools for diagnosis of the disease and strategies for management of bakanae.
ACKNOWLEDGEMENTS
Financial support was provided by the National Academy of Agricultural Science of the Rural Development Administration in the Republic of Korea under project no. PJ008553.
References
- 1.Ou SH. Bakanae disease and foot rot. In: Ou SH, editor. Rice diseases. Surrey, Kew: Commonwealth Mycological Institute; 1985. pp. 262–272. [Google Scholar]
- 2.Sawada K. Beitrage über Formosas-Pilze no. 14. Trans Nat Hist Soc Formosa. 1917;31:31–133. [Google Scholar]
- 3.Ito S, Kimura J. Studies on the 'bakanae' disease of the rice plant. Rep Hokkaido Natl Agric Exp Stn. 1931;27:1–95. [Google Scholar]
- 4.Leslie JF, Summerell BA. The fusarium laboratory manual. Oxford: Blackwell Publishing; 2006. [Google Scholar]
- 5.Nirenberg HI, O'Donnell K. New Fusarium species and combinations within Gibberella fujikuroi species complex. Mycologia. 1998;90:434–458. [Google Scholar]
- 6.Desjardins AE, Manandhar HK, Plattner RD, Manandhar GG, Poling SM, Maragos CM. Fusarium species from Nepalese rice and production of mycotoxins and gibberellic acid by selected species. Appl Environ Microbiol. 2000;66:1020–1025. doi: 10.1128/aem.66.3.1020-1025.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amoah BK, Rezanoor HN, Nicholson P, Mac-Donald MV. Variation in the Fusarium section Liseola: pathogenicity and genetic studies of isolates of Fusarium moniliforme Sheldon from different hosts in Ghana. Plant Pathol. 1995;44:563–572. [Google Scholar]
- 8.Wulff EG, Sørensen JL, Lübeck M, Nielsen KF, Thrane U, Torp J. Fusarium spp. associated with rice Bakanae: ecology, genetic diversity, pathogenicity and toxigenicity. Environ Microbiol. 2010;12:649–657. doi: 10.1111/j.1462-2920.2009.02105.x. [DOI] [PubMed] [Google Scholar]
- 9.Zainudin NA, Razak AA, Salleh B. Bakanae disease of rice in Malaysia and Indonesia: etiology of the causal agent based on morphological, physiological and pathogenicity characteristics. J Plant Prot Res. 2008;48:475–485. [Google Scholar]
- 10.Khanam M, Khanzada AK. Seed-borne fungal diseases of rice in Sindh, Pakistan. Pak J Agric Res. 1989;10:302–305. [Google Scholar]
- 11.Gopalakrishnan C, Kamalakannan A, Valluvaparidasan V. Survey of seed-borne fungi associated with rice seeds in Tamil Nadu, India. Libyan Agric Res Cent J Int. 2010;1:307–309. [Google Scholar]
- 12.Kim JH, Kang MR, Kim HK, Lee SH, Lee T, Yun SH. Population structure of the Gibberella fujikuroi species complex associate with rice and corn in Korea. Plant Pathol J. 2012;28:357–363. [Google Scholar]
- 13.O'Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- 14.O'Donnell K, Cigelnik E, Nirenberg HI. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia. 1998;90:465–493. [Google Scholar]
- 15.Geiser DM, Jiménez-Gasco MM, Kang S, Makalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA, O'Donnell K. FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur J Plant Pathol. 2004;110:473–479. [Google Scholar]
- 16.Ou SH. Rice diseases. 2nd ed. Slough: CAB International; 1987. [Google Scholar]
- 17.Imura J. On the angles between blades and culms in the accelerated rice seedlings caused by Gibberella fujikuroi. Ann Phytopathol Soc Jpn. 1940;10:45–48. [Google Scholar]
- 18.Proctor RH, Brown DW, Plattner RD, Desjardins AE. Co-expression of 15 contiguous genes delineates a fumonisin biosynthetic gene cluster in Gibberella moniliformis. Fungal Genet Biol. 2003;38:237–249. doi: 10.1016/s1087-1845(02)00525-x. [DOI] [PubMed] [Google Scholar]
- 19.Proctor RH, Plattner RD, Brown DW, Seo JA, Lee YW. Discontinuous distribution of fumonisin biosynthetic genes in the Gibberella fujikuroi species complex. Mycol Res. 2004;108(Pt 7):815–822. doi: 10.1017/s0953756204000577. [DOI] [PubMed] [Google Scholar]
- 20.Jurado M, Vázquez C, Marín S, Sanchis V, Teresa González-Jaén M. PCR-based strategy to detect contamination with mycotoxigenic Fusarium species in maize. Syst Appl Microbiol. 2006;29:681–689. doi: 10.1016/j.syapm.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R Core Team. R: A language and environment for statistical computing [Internet] R Foundation for Statistical Computing; 2012. [cited 2013 Aug 1]. Available from: http://www.R-project.org/ [Google Scholar]
- 23.Duncan GH, Koehler B. Age of seed corn in relation to seed infection and yielding capacity. J Am Soc Agron. 1944;36:436–443. [Google Scholar]
- 24.Yan K, Dickman MB. Isolation of a beta-tubulin gene from Fusarium moniliforme that confers cold-sensitive benomyl resistance. Appl Environ Microbiol. 1996;62:3053–3056. doi: 10.1128/aem.62.8.3053-3056.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zitomer NC. Mycotoxicology to accompany phylogenetic revisions to the genus Fusarim [dissertation] State College: Pennsylvania State University; 2006. [Google Scholar]
- 26.Marín P, Megan N, Váquez C, González-Jaén MT. Differential effect of environmental conditions on the growth and regulation of the fumonisin biosynthetic gene FUM1 in the maize pathogens and fumonisin producers Fusarium verticillioides and Fusarium proliferatum. FEMS Microbiol Ecol. 2010;73:303–311. doi: 10.1111/j.1574-6941.2010.00894.x. [DOI] [PubMed] [Google Scholar]
- 27.Glenn AE, Zitomer NC, Zimeri AM, Williams LD, Riley RT, Proctor RH. Transformation-mediated complementation of a FUM gene cluster deletion in Fusarium verticillioides restores both fumonisin production and pathogenicity on maize seedlings. Mol Plant Microbe Interact. 2008;21:87–97. doi: 10.1094/MPMI-21-1-0087. [DOI] [PubMed] [Google Scholar]