Abstract
Fungal pathogens have caused severe damage to the commercial production of Pleurotus eryngii, the king oyster mushroom, by reducing production yield, causing deterioration of commercial value, and shortening shelf-life. Four strains of pathogenic fungi, including Trichoderma koningiopsis DC3, Phomopsis sp. MP4, Mucor circinelloides MP5, and Cladosporium bruhnei MP6, were isolated from the bottle culture of diseased P. eryngii. A species-specific primer set was designed for each fungus from the ITS1-5.8S rDNA-ITS2 sequences. PCR using the ITS primer set yielded a unique DNA band for each fungus without any cross-reaction, proving the validity of our method in detection of mushroom fungal pathogens.
Keywords: Fungal pathogens, ITS primer, Mushroom, Pleurotus eryngii
The king oyster mushroom, Pleurotus eryngii, is one of the most cultivated mushrooms, particularly in East Asia. It has been cultivated with solid substrates in a wide-mouth polypropylene (PP) bottle [1]. Use of the PP bottle has enabled mass production of this mushroom in a confined cultivation facility. However, in return, cultivation in a confined room has resulted in exposure of the whole plant to attack by pathogenic microorganisms when the facility fails to maintain a high degree of sanitation [2]. Maintenance of a clean environment is particularly important in the mycelia development stage because, in general, the mushroom growth rate is significantly lower than that of some pathogenic microorganisms. A few pathogenic spores can easily out-compete the mushroom mycelia. Occasional outbreaks of mushroom diseases have cost significant loss of production for most commercial P. eryngii farms in Korea.
Various pathogens, including virus, bacteria, and fungi, have been identified from cultivation of P. eryngii. PeSV, a mycovirus, was isolated from deformed fruiting bodies and mycelia [3]. Due to loose environmental control, e.g., LIV and MVX to Agaricus bisporus [4, 5] and OMSV to P. ostreatus [6], mycoviruses have often caused massive damage to mushrooms cultivated in open spaces, however, impact of PeSV on P. eryngii has been limited until now because its outbreak has been effectively suppressed by control of environmental conditions. Meanwhile, many bacterial pathogens have been isolated from the diseased mycelia and fruiting bodies [2]. Isolates, belonging to Staphylococcus epidermidis, Enterobacter amnigenus, Bacillus cereus, and Pseudomonas tolaasii [7], inhibit growth of mycelia and thus cause uneven development of mycelia at the early stage of cultivation. When found at the fruiting stage, they cause partial loss of production yield, softening of tissues, and reduction of shelf-life.
Considering their adverse effects on mushroom production, study of fungal pathogens has been inadequate. Species belonging to genera Cladobotryum and Trichoderma have been reported to cause cobweb disease [8] and green mold disease [9] of Pleurotus mushrooms, respectively. Control of fungal pathogens is particularly difficult because pathogenic mycelia can hardly be discriminated from mushroom mycelia. Therefore, development of effective methods for detection of fungal pathogenic infection in mushroom spawns and mycelia cultures is important. Accordingly, in this report, fungal strains were isolated from diseased mushroom cultures and were partially identified using 18S rDNA sequences. Detection of the infected fungi was facilitated by PCR using internal transcribed sequence (ITS)-specific primer sets.
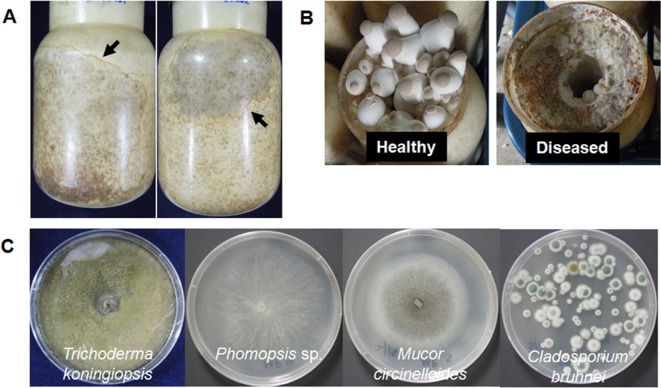
Fungi-infected mushroom cultures were collected from four commercial farms located in Gyeongnam province, Korea. The farms were major providers of P. eryngii KNR2312 culture in Korea, producing 20,000~30,000 bottles of fully developed mushroom culture daily. A typical symptom of fungal infection was out-growth of fungal mycelia over mushroom mycelia, resulting in formation of characteristic sectors of fungal mycelia, which were distinguishable by their color, within the substrate bottle (Fig. 1A). Once the fungal mycelia started to grow, they soon occupied the whole substrate bottle, resulting in total or partial inhibition of primordia formation (Fig. 1B).
Fig. 1.
Symptoms of diseased Pleurotus eryngii KNR2312 caused by fungal pathogens. A, Mycelial culture infected by fungal pathogens. Arrows indicate the area of fungal infection; B, Inhibition of primordia formation by infected fungi; C, Morphological characteristics of isolated fungi.
In order to isolate fungi from the substrate bottle, the inner part of the substrate in the infected bottle was taken and suspended in 1 mL of sterile phosphate-buffered saline buffer. The suspension (100 mL) was spread onto a potato dextrose agar plate (Oxoid Ltd., Basingstoke, UK) and the plate was then incubated at 25℃ until the fungal colonies grew out. Four morphologically distinct fungi were isolated from the infected bottles (Fig. 1C). Results of 18S rDNA analysis after PCR using a forward primer (NS1, 5'-GTAGTCATATGCTTGTCTC-3') and a reverse primer (NS8, 5'-TCCGCAGGTTC ACCTACGGA-3') revealed that they were species of Trichoderma koningiopsis, Phomopsis sp., Mucor circinelloides, and Cladosporium bruhnei, by showing more than 99% identity in the nucleotide sequence (Table 1). Fungal species belonging to Trichoderma have been reported to cause green mold disease in Pleurotus mushrooms; this infection cost up to 78.6% of production yield [10]. M. circinelloides and Phomopsis sp. were reported as phytopathogens causing postharvest decay of tomato [11] and spot disease of grapevine [12], respectively. Phomopsis sp. was also identified as having pathogenicity to P. eryngii [13].
Table 1.
Fungal pathogens isolated from the bottle culture of Pleurotus eryngii
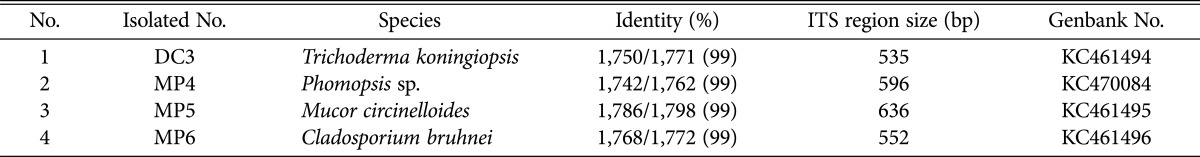
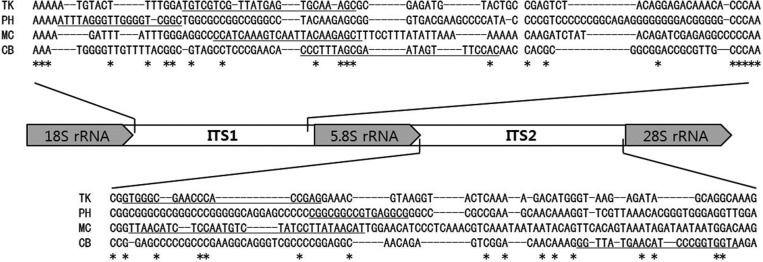
ITS regions of fungal ribosomal RNA are highly variable sequences, which can be useful markers for discrimination of fungal species [14]. The ITS1-5.8S rDNA-ITS regions of the fungal isolates were amplified from the extracted genomic DNA using fungal domain specific primers: ITS1 (5'-TCCGTAGGTGAACCTGCGG-3') and ITS4 (5'-TCCTCCGCTTATTGATATGC-3'). PCR was performed using a Taq DNA polymerase (Enzynomics Co., Daejeon, Korea): 94℃ for 3 min; 25 cycles at 94℃ for 25 sec, 55℃ for 25 sec, and 72℃ for 30 sec; 72℃ for 5 min, yielding 535, 596, 636, and 552 bp for T. koningiopsis, Phomopsis sp., M. circinelloides, and C. bruhnei, respectively (Table 1). The PCR fragments were purified and cloned into a TA cloning vector for determination of their sequences. Results of BLAST analysis of the determined sequences indicated that they contained ITS1 and ITS2 sequences separated by well-conserved 5.8S rDNA. The sequences were deposited in GenBank (NCBI) with the accession numbers shown in Table 1.
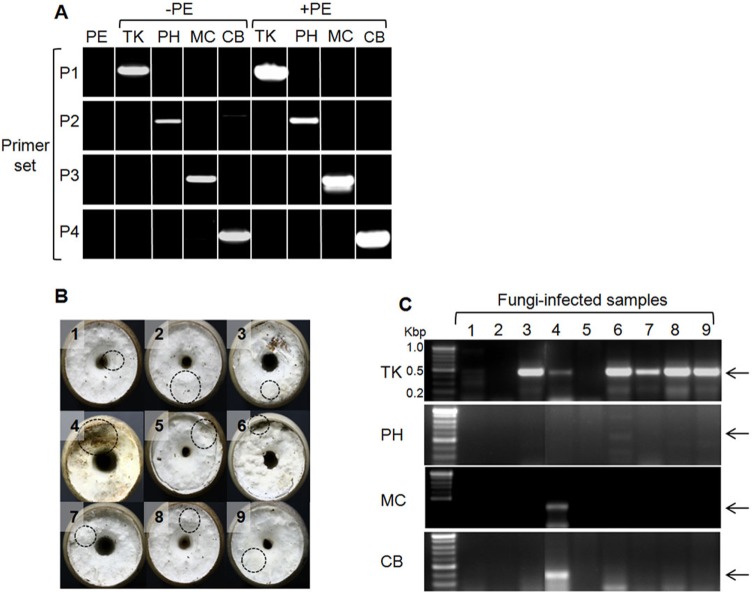
Multiple sequence alignment of the fungal ITS1 and ITS2 sequences showed a high degree of variability (Fig. 2). For development of the species-specific primers, four fungal-specific ITS primer sets based on the species-distinctive sequences were designed (Table 2). The primer sets were employed for detection of fungal pathogens using the following PCR conditions: 94℃ for 5 min; 25 cycles at 94℃ for 30 sec, 60℃ for 30 sec, and 72℃ for 30 sec; 72℃ for 5 min. PCR using each primer set yielded a unique DNA band having the expected size, range 362~439 bp (Fig. 3A). None of the primer sets cross-reacted with the host P. eryngii chromosomes (lane PE and lanes with +PE) (Fig. 3A), indicating that the ITS primer set was exclusively specific to corresponding pathogenic fungi.
Fig. 2.
Multiple sequence alignment of the internal transcribed region (ITS) regions from isolated fungi. The specific primers for each fungal strain were underlined in the sequence.
Table 2.
Sequence of internal trascribed spacer primers
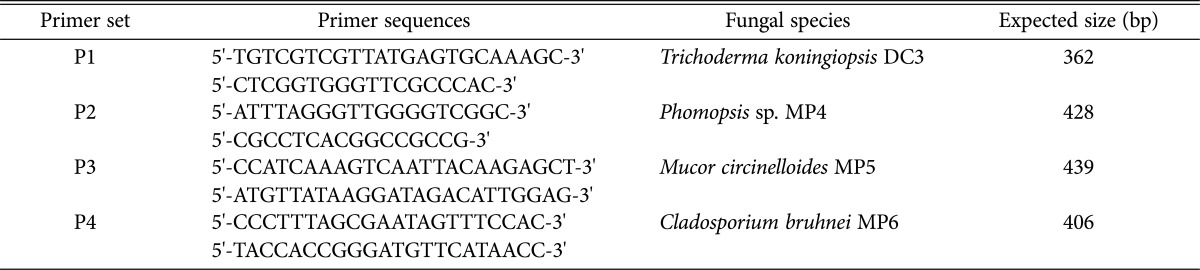
Fig. 3.
Specific detection of fungi using the specific primer sets. A, Specificity of the internal transcribed spacer primer sets. Abbreviations on top of the lanes represent fungal species (PE, Pleurotus eryngii KNR2312; TK, Trichoderma koningiopsis; PH, Phomopsis sp.; MC, Mucor circinelloides; CB, C. bruhnei); B, Morphology of cultivated bottle samples. Dotted circles indicate locations of sampling for PCR analysis; C, PCR analysis with samples from B. Arrows indicate the expected sizes of PCR products.
Next, the PCR detection method using the ITS primer sets was applied to nine field samples collected from a farms near Jinju city, Korea, which failed to produce fruiting bodies. As shown in Fig. 3B, most of the bottles did not show any sign of fungal infection because the mycelial morphology between the pathogens and the mushroom was indistinguishable with bare eyes, particularly at the early stage of infection. However, results of PCR using the ITS primer sets revealed that sample Nos. 3, 4, and 6~9 were infected with T. koningiopsis (Fig. 3C). Due to its rapid rate of growth, Trichoderma has been regarded as one of the most detrimental fungal pathogens. Bottle No. 4, which showed the most severe infection, was found to carry multiple infections with T. koningiopsis, M. circinelloides, and C. bruhnei, validating applicability of the ITS primers in detection of mushroom pathogenic fungi. Three bottles (Nos. 1, 2, and 5) were free of any of the four fungi, meaning that they could be infected with unidentified pathogenic microorganisms.
In conclusion, this study represents an attempt to develop a method for detection of fungal pathogens from P. eryngii mushroom farms. Variability of the ITS sequence region enabled design of species-specific fungal primer sets. PCR analysis using the primer sets can be a useful assay for early detection of fungal infection.
ACKNOWLEDGEMENTS
This work was supported by the Mushroom Export Research Program and Technology Development Program for Agriculture and Forestry, Ministry of Agriculture and Forestry, Korea.
References
- 1.Lee CY, Park JE, Kim BB, Kim SM, Ro HS. Determination of mineral components in the cultivation substrates of edible mushrooms and their uptake into fruiting bodies. Mycobiology. 2009;37:109–113. doi: 10.4489/MYCO.2009.37.2.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim Y, Ryu JS, Shi S, Noh W, Kim E, Le QV, Lee HS, Ro HS. Isolation of bacteria associated with the king oyster mushroom, Pleurotus eryngii. Mycobiology. 2008;36:13–18. doi: 10.4489/MYCO.2008.36.1.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ro HS, Kang EJ, Yu JS, Lee TS, Lee CW, Lee HS. Isolation and characterization of a novel mycovirus, PeSV, in Pleurotus eryngii and the development of a diagnostic system for it. Biotechnol Lett. 2007;29:129–135. doi: 10.1007/s10529-006-9206-4. [DOI] [PubMed] [Google Scholar]
- 4.Goodin MM, Schlagnhaufer B, Romaine CP. Encapsidation of the La France disease-specific double-stranded RNAs in 36-nm isometric viruslike particles. Phytopathology. 1992;82:285–290. [Google Scholar]
- 5.Grogan HM, Adie BA, Gaze RH, Challen MP, Mills PR. Double-stranded RNA elements associated with the MVX disease of Agaricus bisporus. Mycol Res. 2003;107(Pt 2):147–154. doi: 10.1017/s0953756203007202. [DOI] [PubMed] [Google Scholar]
- 6.Yu HJ, Lim D, Lee HS. Characterization of a novel single-stranded RNA mycovirus in Pleurotus ostreatus. Virology. 2003;314:9–15. doi: 10.1016/s0042-6822(03)00382-9. [DOI] [PubMed] [Google Scholar]
- 7.González AJ, González-Varela G, Gea FJ. Brown blotch caused by Pseudomonas tolaasii on cultivated Pleurotus eryngii in Spain. Plant Dis. 2009;93:667. doi: 10.1094/PDIS-93-6-0667B. [DOI] [PubMed] [Google Scholar]
- 8.Back CG, Lee CY, Seo GS, Jung HY. Characterization of species of Cladobotryum which cause cobweb disease in edible mushrooms grown in Korea. Mycobiology. 2012;40:189–194. doi: 10.5941/MYCO.2012.40.3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komon-Zelazowska M, Bissett J, Zafari D, Hatvani L, Manczinger L, Woo S, Lorito M, Kredics L, Kubicek CP, Druzhinina IS. Genetically closely related but phenotypically divergent Trichoderma species cause green mold disease in oyster mushroom farms worldwide. Appl Environ Microbiol. 2007;73:7415–7426. doi: 10.1128/AEM.01059-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi IY, Jung GT, Ryu J, Choi JS, Choi YG. Physiological characteristics of green mold (Trichoderma spp.) isolated from oyster mushroom (Pleurotus spp.) Korean J Mycol. 2003;15(1):110. [Google Scholar]
- 11.Smith WL, Jr, Moline HE, Johnson KS. Studies with Mucor species causing postharvest decay of fresh produce. Postharvest Pathol Mycotoxins. 1979;69:865–869. [Google Scholar]
- 12.Mostert L, Crous PW, Kang JC, Phillips AJ. Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: morphological, cultural, molecular and pathological characterization. Mycologia. 2001;93:146–167. [Google Scholar]
- 13.Ferri F, Ciccarone C. Pests and polluting organisms in Pleurotus eryngii crop. Agric Ric. 1998;20:1–39. [Google Scholar]
- 14.Martin KJ, Rygiewicz PT. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 2005;5:28. doi: 10.1186/1471-2180-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]