Abstract
The ambrosia beetle, Platypus koryoensis, is an economically important pest affecting oak trees in Korea. Candida kashinagacola was isolated from galleries of the beetle in oak wood and identified by analyses of morphology, physiological properties, and nucleotide sequence of the large subunit ribosomal DNA. This is the first report on Candida species associated with oak wilt disease vectored by the ambrosia beetle, Platypus koryoensis, in Korea.
Keywords: Candida kashinagacola, Large subunit ribosomal DNA, Oak wilt disease, Platypus koryoensis
Candida, a genus of basidiomycete yeast, is currently the most common cause of fungal infections worldwide [1]. Many species are found in gut flora, including C. albicans in mammalian hosts, whereas others live as endosymbionts in insect hosts. [2]. Candida species have also been reported as human pathogen and antifungal species in Korea. Some Candida species reported were isolated from soil in Korea [3]. However, there were no reports on Candida species from insect gallery on wood in Korea. We recently surveyed microorganisms in diseased oak trees by Raffaelea quercusmongolicae, the oak wilt disease pathogen transmitted by the ambrosia beetle, Platypus koryoensis [4]. Diverse filamentous fungi, yeasts, and bacteria were found in previous surveys [5, 6]. In this study, we report on identification of some of the yeast isolates as C. kashinagacola.
Several wood disks with a diameter of 20 cm and thickness of 10 cm were cut from oak wilt diseased trees in Seoul and Hanam, Korea for yeast sampling. These sampled wood disks had been infested by P. koryoensis. The wood disks were broken into small wood chips using a sterile hammer and chisel. Insect galleries of P. koryoensis in chips were chopped into finer pieces and ground using a mortar and pestle with sterile distilled water. The ground materials were placed on yeast extract-malt extract (YEME) agar (BD Science, San Jose, CA, USA) plates and incubated at 30℃ for 3~7 days. Single colony isolation was performed at least three times from the grown colonies. Among the yeast isolates obtained, four yeast isolates (DUCC 7040-7043) showing almost the same colony morphology were randomly selected for species identification. These selected isolates were observed for their morphology using an optical microscope (ZEISS AXIOSKOP40; Carl Zeiss, Jena, Germany) and a scanning electron microscope (SEM; Hitachi, Tokyo, Japan). The shape of a streaked culture (DUCC 7040) grown on YEME agar is shown in Fig. 1A. The culture colony was large, irregular, and white or cream colored (Fig. 1A and 1B). Cell shape showed normal yeast cells with bud scar and birth scar, indicating that this species has multiple budding ability (Fig. 1C and 1D). Cell sizes were measured as 2~3 µm by SEM observation (Fig. 1D). The four isolates showed the same cell morphology (data not shown).
Fig. 1.
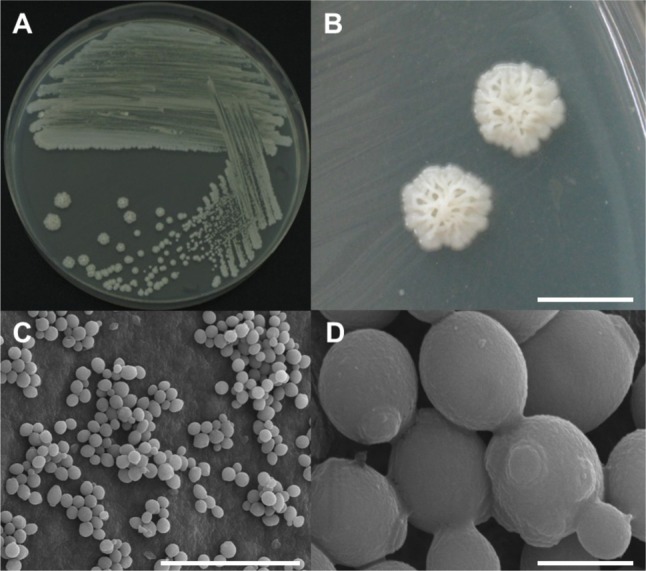
Morphological characterization of Candida kashinagacola (DUCC 7040) observed using a stereoscopic microscope and scanning electron microscope (SEM). A, Growth on a yeast extract-malt extract agar plate; B, Stereoscopic microscope images of colonies; C, D, SEM images of yeast cells (scale bars: B = 5 mm, C = 25 µm, D = 2 µm).
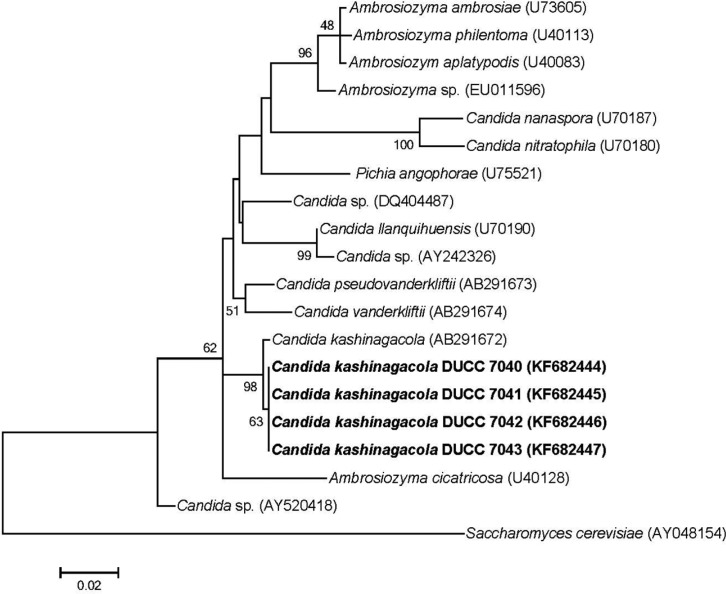
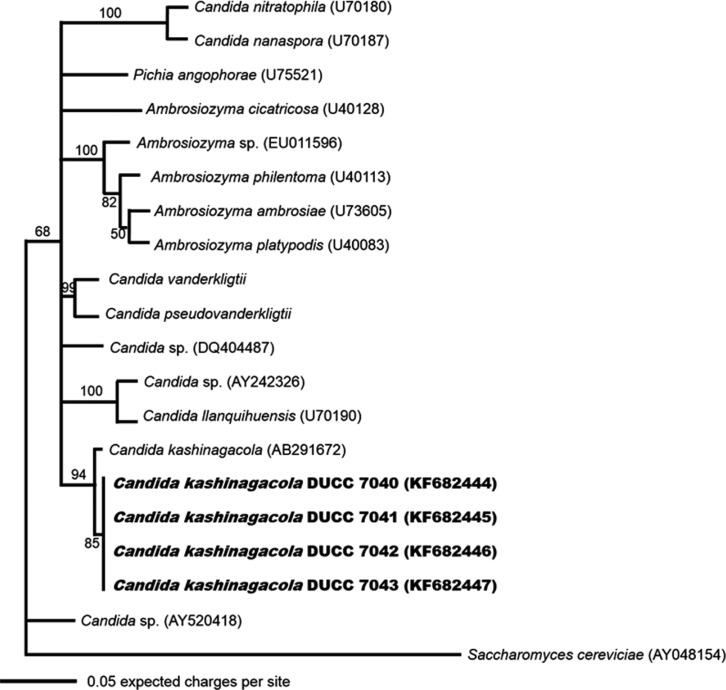
Because the cell morphology was not adequate for identification of yeast species, we analyzed the large subunit (LSU) rDNA D1/D2 region, which has been used widely in identification and evolutional study of yeast species. Yeast genomic DNA was prepared from cells of the four isolates grown in YEME liquid media using the glass bead method [7]. PCR was performed using the primer sets LROR and LR 4 [8, 9]. The PCR amplicons of the LSU rDNA D1/D2 region were sequenced at Macrogen (Seoul, Korea). The determined nucleotide sequences were searched through BLASTN in the GenBank database (http://www.ncbi.nlm.gov/BLAST). The search results showed that the determined sequences had highest homology with known LSU rDNA D1/D2 region sequences of Candida kashinagacola. Thus, we aligned and performed similarity analysis against the determined nucleotide sequences with those of several related taxa downloaded from the GenBank database. All four morphologically similar yeast isolates showed 99% similarity with C. kashinagacola (GenBank accession No. AB291672). In order to further confirm the relationships of the four yeast isolates to Candida kashinagacola and closely related species, phylogenetic analyses were performed using both the Mega 5 program [10] and MrBayes program [11]. Phylogenetic trees were constructed based on LSU rDNA D1/D2 region sequences using the maximum likelihood method. Bootstrap values were generated with 1,000 replicates. The four isolates were closely grouped with C. kashinagacola (Figs. 2 and 3). These results strongly suggest that the four yeast isolates are C. kashinagacola. Consequently, we identified all four isolates as C. kashinagacola and registered the determined LSU rDNA D1/D2 region nucleotide sequences of the four C. kashinagacola isolates in the GenBank database with accession Nos. FK682444-FK682447.
Fig. 2.
Phylogenetic tree of Candida kashinagacola based on nucleotide sequences of large subunit rDNA. The tree was generated using the maximum likelihood method using the Mega 5 program. Saccharomyces cerevisiae was used as the outgroup species.
Fig. 3.
Phylogenetic tree of Candida kashinagacola based on nucleotide sequences of large subunit rDNA. The tree was generated using the Markov chain Monte Carlo (MCMC) method using MrBayes program. Saccharomyces cerevisiae was used as the outgroup species.
Of particular interest, C. kashinagacola (synonym Ambrosiozyma kashinagacola) has recently been reported as a new species from the beetle galleries of Platypus quercivorus in Japan [12]. P. quercivorus is an ambrosia beetle that serves as a vector of Japanese oak wilt disease. Because our isolates were obtained from the beetle galleries of Platypus koryoensis, the insect vector of Korean oak wilt disease, we are certain that C. kashinagacola is a common species that occurs in both Japan and Korea in association with ambrosia beetles that attack oak trees.
In order to obtain further information on the physiological properties of C. kashinagacola, we investigated the ability of utilizing 19 kinds of carbon sources listed in Table 1 using the API 20 C AUX yeast identification kit (BioMérieux, Marcy l'Etoile, France). The four isolates were inoculated on this kit and incubated for 48 hours. The utilizing ability was verified by detection of carbon sources, which was distinguished by change in color and becoming cloudy. The four isolates utilized were D-glucose, D-cellobiose, N-acetyl-D-glucosamine, and D-sorbitol only. The results for the four isolates are shown in Table 1. In comparison of our results with those of C. kashinagacola reported from Japan, the Japanese isolate could not utilize 14 of the 19 carbon sources. There was only one difference in the number of utilized carbon sources (Table 1). Among the 19 carbon sources, D-glucose, D-melezitose, N-acetyl-D-glucosamine, and glycerol were utilized by the Japanese C. kashinagacola [12]. D-Glucose and N-acetyl-D-glucosamine were commonly utilized by both Korean and Japanese C. kashinagacola isolates.
Table 1.
Comparison of the ability to utilize carbon sources between two Candida kashinagacola isolates from Japan and Korea
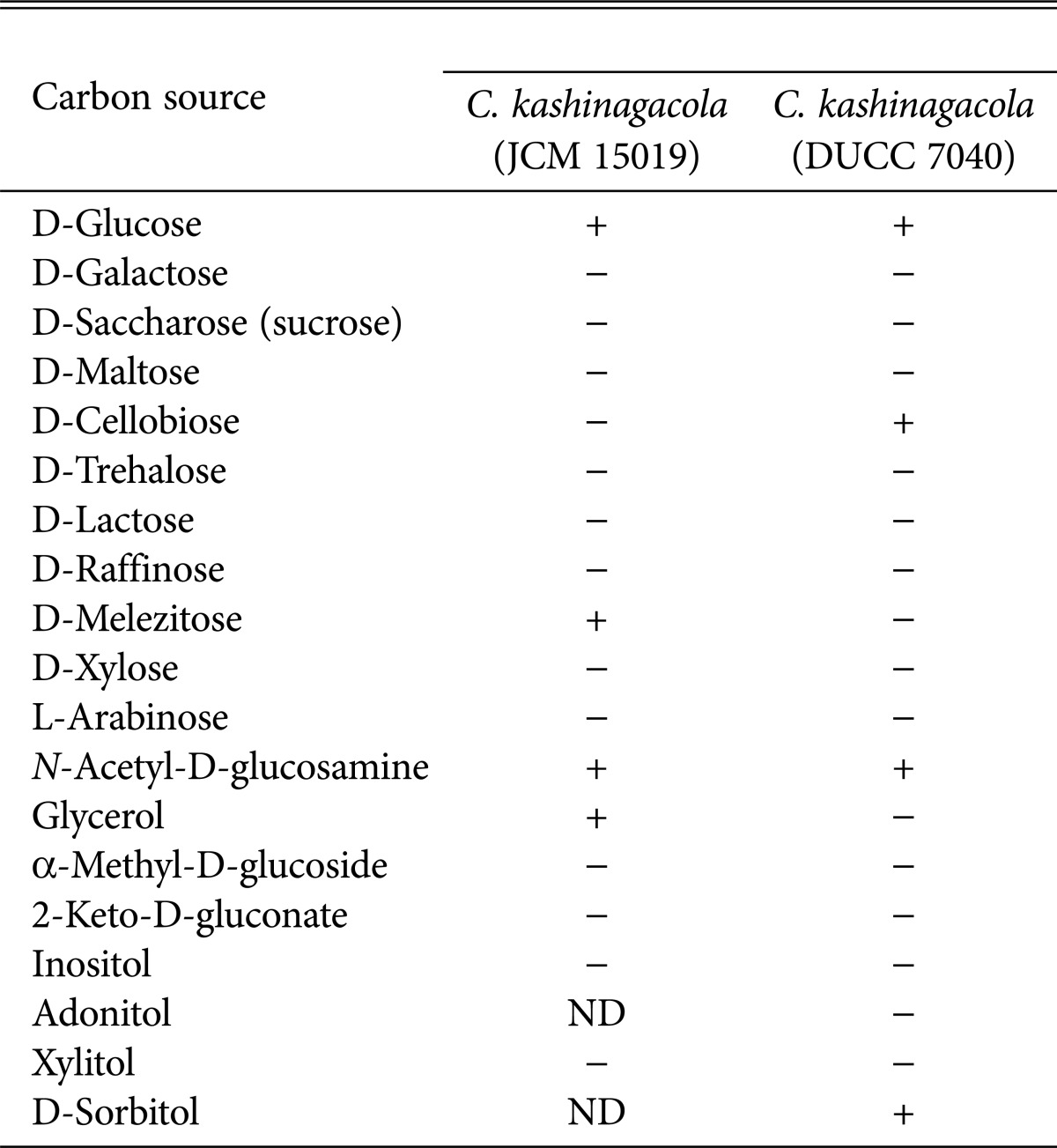
JCM, Japan Collection of Microorganisms; DUCC, Dankook University Culture Collection; ND, no data; +, able to utilize; -, not able to utilize.
In conclusion, we identified and described C. kashinagacola as an unrecorded species in Korea. Because different Platypus beetle species were found in East Asian countries such as Vietnam, Thailand, and China, it would be interesting to investigate the question of whether or not the presence of C. kashinagacola is confined to Korea and Japan.
ACKNOWLEDGMENTS
This research was supported by the Korea Forest Research Institute, National Research Foundation of Korea (616-2011-3-F00002), and the National Institute of Biological Resources, Korea.
References
- 1.Manolakaki D, Velmahos G, Kourkoumpetis T, Chang Y, Alam HB, De Moya MM, Mylonakis E. Candida infection and colonization among trauma patients. Virulence. 2010;1:367–375. doi: 10.4161/viru.1.5.12796. [DOI] [PubMed] [Google Scholar]
- 2.Suh SO, Nguyen NH, Blackwell M. Yeasts isolated from plant-associated beetles and other insects: seven novel Candida species near Candida albicans. FEMS Yeast Res. 2008;8:88–102. doi: 10.1111/j.1567-1364.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- 3.Shin KS, Shin YK, Yoon JH, Park YH. Candida thermophila sp. nov., a novel thermophilic yeast isolated from soil. Int J Syst Evol Microbiol. 2001;51(Pt 6):2167–2170. doi: 10.1099/00207713-51-6-2167. [DOI] [PubMed] [Google Scholar]
- 4.Hong KJ, Kwon YD, Park SW, Lyu DP. Platypus koryoensis (Murayama) (Platypodidae; Coleoptera), the vector of oak wilt disease. Korean J Appl Entomol. 2006;45:113–117. [Google Scholar]
- 5.Suh DY, Hyun MW, Choi IJ, Kim SH, Seo ST, Kim KH. Filamentous fungi and yeasts isolated from Platypus koryoensis and the beetle infested oak tree in Korea; Proceeding of Asian Mycological Congress 2011 & the 12th International Marine and Freshwater Mycology Symposium; 2011 Aug 7-11; Incheon, Korea. Seoul: The Korean Society of Mycology; 2001. p. 196. [Google Scholar]
- 6.Suh DY, Hyun MW, Kim SH, Seo ST, Kim KH. Filamentous fungi isolated from Platypus koryoensis, the insert vector of oak wilt disease in Korea. Mycobiology. 2011;39:313–316. doi: 10.5941/MYCO.2011.39.4.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haugland RA, Brinkman N, Vesper SJ. Evaluation of rapid DNA extraction methods for the quantitative detection of fungi using real-time PCR analysis. J Microbiol Methods. 2002;50:319–323. doi: 10.1016/s0167-7012(02)00037-4. [DOI] [PubMed] [Google Scholar]
- 8.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Geland DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 9.O'Donnell K, Nirenberg HI, Aoki T, Cigelnik E. A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience. 2000;41:61–78. [Google Scholar]
- 10.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 12.Endoh R, Suzuki M, Benno Y, Futai K. Candida kashinagacola sp. nov., C. pseudovanderkliftii sp. nov. and C. vanderkliftii sp. nov., three new yeasts from ambrosia beetle-associated sources. Antonie Van Leeuwenhoek. 2008;94:389–402. doi: 10.1007/s10482-008-9256-9. [DOI] [PubMed] [Google Scholar]