Abstract
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by symptoms related to altered social interactions/communication and restricted and repetitive behaviors. In addition to genetic risk, epigenetic mechanisms (which include DNA methylation/demethylation) are thought to be important in the etiopathogenesis of ASD. We studied epigenetic mechanisms underlying the transcriptional regulation of candidate genes in cerebella of ASD patients, including the binding of MeCP2 (methyl CpG binding protein-2) to the glutamic acid decarboxylase 67 (GAD1), glutamic acid decarboxylase 65 (GAD2), and Reelin (RELN) promoters and gene bodies. Moreover, we performed methyl DNA immunoprecipitation (MeDIP) and hydroxymethyl DNA immunoprecipitation (hMeDIP) to measure total 5-methylcytosine (5-mC) and 5-hydroxymethylcytosine (5-hmC) in the same regions of these genes. The enrichment of 5-hmC and decrease in 5-mC at the GAD1 or RELN promoters detected by 5-hmC and 5-mC antibodies was confirmed by Tet-assisted bisulfite (TAB) pyrosequencing. The results showed a marked and significant increase in MeCP2 binding to the promoter regions of GAD1 and RELN, but not to the corresponding gene body regions in cerebellar cortex of ASD patients. Moreover, we detected a significant increase in TET1 expression and an enrichment in the level of 5-hmC, but not 5-mC, at the promoters of GAD1 and RELN in ASD when compared with CON. Moreover, there was increased TET1 binding to these promoter regions. These data are consistent with the hypothesis that an increase of 5-hmC (relative to 5-mC) at specific gene domains enhances the binding of MeCP2 to 5-hmC and reduces expression of the corresponding target genes in ASD cerebella.
Keywords: autism, cerebellum, epigenetics, gene expression, postmortem
Introduction
Idiopathic ‘Autism Spectrum Disorder' (ASD) is a neurodevelopmental disorder characterized by symptoms associated with social interactions, communication skills and restrictive or repetitive behaviors. The clinical presentation of autism has been reported to overlap with several related neurodevelopmental disorders including Rett syndrome (RTT) and methyl CpG binding protein-2 (MeCP2) duplication syndrome.1,2 Altered MeCP2 levels due to gene deletions or mutations are responsible for the majority of RTT cases.3 Furthermore, an increase in MeCP2 gene dosage found in MeCP2 gene duplication cases4 impacts a critically important epigenetic mechanism, hence altering brain development and leading to an autistic-like phenotype.
MeCP2 is a member of the methyl-binding domain family of proteins that is highly expressed in CNS neurons and is particularly enriched in GABAergic interneurons.5,6 MeCP2 binds with high affinity to modified CpG dinucleotides and is an essential epigenetic regulator operative during human brain development.5,7 The functional importance of MeCP2 in the regulation of synaptic and neuronal plasticity, development of motor skills, cognitive and social behavior is well documented.7 While idiopathic ASD is rarely associated with MeCP2 mutations or duplications, reduced MeCP2 levels are reported in brains of ASD patients.8 This suggests that in addition to gene dose effects, epigenetic mechanisms may also regulate the expression of MeCP2.9, 10, 11
The role of MeCP2 in regulating transcription is more complex than previously proposed and it also appears to act as a positive modulator of gene expression, rather than strictly as a transcriptional repressor.9 Whether MeCP2 acts to repress or activate genes likely depends on post-translational modifications (e.g. phosphorylation, acethylation, ubiquitination) and/or cytosine modification status (i.e. 5-hydroxymethylcytosine (5-hmC) or 5-methylcytosine (5-mC)) of associated target genes.12,13 Any mechanistic explanation for the pleiotropic action of MeCP2 (as either a positive or negative modulator of transcription) is likely to involve the binding affinity of MeCP2 for 5-hmC.13 In other words, the differential action of MeCP2 at 5-hmC- or 5-mC-enriched DNA domains may be associated with complex structural transitions, which impact promoter accessibility to ancillary factors that are either positive or negative.
CpG and non-CpG DNA methylation proximal to promoters or within gene bodies has been generally regarded as a highly stable epigenetic mark that ensures gene expression homeostasis and maintains neuronal phenotype integrity.14 However, recent studies suggest that this epigenetic marking is highly dynamic. It is now believed that steady-state DNA methylation is maintained by the opposing action of DNA-methyltransferases (DNMT) and an active DNA-demethylation pathway.15 DNA demethylation involves the initial hydroxylation of 5-mC to form 5-hmC by members of the TET protein family.16,17 The adult mammalian brain expresses high levels of 5-hmC reaching values of approximately 40% of methylated CpG sites in Purkinje cells.18,19 Compared with the other brain areas, the cerebellum of RTT patients contains enriched amounts of 5-hmC.13,20 Recent studies show that there is an inverse relationship between 5-hmC (enriched) and 5-mC (depleted) abundance in the gene body domains of genes expressed in the cerebella of RTT patients.13,20
Because the levels of 5-hmC in relation to 5-mC have not been extensively studied in the cerebella of ASD patients, we sought to further examine the role of MeCP2 in reading epigenetic marks (5-hmC/5-mC) of active target genes whose expression is altered. We focused on the cerebellum which is anatomically, histologically and functionally a well-defined brain structure that has been associated with ASD pathogenesis.21, 22, 23 One well-established finding in postmortem studies of ASD patients is a decrease in glutamic acid decarboxylase 67 (GAD1), glutamate decarboxylase 65 (GAD2), and reelin (RELN) expression in the cerebellum.22, 23, 24, 25 Hence we selected GAD1, GAD2, and RELN as target genes for these epigenetic studies. We report that MeCP2 binding at the GAD1 and RELN promoters is increased and that this increase is associated with an enrichment of local 5-hmC levels relative to the levels of 5-mC in the cerebella of ASD patients.
Materials and methods
Subjects and cerebellar collection
Blocks of cerebellar cortex from 10 CON and 10 ASD were obtained frozen at −80 °C from the Harvard Brain Tissue Resource Center, McLean Hospital (Belmont, MA, USA) with approval of Autism Speaks (Autism Tissue Program). For each sample, we received a coronal section (0.5 cm block) of cerebellar cortex inferior to the horizontal fissure. From the available neuropathology reports at the level of where the surface was cut, there were no signs of infarction, hemorrhages or inflammatory lesions, with the exception of one ASD subject, which had a large early intermediate stage infarct. Subjects were roughly matched for age, sex, PMI and brain mass. The demographic information associated with each group of subjects is listed in Supplementary Table 1. Patients with Asperger's syndrome, Fragile–X syndrome, RTT, pervasive developmental disorder not otherwise specified, and 15q11-q13 duplication were not included. In addition, all control cases were free of neurological disorders, seizures, mental retardation, dementia, and so on based on medical records.
RNA isolation and quantification of mRNA using real-time PCR
mRNA extraction, purification and measurements of mRNA expression were conducted using a previously published protocol.26 mRNAs corresponding to DNMT1, MeCP2, TET1, GAD67, GAD65 and RELN were measured. ACTB and GAPDH were used as internal controls for sample normalization (see Supplementary Table 2). Each target was run in duplicate and the two housekeeping genes were run in parallel. Values were calculated as relative abundance to the mean of the two housekeeping genes after normalization.
Western blot analysis
For protein quantification we conducted measurements as described in detail elsewhere.26 Anti-MeCP2 polyclonal antibodies (1:2000 dilution, Diagenode, Denville, NJ, USA) were used to detect MeCP2 protein (~75 kDa). Changes in MeCP2 protein levels in ASD vs CON were determined by expressing the data as a percentage of the CON.
Chromatin immunoprecipitation assays
We performed chromatin immunoprecipitation (ChIP) assays based on protocols as previously described.27 The percentage of immunoprecipitated DNA were calculated using the following: % (gene−IP/total input)=2(Ct(10% input)−3.32)−Ct(gene−IP) × 100%. TET1 (Zymo, Irving, CA, USA) and DNMT1 antibodies (Abcam, Cambridge, MA, USA) were previously shown to bind to single immunoreactive bands on western blots.27,28
Methylated and hydroxymethylated DNA immunoprecipitation (MeDIP and hMeDIP)
Methylated/hydroxymethylated DNA immunoprecipitation experiments were performed as previously described29 using 5-mC and 5-hmC monoclonal antibodies (Diagenode, Denville, NJ, USA). The percent methylated or hydroxymethylated DNA/ input DNA was calculated as described for ChIP.
Amplicon-specific TAB pyrosequencing
The distribution of 5-hmC and 5-mC in the GAD1 promoter amplicon was determined by a modification of TAB-seq.30 We followed the method originally described in detail.31 Reagents were obtained from Wisegene (Chicago, IL, USA). Bisulfite conversion was performed using the EZ DNA Lightning Kit (Zymo, Irvine, CA, USA). To determine the efficiency of each enzymatic step, spike-in controls were added and independently scored by conventional Sanger sequencing. Unmethylated λDNA (cI857 Sam7, Promega, Madison, WI, USA) was methylated in vitro with CpG Methyltransferase (New England Biolabs, Ipswich, MA, USA) for a methylated control template. To generate a 5-hmC-containing control, a 275-bp segment corresponding to 1635–1810 bp of pGEM1 (Promega) was amplified using linearized pGEM1 and 5-hydroxymethyl-2'- deoxycytidine-5'-triphosphate (Bioline,Taunton, MA, USA ) during PCR amplication.30,31
DNA from three ASD and three CONs were each immunoprecipitated (three IPs/patient) with either 5-hmC or 5-mC antibodies (as described above). DNA from each patient sample was separately pooled and concentrated. In parallel, three 5-hmC and 5-mC IPs were prepared from each patient for bisulfite conversion directly to determine the total number of Cs present at each position. Following β-glucosyltransferase protection, Tet1-assisted oxidation and bisulfite conversion, DNA was amplified with Pfu Turbo Cx DNA polymerase (Agilent, Santa Clara, CA, USA). Percent 5-hmC and 5-mC at each position along the GAD1 amplicon was measured with pyrosequencing (Epigendx, Hopkinton, MA, USA). The GAD1 bisulfite primers (Epigendx, Assay ADS3737) were tested to ensure the absence of methylation-based amplification bias.
Statistical analysis
The primary test of the difference between CON and ASD for each mRNA, protein, ChIP, hMeDIP and MeDIP assays was an independent two sample t-test for equal and unequal variance as appropriate. Additional analyses were conducted using analyses of variance and covariance controlling for factors such as gender, PMI, pH, presence of medication, type of medication using PASW v.18 software (SPSS, Armonk, NY, USA). Relationships between mRNA and protein expression variables and ChIP level variables were analyzed with Pearson correlations and scatter plots. Two-sided probability levels were used for statistical significance (P<0.05), or trends (P<0.1).
Results
MeCP2 binding to GABAergic and glutamatergic gene promoters and body regions
As shown by comparing Figures 1a and b, MeCP2 binds more efficiently to the gene body regions than to the CpG-rich promoter regions of RELN, GAD1 and GAD2. The binding of MeCP2 to these promoter and gene body regions appears to be selective, as MeCP2 binding is virtually absent (<0.01%) in the promoter region of the housekeeping gene GAPDH (−122 bp to +42 bp). As shown in Figure 1a, there is a 1.5–2-fold increased binding of MeCP2 to both the GAD1 (P=0.04) and RELN (P=0.03) promoters in cerebella of ASD compared with CON. In contrast, MeCP2 binding to the GAD2 promoter region did not show significant changes in ASD vs CON (P=0.32). Furthermore, MeCP2 binding to RELN, GAD1 and GAD2 body regions failed to show significant changes in ASD (Figure 1b).
Figure 1.
MeCP2 binding to RELN, GAD1 and GAD2 promoters and gene bodies and correlations with corresponding mRNA in the cerebella of CON and ASD. (a) MeCp2 binding to the promoter regions of RELN ((−220 to +70), (*P=0.03)) and GAD1 ((−55 to +121), (*P=0.04)) are increased in ASD vs CON, while GAD2 ((−1507 to +1310) (P=0.32)) is unchanged in ASD vs CON. (b) MeCP2 binding to gene body regions of RELN ((+562 to +763), (P=0.2)), GAD1 ((+656 to +856), (P=0.6)) and GAD2 ((+1293 to +1447), (P=0.9)) are unchanged in ASD vs CON. (c) Increased binding of MeCP2 to the RELN promoter was associated with reduced RELN mRNA (Pearson r2=−0.66, *P=0.04, n=10) in ASD. (d) Increased binding of MeCP2 to the GAD1 promoter shows a trend to statistical significance with decreased GAD1 mRNA expression (Pearson r2=−0.53; P=0.08, n=10) in ASD. ASD, autism spectrum disorder; CON, control; GAD1, glutamic acid decarboxylase 67; GAD2, glutamic acid decarboxylase 65; RELN, Reelin.
Increased binding of MeCP2 protein to the RELN promoter correlated with reduced expression of RELN mRNA in ASD (Figure 1c). The increased binding of MeCP2 to the GAD1 promoter was also negatively correlated with GAD1 mRNA expression (Figure 1d), however due to the variability and relatively small number of subjects, this correlation showed only a trend towards statistical significance (P<0.08).
We tested whether background variables or other possible confounding variables may have influenced the binding of MeCP2 to the target genes in the ASD samples. As shown in Supplementary Table 1, average values for the confounding variables were similar in CON and ASD. Furthermore, ANCOVA did not reveal a statistically significant correlation between MeCP2 binding to either the GAD1 or RELN promoters with changes in age (GAD1 vs MeCP2, Pearson r2=−0.31, P=0.19; RELN vs MeCP2, Pearson r2=−0.39, P=0.09), sex (GAD1 vs MeCP2, Pearson r2=0.28, P=0.25; RELN vs MeCP2, Pearson r2=−0.14, P=0.56), PMI (GAD1 vs MeCP2, Pearson r2=−0.3, P=0.23; RELN vs MeCP2, Pearson r2=−0.33, P=0.17) in CON and ASD patients considered either together or separately. In addition we did not find a statistically significant correlation between PMI and GAD1 promoter 5-hmC (Pearson r2=−0.22, P=0.45) or 5-mC enrichment (Pearson r2=−0.033, P=0.914), and RELN promoter 5-hmC (Pearson r2=−0.39, P=0.15), or 5-mC enrichment (Pearson r2=−0.02, P=0.94) in CON and ASD patients. Race was a confounding factor for which we had insufficient information and so was not included in the above analysis. This was particularly the case in the CON group (see ethnicity, Supplementary Table 1). In a subpopulation of ASD subjects with seizure comorbidity (N=4), including one subject with signs of cerebellar infarct, the levels of mRNAs encoding MeCP2, GAD1, RELN; MeCP2 binding to GAD1 and RELN; and 5-hmC enrichment of the GAD1 and RELN promoters, were each similar to the average values obtained from the entire ASD population.
We found no differences in MeCP2 binding between ASD subjects who were treated (antipsychotics, antidepressants or mood stabilizers (n=7)), duration of treatment, and the few ASD (n=3) that apparently were non-treated at the time of death. Hence, the study shows that a diagnosis of ASD is a significant predictor for increased MeCP2 binding to the GAD1 and RELN promoters in cerebellar cortex of ASD patients.
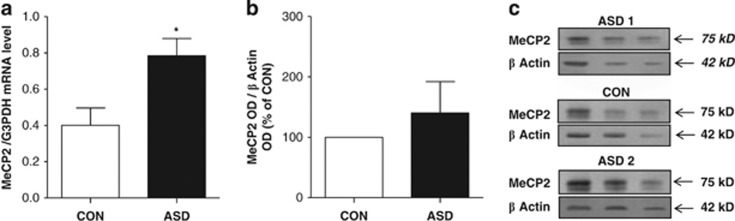
MeCP2 mRNA and protein levels in cerebella of ASD patients
As shown in Figure 2a, the expression of MeCP2 mRNA was increased (by ~70%) in ASD cerebella when compared with CON. Overall, correlation analysis showed a positive correlation between MeCP2 mRNA level and binding of MeCP2 protein to the RELN (Pearson r2=0.54, P=0.04) and GAD1 promoters (Pearson r2=0.59, P=0.01). In contrast, there were no correlations between MeCP2 mRNA expression and binding of MeCP2 protein to the RELN, GAD1 and GAD2 gene body regions.
Figure 2.
MeCP2 mRNA and protein expression in CON vs ASD. (a) MeCP2 mRNA expression is increased in ASD vs CON (*P=0.01). Values were corrected for expression of housekeeping gene GAPDH mRNA. All values are expressed as means±s.e.m. (b) MeCP2 protein levels fail to increase in ASD samples relative to CON (P=0.45, n=10). MeCP2 protein levels in ASD were expressed as percentage relative to CON (100%). (c) Representative immunoblots show a major band of 75 kDa for MeCP2 and 42 kDa for β-actin. ASD, autism spectrum disorder; CON, control.
When measured in cerebellar cortical homogenates, the level of MeCP2 protein was also increased in six and decreased in four of ten ASD vs CON. Hence, the increase in mean MeCP2 protein levels did not reach statistical significance (Figure 2b).
Enrichment of 5-hmC, but not of 5-mC at RELN and GAD1 promoters
As shown in Figure 3a, the levels of 5-hmC detected by 5-hMeDIP, were significantly enriched at the GAD1 and RELN promoters of ASD patients whereas 5-mC (Figure 3b) failed to change. Interestingly, there is a statistically significant inverse correlation between the 5-hmC content at the GAD1 and RELN promoters and their corresponding mRNA levels (Figure 3c) in ASD patients. A similar correlation was not observed between the GAD1 and RELN promoter 5-mC content and the corresponding mRNA levels (Figure 3c). Furthermore, there was no significant enrichment in the level of 5-hmC and 5-mC in the gene body regions of RELN, GAD1 and GAD2 in ASD vs CON (Figures 3a and b). The levels of 5-hmC or 5-mC in the gene body regions of GAD2 were very low and virtually not distinguishable from background (Figures 3a and b). This result indicates a degree of specificity of the observed changes that appear restricted to the promoter regions of the genes analyzed.
Figure 3.
Comparison of 5-hmC and 5-mC enrichment in two different gene regions of the RELN, GAD1, GAD2 and correlations between 5-hmc enrichment in the promoter regions of RELN and GAD1 with the respective mRNA levels in the cerebella of CON and ASD. (a) Increased enrichment of 5-hmC at the promoters of RELN ((−220 to +70 bp) (*P=0.026)) and GAD1 ((−55 to +121 bp), (*P=0.047)), but not GAD2 ((−1507 to −1310 bp), (P=0.36)) in cerebella of ASD vs CON. No changes in the enrichment of 5-hmC at the gene bodies of RELN ((+562 to +763 bp), (P=0.22)), GAD1 ((+562 to +763 bp), (P=0.53)) and GAD2 ((+1293 to +1447 bp), (P=0.46)) in cerebella of ASD vs CON. (b) No changes in the enrichment of 5-mC at the promoter of RELN ((−220 to +70 bp), (P=0.91)), GAD1 ((−55 to +121 bp), (P=0.67)) and GAD2 ((−1507 to −1310 bp), (P=0.36)) in cerebellum of ASD vs CON. There were no changes in the enrichment of 5-mC at body regions of RELN ((+562 to +763 bp), (P=0.16)), GAD1 ((+656 to +856), (P=0.11)) and GAD2 body ((+1293 to +1447 bp), (P=0.46)) in cerebella of ASD vs CON. (c) Statistically significant correlation of 5-hmC in the promoter of GAD1 ((−55 to +121 bp), (Pearson r2=−0.40, *P=0.036)) and RELN ((−220 to +70 bp), (Pearson r2=−0.65, *P=0.01)) with corresponding mRNA levels in ASD samples. No significant correlations between 5-mC content in the promoters of GAD1 ((−55 to +121 bp), (Pearson r2=−0.14, P=0.71)) and RELN ((−220 to +70 bp) vs ASD (Pearson r2=−0.10, P=0.75)) with mRNA levels were evident in ASD (vs CON) samples. ASD, autism spectrum disorder; CON, control; GAD1, glutamic acid decarboxylase 67; GAD2, glutamic acid decarboxylase 65; RELN, Reelin.
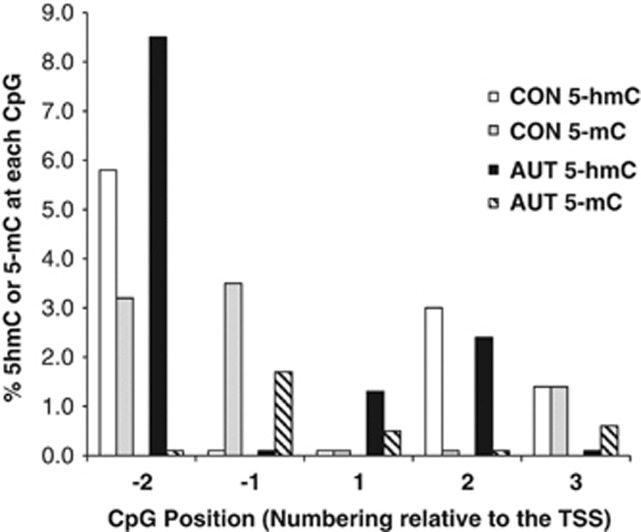
In order to validate the results obtained from the MeDIP and hMeDIP assays, we chose three representative samples for further analysis of the GAD1 promoter enrichment by Tet-assisted bisulfite pyrosequencing with base resolution. Our study shows (Figure 4) that in CON subjects, 5-hmC is preferentially enriched at CpG position −2 of the GAD1 promoter (sequence relative to the TSS: −54 to +69 bp). In the same samples, the levels of 5-mC is low but detectable at positions −2, −1 and 3 and absent in other CpG sites studied. In ASD patients (as compared with CON), 5-hmC is preferentially enriched at CpG position −2 with smaller amounts at position 1 and 2. 5-mC is depleted at this site in the ASD samples but present at these positions in the CON samples (see Figure 4). These methylation changes result in a significant increase in the amounts of 5-hmC relative to 5-mC and the overall 5-hmC/5-mC ratio at the GAD1 promoter (−54 to +69 bp) is 1.2 in CON and 5.5 in ASD patient DNA.
Figure 4.
TAB-seq shows the differential distribution of 5-mC and 5-hmC along the GAD1 promoter. The GAD1 promoter includes regulatory elements proximal to both sides of the transcriptional start site (TSS).32 Pyrosequencing was used to determine the % 5-hmC or 5-mC (y axis) at five CpG sites proximal to the GAD1 TSS. CpGs (x axis) are numbered relative to the TSS (NCBI Ref: NM_000817) and Cs correspond to the following positions: −2=−5 bp, −1=−1 bp, 1=+8 bp, 2=+27 bp and 3=+48 bp. Bisulfite-treated DNA from −54 bp to +69 bp of the GAD1 gene was amplified. While the fragment assayed is shorter, it contains the amplicon used to immunoprecipitate the 5-hmC and 5-mC containing GAD1 fragments (−55 bp to +123 bp, see Supplementary Figure 1). Each bar represents the mean of three sample measurements. Total 5-hmC and 5-mC content is determined by the number of Cs at each position following bisulfite conversion and amplification of the same region. Based on evaluations of the 5-hmC (pGEM1) and the 5-mC (λDNA), we estimate the efficiency of β-glucosylation (protection) to be >95%, while the bisulfite conversion efficiency was greater than 99%. CON, control; GAD1, glutamic acid decarboxylase 67 GAD1.
DNMT1 and TET1 mRNA levels
TET1 mRNA was increased in the cerebella of ASD patients and this is consistent with the increased binding of TET1 to the GAD1 and RELN promoters (Table 1a). This is also consistent with the increased 5-hmC levels observed at the GAD1 and RELN promoters (Figures 3a and b). DNMT1 mRNA was virtually unchanged in the cerebella of ASD patients as was the binding of DNMT1 to GAD1 and RELN promoters (see Table 1b).
Table 1. Summary of mRNA levels and DNA binding of TET1 (a) and DNMT1 (b) to the RELN and GAD1 promoters in cerebella of ASD vs CON.
| (a) |
ChIP (% of DNA input) |
(b) |
ChIP (% of DNA input) |
|||
|---|---|---|---|---|---|---|
| TET1 relative mRNA | TET1 binding to RELN | TET1 binding to GAD1 | DNMT1 relative mRNA | DNMT1 binding to RELN | DNMT1 binding to GAD1 | |
| CON | 0.021±0.010 | 0.011±0.021 | 0.070±0.011 | 0.76±0.25 | 0.20±0.010 | 0.11±0.020 |
| ASD | 0.052 ±0.010 | 0.031±0.064 | 0.100±0.013 | 0.60±0.11 | 0.16±0.030 | 0.13±0.020 |
| P-value | 0.01 | 0.01 | 0.046 | 0.6 | 0.5 | 0.6 |
Abbreviations: ASD, autism spectrum disorder; ChIP, Chromatin ImmunoPrecipitation; CON, control. % of DNA input was calculated as 2(Ct(10% input)−3.32)−Ct(gene−IP) × 100% where IP is the amount of specific gene-immunoprecipitated DNA and input is the amount of the same gene in the starting material. Relative mRNA levels were calculated relative to the amounts of GAPDH mRNA.
Discussion
By measuring the binding of MeCP2 to the CpG-rich promoter and body regions of GAD1 and RELN, we unexpectedly detected a marked and significant increase in MeCP2 binding to the promoters but not the corresponding body regions of these genes in cerebellar cortices of ASD patients. Importantly, demographic data analyses of the patient cohort shows that a diagnosis of ASD, and not other confounding variables, is a significant predictor of increased MeCP2 binding to the GAD1 and RELN promoters. In addition, increased binding of MeCP2 is not restricted to the cerebellum. In fact, a similar increase of MeCP2 binding to the GAD1 and RELN promoters is observed in prefrontal cortices of the same patients.33
The increased binding of MeCP2 to the GAD1 and RELN promoters could be due to increased levels of MeCP2, an increased affinity of MeCP2 (or post-translationally modified MeCP2) to DNA, or an increased presence of methylated or hydroxymethylated cytosines at the corresponding promoters (or a combination of these). We report that, in our cohort, the levels of MeCP2 mRNA and protein are not decreased, but show a tendency to increase in the cerebella of ASD patients. However, an increase of MeCP2 or a post-translationally modified MeCP2 cannot fully explain why the binding of this protein to the GAD1 and RELN promoters is increased. In fact, increased MeCP2 binding to these promoter regions is also not accompanied by increased binding to the respective gene bodies indicating that the levels of MeCP2 (or of a post-translationally modified MeCP2) are not a limiting factor in determining the distribution of MeCP2 at specific gene domains (i.e. promoters vs gene bodies).
A recent study demonstrated that mouse MeCP2 binds to both 5-mC and 5-hmC with high affinity.13 Using MeDIP and hMeDIP with modification-specific antibodies, we found an enrichment of 5-hmC (but not 5-mC) at the GAD1 and RELN promoters and not the promoter of GAD2 in ASD patients. In ASD patients that express comparable amounts of MeCP2 as controls, our data demonstrate that there is more MeCP2 binding to these genes and that the increased binding of MeCP2 to the GAD1 and RELN promoters is likely related to the increased amounts of 5-hmC present.
Both MeCP2 knockout, conditional-mutant, and MeCP2 knock-in mice have been widely used to model various aspects of MeCP2 loss or gain of function in RTT or ASD patients.34, 35, 36 Conditional loss of MeCP2 in vesicular inhibitory amino acid transporter expressing GABAergic mouse neurons results in reduced inhibitory transmitter quantal size, which is accompanied by an ~50% decrease in the levels of Gad1 and Gad2 mRNA and reduced GABA immunoreactivity.5 However, according to recent reviews of MeCP2 function in mutant mice, impaired biological function of MeCP2 occurs as the result of loss of expression, underexpression and also overexpression.2,37 Hence, it is not surprising that ASD cerebella are associated with an increased MeCP2 binding to target gene promoters.
Genome-wide analysis of modified cytosines (5-hmC and 5-mC) across DNA from mouse cerebella shows that 5-hmC is enriched in actively expressed genes while 5-mC is depleted at these same regions.13 The resultant 5-hmC/5-mC ratio is increased and functionally coupled with increased transcriptional activity.13 Our data show that while the ratio of 5-hmC/5-mC has a tendency to be higher at both the GAD1 and RELN promoters and gene bodies in ASD cerebella (compared with CON), the increased ratio did not correlate with increased mRNA expression. In fact, we found that the increased 5-hmC content corresponding to the GAD1 and RELN promoters inversely correlated with gene expression. We should note that the gene body and promoter regions reported in our study are quite small (~200 bps) compared with those used in genome wide studies (>50 kb).13 Our results raise the interesting possibility that the observed increase in TET1 expression leads to elevated levels of 5-hmC, which facilitates increased binding of MeCP2 perhaps preventing transcription. In addition, we demonstrate that DNMT1 (methylation writer) is not changed in the cerebella of ASD patients whereas TET1 is increased significantly in these same samples (see Table 1). Moreover, we show that the binding of TET1 to candidate target promoters (GAD1 and RELN) is increased in ASD cerebella. We previously reported that TET1 mRNA is increased in the cortex of SZ patients and that this correlates with increased levels of 5-hmC at the GAD1 promoter.26,29
Comparisons of wild type and MeCP2 null (knockout) mice showed that MeCP2 has no role in establishing the levels of 5-hmC and that regional 5-hmC/5-mC ratios occur by a mechanism independent of MeCP2 binding or expression.13 Instead, the observed increase in TET1 expression in ASD cerebella is associated with an enrichment of 5-hmC at selected promoters, including GAD1 and RELN. Hence, our data suggest that the increased binding of MeCP2 to the GAD1 and RELN regulatory domains may be mediated by an enrichment of 5-hmC. Taken together, these findings extend a role for MeCP2 from MeCP2 duplication syndrome to idiopathic ASD patients. Moreover, these data underscore the importance of regional variations in 5-hmC and 5-mC content in the regulation of GAD1 and RELN expression. It seems plausible to suggest that these regional variations may lead to alterations in the dissociation of MeCP2 from specific gene targets in ASD cerebella. Collectively, we believe that recent studies13,20 (including our own) warrant additional investigations into the role of 5-hmC in modulating differential transcription in ASD brains.
Acknowledgments
We would like to acknowledge the Autism Tissue Program and the Harvard Brain Tissue Resource Center (supported in part by NIH R24 MH-068855) for providing human brain tissue samples. This work was supported in part by NIH P50 HD055751 and 5R01 MH093348 to EHC and AG, respectively. We would also like to thank the families for their sacrifice and contribution to the research.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Neul JL. The relationship of Rett syndrome and MECP2 disorders to autism. Dialogues Clin Neurosci. 2012;14:253–262. doi: 10.31887/DCNS.2012.14.3/jneul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na ES, Nelson ED, Kavalali ET, Monteggia LM. The impact of MeCP2 loss- or gain-of- function on synaptic plasticity. Neuropsychopharmacology. 2013;38:212–219. doi: 10.1038/npp.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro J, Mellios N, Sur M. Mechanisms and therapeutic challenges in autism spec trum disorders: insights from Rett syndrome. Curr Opin Neurol. 2013;26:154–159. doi: 10.1097/WCO.0b013e32835f19a7. [DOI] [PubMed] [Google Scholar]
- Shimada S, Okamoto N, Ito M, Arai Y, Momosaki K, Togawa M, et al. MECP2 duplication syndrome in both genders. Brain Dev. 2013;35:411–419. doi: 10.1016/j.braindev.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493:327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics. 2006;1:e1–11. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales ML, LaSalle JM. The role of MeCP2 in brain development and neurodevelopmental disorders. Curr Psychiatry Rep. 2012;12:127–134. doi: 10.1007/s11920-010-0097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan RP, Patzel KA, Martin M, Yasui DH, Swanberg SE, Hertz-Picciotto I, et al. Autism Res. 2008. pp. 169–178. [DOI] [PMC free article] [PubMed]
- Gonzales ML, Adams S, Dunaway KW, LaSalle JM. Phosphorylation of distinct sites in MeCP2 modifies cofactor associations and the dynamics of transcriptional regulation. Mol Cell Biol. 2012;32:2894–2903. doi: 10.1128/MCB.06728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology. 2013;38:138–166. doi: 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5- methylcytosine. Cell Res. 2010;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Pan Q, Lin L, Szulwach KE, Song CX, He C, et al. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum Mol Genet. 2012;21:5500–5510. doi: 10.1093/hmg/dds394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009;18:2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip J, Soghomonian JJ, Blatt GJ. Decreased GAD67 mRNA levels in cerebellar Purkinje cells in autism: pathophysiological implications. Acta Neuropathologica. 2007;113:559–568. doi: 10.1007/s00401-006-0176-3. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Aldinger KA, Ashwood P, Bauman ML, Blaha CD, Blatt GJ, et al. Consensus paper: pathological role of the cerebellum in autism. Cerebellum. 2012;11:777–807. doi: 10.1007/s12311-012-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Halt AR, Stary JM, Kanodia R, Schulz SC, Realmuto GR. Glutamic acid decarboxylase 65 and 67 kDa proteins are reduced in autistic parietal and cerebellar cortices. Biol Psychiatry. 2002;52:805–810. doi: 10.1016/s0006-3223(02)01430-0. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Stary JM, Halt AR, Realmuto GR. Dysregulation of Reelin and Bcl-2 proteins in autistic cerebellum. J Autism Dev Disord. 2001;31:529–535. doi: 10.1023/a:1013234708757. [DOI] [PubMed] [Google Scholar]
- Dong E, Gavin DP, Chen Y, Davis J. Upregulation of TET1 and downregulation of APOBEC3A and APOBEC3C in the parietal cortex of psychotic patients. Transl Psychiatry. 2012;2:e159. doi: 10.1038/tp.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Nelson M, Grayson DR, Costa E, Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci USA. 2008;105:13614–13619. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadriu B, Guidotti A, Chen Y, Grayson DR. DNA methyltransferases1 (DNMT1) and 3a (DNMT3a) colocalize with GAD67-positive neurons in the GAD67-GFP mouse brain. J Comp Neurol. 2012;520:1951–1964. doi: 10.1002/cne.23020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A. Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology. 2012;37:531–542. doi: 10.1038/npp.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Jin P, Ren B, et al. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat Protoc. 2012;7:2159–2170. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dong E, Grayson DR. Analysis of the GAD1 promoter: trans-acting factors and DNA methylation converge on the 5' untranslated region. Neuropharmacology. 2011;60:1075–1087. doi: 10.1016/j.neuropharm.2010.09.017. [DOI] [PubMed] [Google Scholar]
- Zhubi A, Chen Y, Guidotti A, Grayson D.Epigenetic mechanisms regulate REELIN and GAD67 expression in autism spectrum disorderProgram No. 56.11. 2012 Neuroscience Meeting Planner. New Orleans, LA, USA: Society for Neuroscience, 2012. Online..
- Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annu Rev Cell Dev Biol. 2011;27:631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Zoghbi HY. Rett syndrome and MeCP2: linking epigenetics and neuronal function. Am J Hum Genet. 2002;71:1259–1272. doi: 10.1086/345360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HT, Zoghbi HY. MeCP2: only 100% will do. Nat Neurosci. 2012;15:176–177. doi: 10.1038/nn.3027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.