Abstract
Development of three dimensional (3D) microenvironments that direct stem cell differentiation into functional cell types remains a major challenge in the field of regenerative medicine. Here, we describe a new platform to address this challenge by utilizing a robotic microarray spotter for testing stem cell fates inside various miniaturized cell-laden gels in a systematic manner. To demonstrate the feasibility of our platform, we evaluated the osteogenic differentiation of human mesenchymal stem cells (hMSCs) within combinatorial 3D niches. We were able to identify specific combinations, that enhanced the expression of osteogenic markers. Notably, these ‘hit' combinations directed hMSCs to form mineralized tissue when conditions were translated to 3D macroscale hydrogels, indicating that the miniaturization of the experimental system did not alter stem cell fate. Overall, our findings confirmed that the 3D cell-laden gel microarray can be used for screening of different conditions in a rapid, cost-effective, and multiplexed manner for a broad range of tissue engineering applications.
The differentiation of stem cells into specialized cell types is governed by microenvironmental cues from the surrounding extracellular matrix (ECM)1,2,3, soluble factors1, matrix stiffness2,3, substrate topography4,5 and direct cell-cell contact. These components commonly act in a synergistic manner to regulate stem cell fate and promote the formation of functional tissues. In particular, the three-dimensional (3D) nature of the ECM plays a crucial role in regulating cell behavior6,7. In fact, many studies have confirmed that cellular functions substantially deviate on 2D substrates compared to 3D microenvironments8,9,10,11. Therefore, it is important to develop methods of studying stem cell responses in 3D settings while controlling the presentation of other microenvironmental signals. An important step to meet these requirements, is the development of 3D combinatorial platforms to simultaneously study stem cell differentiation in response to various cues12.
Multiwell-based assays have been well accepted for screening of stem cell fate inside combinatorial environments7,13,14,15. These, platforms are typically generated by adding stem cell encapsulated hydrogels and ECM proteins into standard multiwell plates13. Other approaches have relied on the deposition and freeze drying of polymeric solutions into multiwell plates to generate scaffold libraries capable of screening cell-matrix interactions within 3D microenvironments14,15. Despite, the wide application of multiwell-based combinatorial platforms, such assays face throughput limitations due to the scarce supply of primary cells and biological signals as well as high reagent costs12.
Recent advancements in robotic microarray technologies have enabled the development of versatile and cost-efficient platforms that can address the limitations of conventional screening assays16,17,18,19,20. These technologies have significantly contributed to our understanding of cell adhesion, proliferation and differentiation on 2D surfaces21,22,23,24,25,26,27,28,29. Although the 2D microarray systems have provided valuable insight regarding the synergetic effects of ECM proteins on stem cell differentiation21,22,28, they do not precisely mimic the in vivo tissue architecture. Microarray technology is currently being utilized to fabricate 3D miniaturized cellular platforms for drug discovery and toxicology research30,31 with limited focus directed toward stem cell differentiation32. For instance, a 3D cellular microgel array was previously created to study the effects of fibroblast growth factor-4 and tretinoin on embryonic stem cell pluripotency32. Due to the few tested conditions, this approach did not embrace the multiplexed screening potential of the microarray technology. Therefore, the development of miniaturized platforms that enables the analysis of stem cell differentiation within 3D combinatorial microenvironments still needs to be fully explored12.
In this work, we present a 3D cell-laden gel microarray platform for combinatorial screening of human mesenchymal stem cells (hMSCs) differentiation in response to multiple ECM and growth factors components. An automated printing strategy, which utilizes 1000-fold less materials and cells, compared to conventional multiwell-based assays was employed to generate arrays of miniaturized cell-laden hydrogel constructs. Each microgel unit, composed of methacrylated gelatin (GE), contained living hMSCs along with ECM proteins and was exposed to osteogenic bone morphogenic proteins (BMPs). From the microarray analysis, we identified ECM combinations, which induced a 2-fold increase in Alkaline Phosphatase (ALP) expression. Furthermore, we evaluated the relevance of our platform within macroscale settings to investigate its translational potential. By utilizing our 3D microarray platforms, it is possible to efficiently screen ECM and growth factor combinations, which promote stem cell differentiation. We envision that, our cell-laden gel microarray platform could potentially accelerate the development of innovative and biomimetic materials for a wide range of applications from tissue engineering to stem cell bioengineering and drug screening.
Results
Fabrication of the cell-laden hydrogel microarray platform
The 3D microarray platform was fabricated with a robotic microarray spotter equipped with four pins, enabling rapid printing of the gel spots (400 spots) in a few minutes. The schematic for the microarray fabrication process is shown in Figure 1a. Primarily, prepolymer solutions consisting of GE hydrogels, hMSCs and different combinations of ECM proteins were premixed in a 384 well plate. Then, the robotic microspotting system was used to print the prepolymer solutions on 3-(trimethoxysilyl) propyl methacrylate (TMSPMA) treated glass slides (25 mm × 75 mm) providing long-term surface stability of the deposited hydrogel droplets (Figure 1a). Subsequently, the deposited droplets were exposed to ultraviolet (UV) light to develop the miniaturized cell-laden constructs (Suppl. Figure 1a, b). Fluorescence imaging revealed a uniform distribution of hMSCs inside 75 μm thick hydrogel constructs (Suppl. Figure 1b), with high cellular viability after day 1 (i.e. 91 ± 6.2% and 78 ± 6.3%) and a slightly lower viability after 7 days of culture (Suppl. Figure 2a, b). The lower viability at day 7 is mainly attributed to the high crosslinking density of the 3D hydrogel matrix.
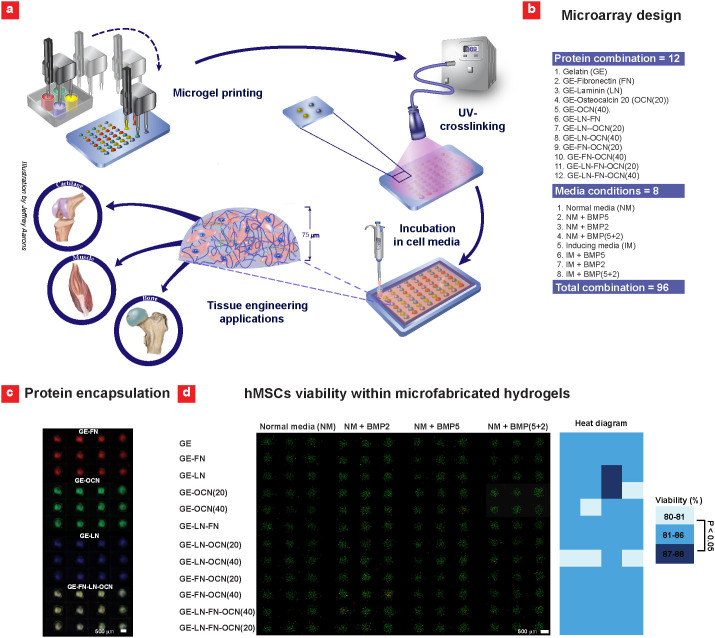
Figure 1. Fabrication of 3D hMSC-laden gel microarray.
(a) A robotic microarray spotter was used to rapidly print droplets consisting of hMSCs, gelatin methacrylate (GE)-based prepolymer solution and various ECM proteins on TMSPMA functionalized glass slide. The printing step was followed by a 15 sec UV light exposure to form the miniaturized cell-laden constructs. Following printing, cell-laden gel microarrays were placed inside sealed chambers (Illustration made by Jeffrey Aarons). (b) Various combinations of ECM proteins and media formulations were used to conduct the microarrays experiments. The concentration of LN and FN was selected to be 40 μg/ml while OCN was printed at two concentrations of 20 μg/ml and 40 μg/ml. (c) Fluorescence images of the encapsulated proteins within the hydrogel constructs after 24 hours in solution. (d) hMSCs viability within 48 combinatorial 3D microenvironments in normal (control) media after 7 days of culture along with color-diagram displaying the quantified cell viability (n = 3–9).
To investigate the effect of ECM components on hMSCs behavior, a range of proteins, including fibronectin (FN, 40 μg/ml), laminin (LN, 40 μg/ml) and osteocalcin (OCN, 20 μg/ml & 40 μg/ml) were entrapped within the GE hydrogels (Figure b). The selected proteins are well known for their roles in osteogenic differentiation of hMSCs33,34,35,36,37,38 and their affinity towards collagen I and IV39,40,41, from which GE is derived. We also examined the effects of growth factors, using bone morphogenic proteins, (BMP2 and & BMP5, 50 ng/ml), which promote osteogenesis via the hedgehog pathway42. Fluorescent labeling (FN-FITC, LN-Rhodamine, and OCN-Cy5) confirmed the presence of FN, LN and OCN in different combinations of the GE microgels tested after 24 hours of encapsulation (Figure 1c). Subsequent viability analysis demonstrated high hMSCs survival (i.e. 80–88%) within miniaturized 3D hydrogel constructs exposed to BMP2 and BMP5 (Figure 1d). Overall, our preliminary results suggested that hMSCs can be uniformly cultured with a high cell survival inside the spotted 3D combinatorial microenvironments.
Osteogenic potential of hMSCs within 3D microarrayed hydrogels
The osteogenic differentiation of hMSCs was evaluated by analyzing the expression of alkaline phosphatase (ALP) in normal (control) and osteogenic media using an automated high content imaging technique (Figure 2a). The images shown in Figure 2a suggested that combinations consisting of multiple ECM proteins lead to the highest ALP expression (Figure 2a). In particular, it was evident that the GE-LN-FN-OCN gels resulted in the most pronounced ALP up-regulation in both control and inducing media compared to the GE condition. Furthermore, the addition of BMP2 and BMP5 had less impact on the ALP expression levels compared to combinatorial ECM proteins (Figure 2a, b).
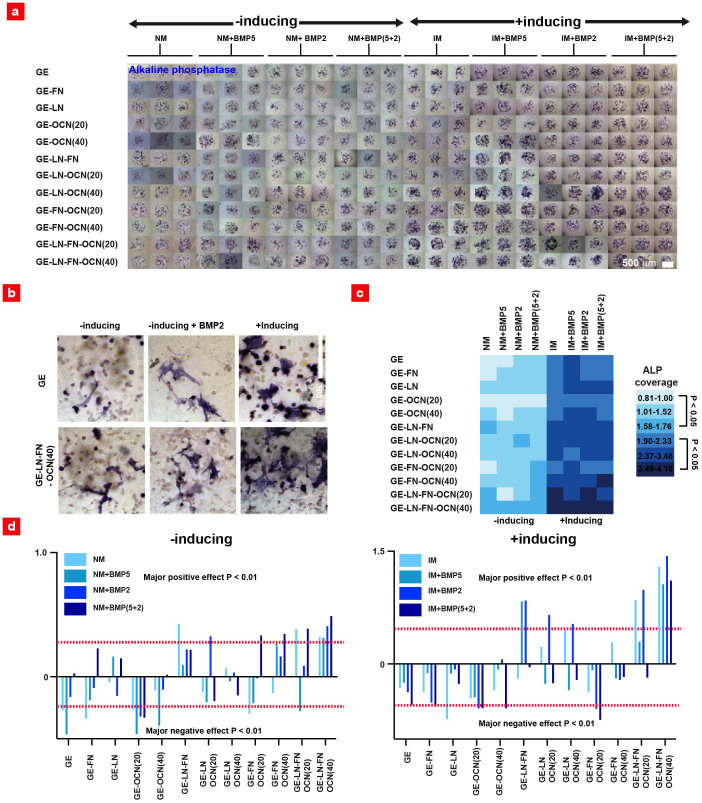
Figure 2. ALP expression within the 3D hMSC-laden gel microarray platform.
(a) Optical images of ALP expression of hMSCs within various 3D combinatorial microenvironments. (b) High magnification images indicating ALP expression inside the representative constructs. (c) Color-diagram representing the normalized ALP coverage within each 3D microenvironment. Groups with the same color code were not statically different from one another (n = 9–22) (d) The scaled estimates of major effects on ALP expression levels calculated using one-way ANOVA analysis.
Quantitative ALP expression analysis, depicted in a color coded diagram, showed that single protein conditions did not significantly promote the differentiation of hMSCs (Figure 2c). On the other hand, hydrogel constructs consisting of combinatorial ECM proteins (GE-FN-OCN and GE-LN-OCN-FN) induced a 2 to 4-fold higher ALP coverage than control condition (GE) depending on the specific media formulation (p < 0.05). Moreover, increasing the concentration of OCN (20 μg/ml to 40 μg/ml) did not have a significant influence on ALP levels.
A one-way ANOVA analysis was performed to statistically compare all available groups for major effects on osteogenic differentiation of hMSCs (Figure 2d). This analysis indicated that out of 96 microenvironments, 20 resulted in a major positive effect on ALP expression (p < 0.01). The majority of these microenvironments contained more than one ECM protein, thus confirming the ALP expression trends observed from the optical images (Figure 2a, b) and the color diagram (Figure 2c). Furthermore, the statistical analysis indicated that nearly 80% of the conditions with major positive effect on ALP expression, contained BMP2 and BMP5 (p < 0.01). This further indicates that although BMP2 and BMP5 can alter the osteogenic fate of hMSCs, their effect on up-regulation of ALP is less than combinatorial ECM microenvironments.
Osteogenic potential of specific combinations within macroscale hydrogels
A key aspect of our 3D cell-laden gel microarray platform is to predict the cell responses in larger constructs (centimeter scale) that can be used for regenerative medicine applications. To this end, specific gel compositions were selected to validate the results of the microarray experiments in macroscale gels. In particular, we chose to validate the following microarray results: a) No significant ALP difference was observed between GE-LN and GE gels, b) GE-LN-FN-OCN gels led to the highest ALP expression, and C) the addition of BMP2 to GE-LN-FN-OCN gels did not have a major impact on ALP expression. This was accomplished by examining the osteogenic potential of the combinations GE, GE-LN, GE-LN-OCN-FN, GE-LN-OCN-FN + BMP2 in control and inducing media using conventional macroscale encapsulation approaches.
Optical imaging indicated an up-regulation of ALP expression in GE-FN-LN-OCN constructs compared to GE ones in both media formulations. Alternatively, in control media, the addition of LN to GE and BMP2 to GE-FN-LN-OCN hydrogels did not change the ALP expression (Figure 3a). These trends were further validated from high-magnification images of the hydrogel constructs as demonstrated in supplementary Figure 3.
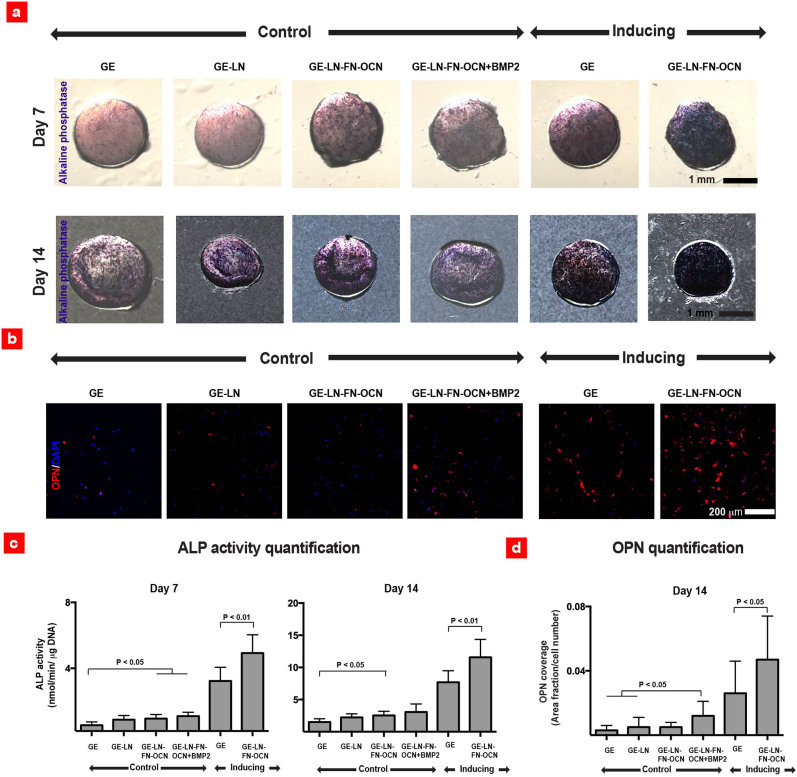
Figure 3. Microarray hit combinations stimulates early osteogenic differentiation of hMSCs within the macroscale GE gels.
(a) ALP expression after 7 and 14 days of culture. (b) Osteopontin (OPN) expression and DAPI staining of cell nuclei along with (c) quantified ALP activity after 7 and 14 days of culture (n = 7–15). (d) Quantification of the normalized OPN coverage after 14 days of culture (n = 9–15).
Osteogenic differentiation of hMSCs can also be identified by production of bone-related extracellular proteins such as osteopontin (OPN)43. After 14 days of culture, the GE-LN-OCN-FN and GE-LN-OCN-FN + BMP2 constructs showed a higher OPN coverage compared to the GE gels in both media formulations (Figure 3b). Consistent with these observations, the quantification of ALP activity and OPN coverage demonstrated that the GE-LN-FN-OCN gels led to a significantly higher ALP expression, while GE-LN-OCN-FN + BMP2 in control media up-regulated OPN expression (Figure 3d). Notably, the ALP activity observed in macroscale setting (Figure 3c) was closely correlated with the microarray results as confirmed by a linear regression fit (Suppl. Figure 4).
Another hallmark of osteogenic differentiation is the production of calcified matrix44. Therefore, we conducted further analysis to identify the ECM combinations, which promoted matrix calcification. As expected, there was no notable calcification in the tested conditions under control medium when compared to the inducing medium, as detected by Alizarin Red staining (Figure 4a). However, the production of calcified matrix in GE-LN-FN-OCN was significantly higher compared to GE hydrogels in inducing media after 14 and 25 days of culture. After 25 days of culture, the GE-LN-FN-OCN constructs displayed dense microstructures that were heavily stained by Alizarin Red in accordance with formation of bone-like calcified structures (Figure 4a). Theses observations were further confirmed through quantification of Alizarin Red stain as shown in Figure 4b.
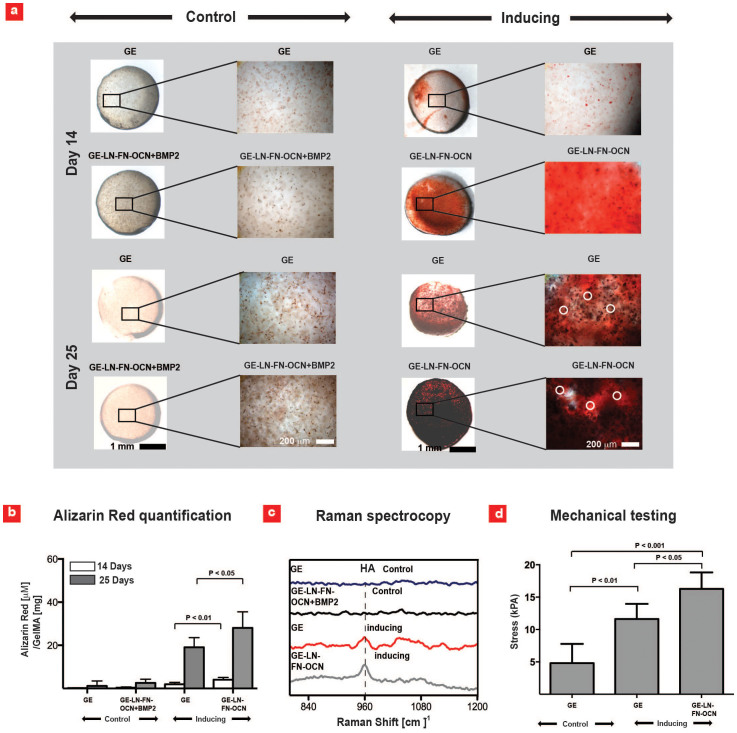
Figure 4. Microarray hit combinations stimulates macroscale bone-mineralization.
(a) Formation of calcified matrix by hMSCs investigated using Alizarin Red S staining after 14 and 25 days of culture; white circles denotes regions of intensified Alizarin Red S staining. (b) Quantified amount of calcified matrix evaluated by Alizarin Red S content inside the hydrogels using a colorimetric assay (n = 4–8). (c) Raman spectra of the hydrogel constructs after 25 days of culture. (d) Unconfined mechanical testing to evaluate the compressive modulus (kPa) of the macroscale hydrogels upon formation of mineralized matrix (n = 4).
Earlier studies have demonstrated that production of mineralized matrix also induce a transformation from transparent into translucent/opaque hydrogel structures45. Consistently, the hydrogels cultured in non-inducing media were nearly transparent, whereas hydrogels in inducing media displayed opaque-white microscale features after 25 days in culture (Suppl. Figure 5). In particular, GE-LN-FN-OCN gels exhibited highest level of white coloration (Suppl. Figure 5) caused by clusters of micrometer-sized nodules, similar to the calcified microparticles seen in Figure 4a.
To investigate the mineral phase of the deposited matrix, a series of Raman spectra was acquired from the macroscale constructs. As shown in Figure 4c, a sharper and more intense hydroxyapatite (HA) peak was detected in GE-LN-FN-OCN compared to GE gels in inducing media confirming the formation of crystalline HA46. Alternatively, there was no HA peak present in the spectral analysis conducted on the control conditions. Matrix mineralization is well known to enhance the mechanical properties of hydrogels45. Therefore, an unconfined compression test was performed to evaluate the mechanical properties of our engineered tissue constructs after 25 days of culture. The results indicated a significant increase in the compressive modulus of GE-LN-FN-OCN gels (16.3 kPA) compared to the GE gels (4.8 kPA) in inducing mediuma (Figure 4d). In addition, all the constructs tested in the inducing media exhibited a significantly higher compressive modulus compared to the samples cultured in control media. In summary, our study confirmed that GE-LN-FN-OCN stimulates osteogenic differentiation of hMCSs and promotes bone mineralization as predicted by the 3D cell-laden gel microarray platform.
Discussion
Fabrication of a miniaturized cell-laden gel microarray platform requires an optimal printing technique, appropriate biomaterials (i.e. hydrogels) and functionalized substrates to ensure high cell survival and experimental stability for long-term studies. We utilized a robotic microspotter system equipped with multiple pins for rapid deposition of the cell-laden hydrogel constructs inside a temperature and humidity controlled chamber to keep the cells viable during the fabrication process. Furthermore, TMSPMA functionalized substrates and GE prepolymer solution were used to ensure high cellular viability, successful ECM protein and cellular encapsulation and platform stability up to 7 days of culture. We noted that it was challenging to maintain the stability of our microarray platform beyond 7 days of culture due to hydrogel degradation most likely caused by hMSC spreading, migration and collagenase secretion of hMSCs. We could potentially improve this by adding naturally derived hydrogels with low degradation rate (i.e. alginate, hyaluronic acid) to the prepolymer solution.
To date, significant efforts have been devoted to develop 2D microarray platforms. However, by using 2D settings, it is difficult to fully understand the role of matrix cues on stem cell differentiation inside native-like 3D microenvironments6. Moreover multiwell-based arrays do not enable high-throughput studies. Therefore, we believe that the 3D microarray platform presented here better simulates in vivo responses of stem cells, while allowing cost-efficient and high-throughput analysis. In fact, there is growing evidence that stem cells elucidate more biomimetic behavior inside 3D microenvironments compared to 2D surfaces8,9,10,11. Specific examples include osteogenic8, and hepatic differentiation9 of embryonic stem cells (ESCs) within 3D microenvironments.
Our interest in bone tissue engineering prompted us to focus on ECM proteins and growth factors that are well known to induce osteogenic differentiation of hMSCs. Both FN and LN promote osteogenic differentiation through binding to integrins α5β1 and α3β1 located on the membrane of hMSCs34,35. Similarly, OCN plays an important role in the formation of mineralized matrix through modulation of HA crystal morphology and growth33. Our findings indicated that, combinations of FN, OCN and LN resulted in a higher ALP expression compared to individual incorporation of ECM proteins (Figure 2). It is widely accepted that functional properties of ECM proteins can be changed through structural alterations directed by protein-protein interactions39,47,48. Such structural alterations could ultimately expose hidden osteogenic regions on FN and LN, thereby enhancing the osteogenic differentiation of hMSCs. Therefore, it is reasonable to assume that these synergistic interactions may be responsible for the up-regulation of ALP observed in the microarray experiments. Similar synergistic interactions were also observed by Flaim21 and Fang13 et al. upon screening of ESCs responses to different ECM combinations on 2D micro- and 3D multiwell-based arrays, respectively21. They reported that the crosstalk between collagen, LN and FN could lead to up-regulation of stem cell markers such as albumin (hepatic marker), osteocalcin (bone marker) and von Willebrand factor (endothelial marker). These studies reinforce the fact that ECM proteins significantly influence the biological functionality of stem cells through structural cues that are directed by protein-protein interactions.
The applicability of our developed 3D microarray platform was further investigated by evaluating the osteogenic differentiation of hMSCs within macroscale gels containing ECM proteins. Significantly higher ALP expression was observed in GE-LN-FN-OCN compared to GE gels after 7 and 14 days of culture. The increase in ALP expression is known to promote bone mineralization by removing extracellular inorganic pyrophosphate (ePPi), a calcification inhibitor, and releasing inorganic phosphate49. Calcium quantification and Raman spectral analysis of the gel constructs along with mechanical tests performed on the GE-LN-FN-OCN gels further confirmed that ‘hit' combinations retrieved from the proposed microarray platform could potentially enhance macroscale bone formation. In summary, our findings demonstrated that the proposed 3D microarray platform provide a unique setting to screen cell-matrix interactions in a high throughput manner for tissue engineering applications (i.e. bone) with potential translational impact.
Conclusion
In this work, a microarray platform consisting of miniaturized hMSC-laden hydrogel constructs was developed for high throughput and cost efficient screening of stem cell-matrix interactions. The microarray system allowed for rapid printing of hMSCs inside various 3D milieus with high cell survival rate. The utility of the microarray platform was further demonstrated by evaluating the osteogenic potential of hMSCs inside 96 different miniaturized 3D niches. A range of ECM proteins were identified, which induced significantly higher ALP expression in combinatorial settings compared to constructs containing single protein. The hit combinations, identified using the microarray platform, were reinvestigated in macroscale setting confirming their potential in the formation of mineralized bone tissue. Our findings indicated that the 3D miniaturized stem cell niches could be used to predict cellular responses and explore appropriate matrix components for a wide range of tissue engineering applications.
Methods
hMSCs cell culture
Bone-marrow derived hMSCs were purchased at passage 2 from a commercial source (Lonza, MD) and expanded in a T-150 flask in mesenchymal stem cell growth media (MSCGM) (Lonza, MD) to passage 3–5 before use. Cells where passaged when 80–90% confluent in accordance with the manufactures instructions and the growth medium was changed twice a week. For osteogenic differentiation studies, the cells were cultured in osteogenic induction media (Lonza, MD) containing mesenchymal cell growth media supplemented with Dexamethasone, L-Glutamine, Ascorbate, Pen/Strep and β-Glycerophosphate.
Methacrylated gelatin synthesis
GE was synthesized according to previously published method50. Briefly, 10 g of gelatin (Sigma) was diluted in 100 mL PBS at 50°C. Then, methacrylation was induced by slowly adding 8% (v/v) methacrylic anhydride to the gelatin solution and reacting at 50°C for 3 h. The final GE solution was dialyzed using a 12–14 kDa dialysis membrane for one week. Following the dialysis process, the solution was frozen at −80°C and later lyophilized for 4 days before the final use for the microarray experiments.
Microarray printing
The microarray printing process was performed on glass slides functionalized with 3-(trimethoxysilyl)propyl methacrylate (TMSPMA) (Sigma, MO). Prior to printing, different combinations of proteins and cell contained hydrogel solution were mixed and injected into 384-well plates. Specifically, the mixtures contained Phosphate Buffered Saline (PBS) (Invitrogen, NY), hMSCs, GE prepolymer solution and various combinations of ECM proteins including FN (Abcam, MA), OCN (Abcam, MA) and LN (BD Biosciences, CA). hMSCs were trypsinized and suspended in a solution containing 5% (w/v) GelMA and 0.5% (w/v) Irgacure 2959 photoinitiator ((2-hydroxy-1-(4-(hydroxyethoxy) phenyl)-2-methyl-1-propanone) (CIBA Chemicals, CH) to reach a final cell concentration of 8 × 106 cell/ml of hydrogel solution. FN and LN concentrations were selected to be 40 μg/ml, while two different concentrations of 40 μg/ml and 20 μg/ml were used for OCN. The mixtures were printed on the TMSPMA coated glass slides using a SpotBot 3 contact microarrayer with four pinheads (Arrayit, CA). During the printing process, the temperature and humidity inside the microarray chamber were kept above 90% and 22°C respectively. The high level of humidity was used to avoid polymer evaporation from the printed spots and potential cell death. The deposited microspots were subsequently crosslinked using UV light (320–500 nm, 800 mW power, 10 cm height) for 15 seconds. Following the printing process, the samples were cultured in hybridization chambers (Grace Bio, OR) containing MSCGM media. After 24 hours, 4 different combinations of BMP2 and BMP5 (Abcam, MA) were added to each chamber. For osteogenic differentiation experiments, the MSCGM media was replaced with osteoinductive media after 24 hours of culture. We assured that the results obtained from the microarray platform was not influenced by cross-communication between the neighboring gels, by printing each microgel combination in isolated 3 × 3 spot quadrants. Moreover, the experiments conducted on glass slides containing scrambled microgel blocks also resulted in similar findings (data not shown), which may have minimized the spot-to-spot crosstalk. Images of the printed microarrays were acquired at 5× magnification using a Zeiss Axio Observer Z1 microscope equipped with a programmable motorized stage for high-content imaging. Images analysis was performed in a high throughput fashion using National Institute of Health (NIH) ImageJ software (v. 1. 4).
Confocal imaging
Confocal images were acquired using a Leica TCS SP5 microscope equipped with a 405 nm diode and a white light laser tuned to 495 nm, a UV corrected HCX PL APO CS 20.0×/0.70N.A. air objective, and Leica hybrid detectors collecting at 415–485 nm and 505–596 nm, respectively, with 1.51 × 1.51 × 2.9 μm voxels (Leica Microsystems, IL). The 3D constructed images were acquired using Imaris v7.6.1 software (Bitplane Inc., CT).
Viability analysis
A Live/Dead Kit (Invitrogen) was used to quantify the percentage of live cells after 1, 3, 5 and 7 days of culture by exposing the printed microarrays slides to calcein AM (Cl) (0.5 μl/ml) and ethidium homodimer (ETD) (2 μl/ml) for 30 min. The stained slides were imaged at 5× magnification using an inverted fluorescence microscope (Zeiss Axio Observer Z1, Zeiss, USA). The obtained images were then analyzed using ImageJ v. 1. 4 software.
Histology
Hydrogel samples were fixed in 4% (v/v) paraformaldehyde (PF) (Electron Microscopy Sciences, PA) and placed in 30% (w/v) sucrose overnight. Samples were then embedded in OCT and cryosectioned (Leica Cryostat) at 20 μm. Each hydrogel section was then prepped according to the subsequent immunostaining procedure provided by the manufacturer.
Osteogenic differentiation
After fixing the samples in 4% (v/v) PF for 20 min, the osteogenic differentiation of the cells was examined using ALP and Osteopontin (OPN) expression as the early differentiation markers. Alizarin Red S was further used to define the matrix calcification as a late marker. A standard substrate containing BCIP/NBT (MP biomedical, OH) was used to evaluate ALP expression. We gently added the solutions to each slide and incubated them for 90 min. The slides were then rinsed three times in PBS for subsequent imaging. The ALP activity was evaluated using a commercially available colorimetric Assay kit (Abcam, MA). Briefly, the gels were digested with 100 μg/ml dispase II (Sigma, USA) and added to a 96-well plate and optical density (O.D.) measurement was performed at 405 nm using a SpectraMax M5 (Molecular devices, CA) spectrometer. From standard curves, we calculated the ALP activity within each representative hydrogel construct. The ALP activity was finally normalized to the DNA content using a Quant-iT PicoGreen kit (Invitrogen, NY) according to the manufacturer's protocols. For OPN staining, the samples were primarily blocked for 2 hours using 2.5% (v/v) goat serum (Invitrogen, NY), 1% (w/v) bovine serum albumin (BSA)(Sigma) and 0.3% (v/v) Triton (Sigma). The samples were then exposed to 1:200 dilution of primary polyclonal rabbit anti-OPN antibody (Abcam, MA) in 1% goat serum in PBS for overnight at 4°C. Following, a 1:200 dilution of Alexa Fluor-555 conjugated goat anti-rabbit secondary antibody (Invitrogen, NY) in 1% goat serum in PBS was added to the slides for 2 hours at room temperature. Between each step the slides were gently washed three times in PBS. The samples were washed repeatedly for three hours in ddH2O and afterwards incubated in ddH2O overnight to remove excess stains. Quantitative analysis of Alizarin Red S was performed by dissolving the stained deposits in 10% (v/v) acetic acid for 30 min followed by 30 min in 10% ammonium hydroxide (w/v) to ensure the pH-value was maintained in the range 4.1–4.3. Finally the OD405 was measured with a SpectraMax M5 (Molecular devices, CA) and compared with the standard curve to extract the Alizarin Red S content from each sample. In all the differentiation readouts we used GE constructs cultured in non-inducing medium as our negative control.
Raman spectroscopy
Mineralization of the matrix was investigated using R200-L SENTERRA Raman microscope equipped with Olympus BX51 microscope stand. The sample was exposed to He–Ne laser at 785 nm wavelength with a power of 5 mW as the excitation source. The final spectrum was collected through averaging of five samples with 8 seconds in between acquisition time.
Mechanical testing
Mechanical properties of hydrated macroscale hydrogel constructs were determined by unconfined compression testing using Instron 5943 Materials Testing System Capacity (Norwood, MA, USA) equipped with a 50 N load cell. The uniaxial compression testing was performed with a crosshead speed of 1 mm/min on circular samples with 8 mm diameter and 1.0–1.4 mm thickness until point of failure. The 5–15% strain region was used to measure the compressive modulus of the samples.
Statistical analysis
To assess the level of significant differences between different experimental groups in the microarray analysis, a one-way ANOVA was performed followed by a post hoc test. We used a Tukey-Kramer all pairs post hoc test when the groups followed a normal distribution and had equal variances, otherwise a Steel-Dwars all pairs non-parametric post hoc test was used. Groups, which were not statistically different, were organized in the same color group represented in a color diagram. To reveal combinations, which had a major osteogenic effect, we carried out a scaled effect screening within various combinations using a one-way ANOVA analysis.
For the macroscale experiments, we followed the same approach when comparing more than three groups. Otherwise either a two-tailed t-test was performed, when the data was normally distributed, or a non-parametric Mann-Whitney test was carried out. In general, averages were considered to be significantly different when the p-value was less than 0.05. All the data were presented as average ± standard deviation (SD). All the statistical analysis was carried out using JMP software (vs. 10).
Author Contributions
A.K. and A.D.P. outlined the research plan. A.D.P. participated/performed all the experiments in the research paper, A.G. helped out with chemical/mechanical characterization of the hydrogels, B.H. with histology, T.F. with confocal microscopy, E.G. and H.A. with optimizing the printing of cell-laden gels, M.N. and G.C.U. with generating the GE hydrogels. M.N., A.K., B.H., G.C.U, M.F. and D.I. contributed to the writing of the paper.
Supplementary Material
A combinatorial cell-laden gel microarray for inducing osteogenic differentiation of human mesenchymal stem cells
Acknowledgments
The authors would like to acknowledge the Office of Naval Research Young National Investigator Award, the Presidential Early Career Award for Scientists and Engineers (PECASE), the National Institutes of Health (HL092836, HL099073, DE019023, DE019024, AR057837), the National Science Foundation CAREER Award (DMR 0847287), The Danish Council for Independent Research (Technology and Production Sciences, 10-100118), and the Wyss Institute for Biologically Inspired Engineering at Harvard University for funding. We also thank Mr. Jeffrey Aarons for making the illustration presented Figure 1(a).
References
- Schuldiner M., Yanuka O., Itskovitz-Eldor J., Melton D. A. & Benvenisty N. Effects of eight growth factors on the differentiation of cells derived from human embryonic stem cells. Proc Natl Acad Sci 97, 11307–11312, 10.1073/Pnas.97.21.11307 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A. J., Sen S., Sweeney H. L. & Discher D. E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689, 10.1016/J.Cell.2006.06.044 (2006). [DOI] [PubMed] [Google Scholar]
- Discher D. E. Matrix elasticity directs stem cell lineage - Soluble factors that limit osteogenesis. Bone 44, S205–S206, 10.1016/J.Bone.2009.03.025 (2009). [Google Scholar]
- Dalby M. J. et al. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat Mater 6, 997–1003, 10.1038/Nmat2013 (2007). [DOI] [PubMed] [Google Scholar]
- Markert L. D. et al. Identification of Distinct Topographical Surface Microstructures Favoring Either Undifferentiated Expansion or Differentiation of Murine Embryonic Stem Cells. Stem Cells Dev 18, 1331–1342, 10.1089/Scd.2009.0114 (2009). [DOI] [PubMed] [Google Scholar]
- Lund A. W., Yener B., Stegemann J. P. & Plopper G. E. The Natural and Engineered 3D Microenvironment as a Regulatory Cue During Stem Cell Fate Determination. Tissue Eng Part B-Re 15, 371–380, 10.1089/Ten.Teb.2009.0270 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A., Brey D. M. & Burdick J. A. High-Throughput and Combinatorial Technologies for Tissue Engineering Applications. Tissue Eng Part B-Re 15, 225–239, 10.1089/Ten.Teb.2009.0049 (2009). [DOI] [PubMed] [Google Scholar]
- Tian X. F. et al. Comparison of osteogenesis of human embryonic stem cells within 2D and 3D culture systems. Scand J Clin Lab Invest 68, 58–67, 10.1080/00365510701466416 (2008). [DOI] [PubMed] [Google Scholar]
- Baharvand H., Hashemi S. M., Ashtian S. K. & Farrokhi A. Differentiation of human embryonic stem cells into hepatocytes in 2D and 3D culture systems in vitro. Int J Dev Biol 50, 645–652, 10.1387/Ijdb.052072hb (2006). [DOI] [PubMed] [Google Scholar]
- Jabbari E. Role of Substrate Microstructure on Osteogenic Differentiation of Mesenchymal Stem Cells. IEEE Eng Med Bio, 3543–3545, 10.1109/Iembs.2010.5627489 (2010). [DOI] [PubMed] [Google Scholar]
- Jarrahy R. et al. Osteogenic differentiation is inhibited and angiogenic expression is enhanced in MC3T3-E1 cells cultured on three-dimensional scaffolds. Am J Physiol-Cell Ph 289, C408–C414, 10.1152/Ajpcell.00196.2004 (2005). [DOI] [PubMed] [Google Scholar]
- Mei Y. Microarrayed materials for stem cells. Mater Today 15, 444–452 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F. et al. Combinatorial extracellular matrices for human embryonic stem cell differentiation in 3D. Biomacromolecules 11, 1909–1914, 10.1021/bm100357t (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. et al. Combinatorial polymer scaffold libraries for screening cell-biomaterial interactions in 3D. Adv Mater 20, 2037, 10.1002/Adma.200702088 (2008). [Google Scholar]
- Simon C. G. & Lin-Gibson S. Combinatorial and High-Throughput Screening of Biomaterials. Adv Mater 23, 369–387, 10.1002/Adma.201001763 (2011). [DOI] [PubMed] [Google Scholar]
- Revzin A. et al. Designing a hepatocellular microenvironment with protein microarraying and poly(ethylene glycol) photolithography. Langmuir 20, 2999–3005, 10.1021/La035827w (2004). [DOI] [PubMed] [Google Scholar]
- MacBeath G. & Schreiber S. L. Printing proteins as microarrays for high-throughput function determination. Science 289, 1760–1763 (2000). [DOI] [PubMed] [Google Scholar]
- Falsey J. R., Renil M., Park S., Li S. J. & Lam K. S. Peptide and small molecule microarray for high throughput cell adhesion and functional assays. Bioconjugate Chem 12, 346–353, 10.1021/Bc000141q (2001). [DOI] [PubMed] [Google Scholar]
- Akinc A. et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol 26, 561–569, 10.1038/Nbt1402 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook A. L. et al. Combinatorial discovery of polymers resistant to bacterial attachment. Nat Biotechnol 30, 868–899, 10.1038/Nbt.2316 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaim C. J., Chien S. & Bhatia S. N. An extracellular matrix microarray for probing cellular differentiation. Nat Methods 2, 119–125, 10.1038/Nmeth736 (2005). [DOI] [PubMed] [Google Scholar]
- Flaim C. J., Teng D., Chien S. & Bhatia S. N. Combinatorial signaling microenvironments for studying stem cell fate. Stem Cells Dev 17, 29–39, 10.1089/Scd.2007.0085 (2008). [DOI] [PubMed] [Google Scholar]
- Gupta N. et al. A versatile approach to high-throughput microarrays using thiol-ene chemistry. Nat Chem 2, 138–145, 10.1038/nchem.478 (2010). [DOI] [PubMed] [Google Scholar]
- Anderson D. G., Levenberg S. & Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotechnol 22, 863–866, 10.1038/Nbt981 (2004). [DOI] [PubMed] [Google Scholar]
- Klim J. R., Li L. Y., Wrighton P. J., Piekarczyk M. S. & Kiessling L. L. A defined glycosaminoglycan-binding substratum for human pluripotent stem cells. Nat Methods 7, 989–972, 10.1038/Nmeth.1532 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brafman D. A., Chien S. & Willert K. Arrayed cellular microenvironments for identifying culture and differentiation conditions for stem, primary and rare cell populations. Nat Protoc 7, 703–717, 10.1038/Nprot.2012.017 (2012). [DOI] [PubMed] [Google Scholar]
- Mei Y. et al. Combinatorial development of biomaterials for clonal growth of human pluripotent stem cells. Nat Mater 9, 768–778, 10.1038/Nmat2812 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soen Y., Mori A., Palmer T. D. & Brown P. O. Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments. Mol Syst Biol 2, 10.1038/Msb4100076 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobaa S. et al. Artificial niche microarrays for probing single stem cell fate in high throughput. Nat Methods 8, 949–955, 10.1038/nmeth.1732 (2011). [DOI] [PubMed] [Google Scholar]
- Fernandes T. G., Diogo M. M., Clark D. S., Dordick J. S. & Cabral J. M. S. High-throughput cellular microarray platforms: applications in drug discovery, toxicology and stem cell research. Trends Biotechnol 27, 342–349, 10.1016/J.Tibtech.2009.02.009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. Y. et al. Three-dimensional cellular microarray for high-throughput toxicology assays. Proc Natl Acad of Sci 105, 59–63, 10.1073/Pnas.0708756105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes T. G. et al. Three-Dimensional Cell Culture Microarray for High-Throughput Studies of Stem Cell Fate. Biotechnol Bioeng 106, 106–118, 10.1002/Bit.22661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang Q. Q., Sicheri F., Howard A. J. & Yang D. S. C. Bone recognition mechanism of porcine osteocalcin from crystal structure. Nature 425, 977–980, 10.1038/Nature02079 (2003). [DOI] [PubMed] [Google Scholar]
- Klees R. F. et al. Laminin-5 induces osteogenic gene expression in human mesenchymal stem cells through an ERK-dependent pathway. Mol Biol Cell 16, 881–890 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino M. M. et al. Controlling integrin specificity and stem cell differentiation in 2D and 3D environments through regulation of fibronectin domain stability. Biomaterials 30, 1089–1097, 10.1016/J.Biomaterials.2008.10.047 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoslahti E. Fibronectin in Cell-Adhesion and Invasion. Cancer Metast Rev 3, 43–51, 10.1007/Bf00047692 (1984). [DOI] [PubMed] [Google Scholar]
- Kikkawa Y. et al. Laminin isoforms differentially regulate adhesion, spreading, proliferation, and ERK activation of beta 1 integrin-null cells. Exp Cell Res 300, 94–108, 10.1016/J.Yexcr.2004.06.031 (2004). [DOI] [PubMed] [Google Scholar]
- Chen M. et al. NC1 domain of type VII collagen binds to the beta 3 chain of laminin 5 via a unique subdomain within the fibronectin-like repeats. J Invest Dermatol 112, 177–183, 10.1046/J.1523-1747.1999.00491.X (1999). [DOI] [PubMed] [Google Scholar]
- Prigodich R. V. & Vesely M. R. Characterization of the complex between bovine osteocalcin and type I collagen. Archives of Biochemistry and Biophysics 345, 339–341, 10.1006/Abbi.1997.0254 (1997). [DOI] [PubMed] [Google Scholar]
- Dzamba B. J., Wu H., Jaenisch R. & Peters D. M. Fibronectin-Binding Site in Type-I Collagen Regulates Fibronectin Fibril Formation. J Cell Biol 121, 1165–1172, 10.1083/Jcb.121.5.1165 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charonis A. S., Tsilibary E. C., Yurchenco P. D. & Furthmayr H. Binding of Laminin to Type-Iv Collagen - a Morphological-Study. Ann Ny Acad Sci 460, 401–403, 10.1111/J.1749-6632.1985.Tb51191.X (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykaras N. & Opperman L. A. Bone morphogenetic proteins (BMPs): how do they function and what can they offer the clinician? J Oral Sci 45, 57–73 (2003). [DOI] [PubMed] [Google Scholar]
- Perrien D. S. et al. Immunohistochemical study of osteopontin expression during distraction osteogenesis in the rat. J Histochem Cytochem 50, 567–574 (2002). [DOI] [PubMed] [Google Scholar]
- Jaiswal N., Haynesworth S. E., Caplan A. I. & Bruder S. P. Osteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitro. J Cell Biochem 64, 295–312, 10.1002/(Sici)1097-4644(199702)64:2<295::Aid-Jcb12>3.0.Co;2-I (1997). [PubMed] [Google Scholar]
- Phadke A., Shih Y. R. V. & Varghese S. Mineralized Synthetic Matrices as an Instructive Microenvironment for Osteogenic Differentiation of Human Mesenchymal Stem Cells. Macromol Biosci 12, 1022–1032, 10.1002/Mabi.201100289 (2012). [DOI] [PubMed] [Google Scholar]
- Tsuji T., Onuma K., Yamamoto A., Iijima M. & Shiba K. Direct transformation from amorphous to crystalline calcium phosphate facilitated by motif-programmed artificial proteins. Proc Natl Acad Sci 105, 16866–16870, 10.1073/Pnas.0804277105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblinski J. E., Wu M., Demeler B., Jacob K. & Kleinman H. K. Matrix cell adhesion activation by non-adhesion proteins. J Cell Sci 118, 2965–2974, 10.1242/Jcs.02411 (2005). [DOI] [PubMed] [Google Scholar]
- Sottile J. et al. Fibronectin-dependent collagen I deposition modulates the cell response to fibronectin. Am J Physiol-Cell Ph 293, C1934–C1946, 10.1152/Ajpcell.00130.2007 (2007). [DOI] [PubMed] [Google Scholar]
- Orimo H. The mechanism of mineralization and the role of alkaline phosphatase in health and disease. J Nippon Med Sch 77, 4–12 (2010). [DOI] [PubMed] [Google Scholar]
- Ramon-Azcon J. et al. Gelatin methacrylate as a promising hydrogel for 3D microscale organization and proliferation of dielectrophoretically patterned cells. Lab Chip 12, 2959–2969, 10.1039/C2lc40213k (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A combinatorial cell-laden gel microarray for inducing osteogenic differentiation of human mesenchymal stem cells