Abstract
Background
Mechanical stimulation is necessary for regulating correct formation of the skeleton. Here we test the hypothesis that mechanical stimulation of the embryonic skeletal system impacts expression levels of genes implicated in developmentally important signalling pathways in a genome wide approach. We use a mutant mouse model with altered mechanical stimulation due to the absence of limb skeletal muscle (Splotch-delayed) where muscle-less embryos show specific defects in skeletal elements including delayed ossification, changes in the size and shape of cartilage rudiments and joint fusion. We used Microarray and RNA sequencing analysis tools to identify differentially expressed genes between muscle-less and control embryonic (TS23) humerus tissue.
Results
We found that 680 independent genes were down-regulated and 452 genes up-regulated in humeri from muscle-less Spd embryos compared to littermate controls (at least 2-fold; corrected p-value ≤0.05). We analysed the resulting differentially expressed gene sets using Gene Ontology annotations to identify significant enrichment of genes associated with particular biological processes, showing that removal of mechanical stimuli from muscle contractions affected genes associated with development and differentiation, cytoskeletal architecture and cell signalling. Among cell signalling pathways, the most strongly disturbed was Wnt signalling, with 34 genes including 19 pathway target genes affected. Spatial gene expression analysis showed that both a Wnt ligand encoding gene (Wnt4) and a pathway antagonist (Sfrp2) are up-regulated specifically in the developing joint line, while the expression of a Wnt target gene, Cd44, is no longer detectable in muscle-less embryos. The identification of 84 genes associated with the cytoskeleton that are down-regulated in the absence of muscle indicates a number of candidate genes that are both mechanoresponsive and potentially involved in mechanotransduction, converting a mechanical stimulus into a transcriptional response.
Conclusions
This work identifies key developmental regulatory genes impacted by altered mechanical stimulation, sheds light on the molecular mechanisms that interpret mechanical stimulation during skeletal development and provides valuable resources for further investigation of the mechanistic basis of mechanoregulation. In particular it highlights the Wnt signalling pathway as a potential point of integration of mechanical and molecular signalling and cytoskeletal components as mediators of the response.
Keywords: Skeletal development, Mechanical stimulation, Transcriptome, Mechanosensitive genes, Wnt signalling, Spd, Cytoskeleton
Background
Mechanical stimulation plays an important role in skeletal growth and repair reviewed in [1] and, although much less well studied, it is also required for normal skeletal development. This was initially indicated by observations that infants who experience decreased foetal movement in utero due to neuromuscular disorders present a range of skeletal anomalies including multiple joint fusions, craniofacial abnormalities and thin hypo-mineralised bones [2,3]. Direct evidence that mechanical stimuli generated by embryonic muscle contractions impacts skeletal development comes from a variety of experimental animal models that show similar abnormalities in ossification and joint formation, for example following muscle immobilisation in chick embryos, and in mouse embryos lacking muscle or with reduced or immobile muscle reviewed in [4,5]. However little is known about the molecular mechanisms through which mechanical stimuli influence cellular events during skeletal development. The interplay between biophysical stimuli and gene regulation in differentiating cells is emerging as an important phenomenon in multiple developmental systems [6,7].
A number of different strains of mutant mice have been studied that phenotypically lack limb muscle or show reduced stimuli from muscle contraction during development [8-10], including Splotch (Pax3Sp) and Splotch-delayed (Pax3Spd), where muscle precursor cells fail to migrate to the developing limbs and no limb muscle forms [11,12]. Common defects in muscle-less and immobilised embryos include abnormal initiation and/or progression of ossification [9,13], loss of definition of tissue territories in the joint region [8] and altered rudiment morphology [9], associated with reduced local cell proliferation [14]. Therefore, mechanical stimuli impact a variety of developmental processes and presumably must influence or integrate with signalling pathways and molecular changes known to guide these events. One clue to a signalling pathway impacted by mechanical stimulation comes from the work of Kahn et al.[8] who showed that canonical Wnt signalling is altered in the elbow joint of Splotch-delayed embryos. Several regulatory genes have been shown to have dramatically altered expression patterns in reduced mechanical stimuli including, Ihh and ColX at the site of ossification [10,15] and Bmp2, Fgf2, and Pthlp at the joint line [16,17]. Whether expression of these genes is directly affected by the mechanical environment or as a more indirect consequence of altered cell behaviour is not known; a genome-wide, open ended screen is required to know more about the spectrum of molecular changes that occur when mechanical stimuli are altered.
Gene expression profiling to identify genome-wide changes under altered mechanical environments has been carried out on cells in culture using microarray technology, including osteoblast cell-lines subjected to weightlessness or microgravity conditions [18], chondrocyte-laden constructs and murine cartilage explants to which dynamic compression was applied [19] and chondrocyte cell lines exposed to hydrostatic pressure [20]. Gene expression profiling has the potential to uncover hundreds of genes that respond to mechanical stimuli simultaneously (mechanosensitive genes); however no direct analyses of in vivo changes in gene expression during skeletal development following alteration of the mechanical environment have been performed. This is required to begin to assemble a picture of the molecular landscape impacted by mechanical stimuli in a developmental context.
In this study we analysed the transcriptional changes in the developing humerus and associated joints at Theiler stage (TS) 23 [21] (typically embryonic day (E) 14.5) in muscle-less (Splotch-delayed) compared to phenotypically normal littermate controls. We previously established that the humerus is the most strongly affected rudiment and TS23 the earliest time point at which the specific effects on ossification and joint line reduction in the elbow and shoulder regions are detected [9,10]. We hypothesise that mechanical stimulation of the embryonic skeletal system impacts expression levels of genes implicated in a variety of regulatory pathways and biological processes, as would be expected when an integrated regulatory system is disturbed. The genes that show altered expression would include direct and indirect targets of mechanical stimulation. Therefore, a genome wide analysis of altered transcript levels is required to indicate the principal molecular mechanisms disturbed and the most likely candidates for direct regulation. We have used both RNA whole transcriptome sequencing analysis (RNA-seq) and Microarray technology to allow a comprehensive investigation of the altered transcriptome. Microarray analysis is a more established technique [22], but RNA-seq offers the potential of greater sensitivity [23] and analysing the same tissues in parallel allows direct comparison of the two assays and integration of the data sets. We also used RNA-seq analysis of the normal developing humerus to explore the transcriptome at this specific stage of development. The humerus developing in the absence of muscle generated stimulation showed both up and down-regulation of gene expression. We reveal alteration of genes encoding components and targets of specific signalling pathways, in particular the Wnt signalling pathway. Genes associated with cytoskeletal rearrangement and extracellular matrix components are also affected. This analysis has allowed us to profile genome wide transcriptional changes giving an overview of the molecular processes and pathways most affected and identifying a set of putative direct target genes responding to mechanical stimulation during ossification and joint formation.
Results
Transcription profiling of the developing humerus during early ossification and joint formation (TS23)
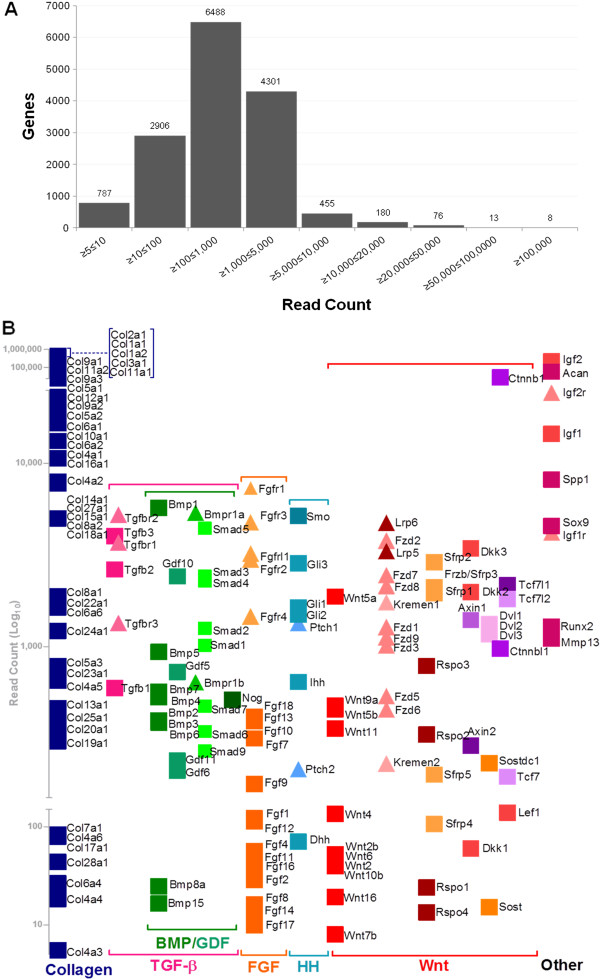
RNA sequencing of control humeri at TS23 gives an insight into the transcriptome at this key stage of development when the rudiment is undergoing early stages of ossification and tissue zones in the joint are being defined (Figure 1). Transcripts from 15,214 individual genes (n = 3 biological replicates, Figure 2; data submitted to EMBL-EBI ArrayExpress repository: [E-MTAB-1745]) were detected in this tissue at this stage. A minimum of 5 transcript reads (on average), with at least one read from each replicate sample, was chosen as the cut off point to reliably indicate reproducible expression across biological replicates [24]. Expressed genes were divided into groups according to their relative expression level (Figure 2A). 787 genes showed the lowest level of expression represented by between 5 and 10 read counts, while the majority of genes (71%; 10,789/15,214) showed between 100 and 5000 read counts. Only 732 genes are in the most highly expressed categories (≥5,000 read counts) (Figure 2A), representing 4.8% of expressed genes. Expression levels of selected individual genes are represented in Figure 2B. The most abundantly transcribed gene is Col2a1 (collagen 2, alpha 1) with a read count of 452,576, and among the 8 genes with read counts of more than 100,000, there are 4 other collagen encoding genes (Figure 2A and B). In total 41 collagen subtype genes are expressed (Figure 2B).
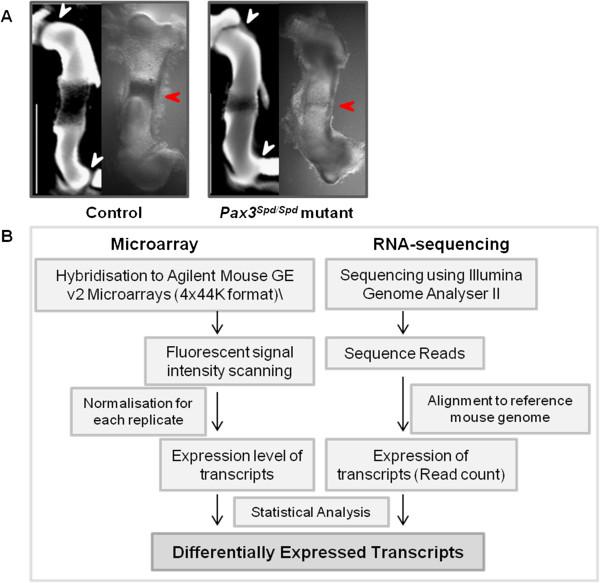
Figure 1.
Overview of gene expression profiling approach used. A) Images of control and Pax3Spd/Spd mutant humeri at Theiler stage 23 dissected prior to RNA extraction (right hand image); the images on the left are external views of stage and genotype matched 3D scanned specimens stained to reveal the morphology and ossification more clearly. White arrow heads indicate the elbow joint and shoulder joint line and red arrow heads the ossification site, a visible reduction is apparent in the mutant compared to the control in each case. B) Work-flow of Microarray and RNA-sequencing data analysis.
Figure 2.
Transcriptome Profiling of the developing humerus at TS23 by RNA-seq, (A) The number of genes (y-axis) with increasing relative expression levels represented by transcript read counts as indicated. (B) The relative expression of selected genes represented by read counts (Log10) (y-axis). Collagen genes and signalling pathways genes are grouped: TGFβ (including TGFβ, BMP and GDF ligands, receptors and agonists), FGF (ligands and receptors), HH (ligands, receptors and intracellular modulators), Wnt (Wnt ligands and agonists (Wnt, Rspo), receptors (Fzd) and co-receptors (Kremen, Lrp), antagonists (Sfrp, Dkk) Intracellular components (Sost, Dvl, Ctnnb) and nuclear components (Tcf, Lef)) and a selection of ‘other’ genes associated with skeletal development. Squares indicate ligands and modulators, triangles indicate receptors.
The relative levels of expression of genes associated with signalling pathways involved in regulating skeletal development reviewed in [25,26] are highlighted in Figure 2B. This shows the potential components that can contribute to these signalling pathways at this stage of skeletal development. For example the hedgehog (HH) pathway is known to play an important role in ossification through the action of Ihh (640 reads), binding to its receptor Ptch1 (1,342 reads) activating Smo (5,154 reads). Similarly, 14 FGF ligand encoding genes and 5 FGF receptor encoding genes were detected, highlighting the potential for multiple FGF signalling interactions. The BMP signalling pathway genes also reveal potential for multiple signalling interactions with 9 BMP encoding genes expressed. Bmp1 is by far the most highly expressed (5719 reads); although it is not previously reported in this tissue at this stage in gene expression databases (http://www.informatics.jax.org/). The relative expression levels of Wnt ligand (Wnt), Wnt receptor (Fzd, Lrp, Kremen), extracellular Wnt interactor (Sfrp, Dkk, Rspo, Sost), intracellular Wnt pathway component (Dvl, Axin, Ctnnb) and Wnt pathway transcription factor (Tcf, Lef) encoding genes are represented. The detection of previously unreported Wnt gene expression in the humerus (e.g. Wnt2 and Wnt2b) opens up new considerations for functional roles, especially as both genes are up-regulated in muscle-less rudiments (described below). The low density lipoprotein receptor-related genes Lrp5 and Lrp6, which are Wnt co-receptors, are most highly expressed among the Lrp gene family (4,727 and 3,310 reads respectively). Interestingly eight Fzd receptor encoding genes are detected. Other genes known to be involved in skeletal development are highly expressed; Sox9 (4,550 reads), Runx2 (1,285reads), Spp1 (8,155 reads) and Mmp13 (1,103 reads).
Identification of differentially expressed genes in muscle-less versus control developing humeri and associated joints
Microarray analysis of RNA extracted from control and Pax3 Spd/Spd muscle-less humeri (n = 4 biological replicates) detected expression of a similar proportion of individual genes on the array; 20,697 independent genes from the control and 20,949 from the muscle-less humeri (data submitted to EMBL-EBI ArrayExpress repository: E-MTAB-1744). Comparing hybridisation intensity between control and mutant derived cDNAs, using cut off points of at least a 2-fold change and corrected p-value ≤0.05 [27] for significance across replicates, identified 374 independent genes as differentially expressed (DE). Of these, 284 genes (75.9%) were down-regulated and 90 genes (24.1%) were up-regulated (Table 1). RNA-seq analysis (n = 3 biological replicates) detected 15,031 independent genes (with ≥5 read counts across replicates, as described above) in muscle-less humeri, compared to 15,214 in control tissue (data submitted to EMBL-EBI ArrayExpress repository: E-MTAB-1746). To determine differential expression, the same cut off points of a corrected p-value ≤0.05 and at least a 2-fold change were applied to the RNA-seq data-set, identifying 1,037 genes as DE across replicates. Of these, 618 genes (59.6%) were down-regulated and 419 genes (40.4%) were up-regulated in the muscle-less humeri and associated joints compared to that of phenotypically normal littermate control humeri (Table 1).
Table 1.
Differentially expressed genes (p ≤0.05) with fold change values ≥ 2 revealed by Microarray and RNA-Seq analysis and Combined
| Microarray | RNA-Seq | Combined | ||||
|---|---|---|---|---|---|---|
| Total number of differentially expressed transcripts |
374 |
1,037 |
1,132 |
|||
| |
Down-regulated |
Up-regulated |
Down-regulated |
Up-regulated |
Down-regulated |
Up-regulated |
| Independent named genes |
282 |
87 |
419 |
618 |
680 |
452 |
| Putative uncharacterized proteins/unknown genes |
2 |
3 |
- |
- |
- |
- |
| 284 | 90 | 419 | 618 | |||
The Venn diagrams in Figure 3 represent overlap of DE genes identified by the two platforms. In total 1,132 independent genes were identified as DE in the muscle-less mutant compared to its phenotypic control (Figure 3, Table 1). Of the 374 genes identified by microarray (Table 1), 73.2% of these genes were also represented in the RNA-seq data. RNA-seq detected a greater total number of DE genes than the microarray (1,037 versus 374, respectively, using the standard cut off criteria). In addition if the stringency for DE gene selection is weakened for the microarray data by moving the cut off point to corrected p-value of ≤0.08, the number of genes detected as DE in common by the two platforms increases to 426 from 274 (not shown). This suggests that the RNA-seq approach was more sensitive in detecting differential expression. Grouping genes based on the degree of change showed that the spread of fold change values was similar across platforms although there were differences in the number of genes in different categories (Additional file 1: Table S1). Additional file 1: Table S2 and S3 list all DE genes and fold change values estimated by Microarray and RNA-seq.
Figure 3.
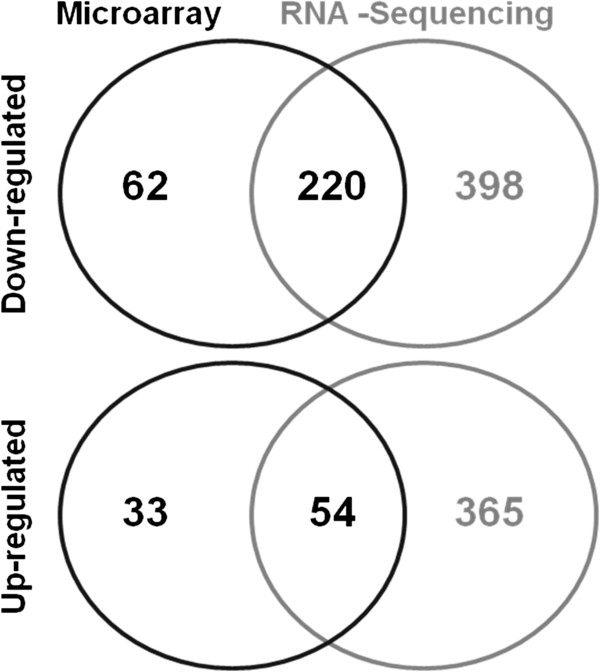
Venn diagrams representing the overlap of independent genes detected as differentially expressed (up-regulated and down-regulated; cut off criteria p ≤ 0.05, and fold change ≥2) from the microarray and RNA seq analysis, as indicated.
Verification of fold change values for DE genes was performed using qRT-PCR (Table 2). The DE genes chosen (Fgf4, Cilp, Rxrg, Dll1, Spp1, Vstm2a, Figf, Fgf10 and Sfrp2) include both down-regulated and up-regulated genes. The direction and degree of fold changes were similar in all cases for the microarray and the RNA-seq. For all genes analysed there was a good correspondence across all platforms (Microarray, RNA-seq and RT-PCR), although greater fold change differences were detected for the down-regulated gene Rxrg and the up-regulated gene Vstm2a by qRT-PCR.
Table 2.
Fold change in expression level of selected genes in Pax3 Spd / Spd mutant humeri compared to control humeri revealed by Microarray, RNA seq and qRT-PCR
| Gene symbol | Gene name | Regulation status | Microarray (Fold change) | RNA-seq (Fold change) | qRT-PCR (Fold change) |
|---|---|---|---|---|---|
|
Fgf4 |
Fibroblast growth factor 4 |
Down-regulated |
41.05 |
#No numerical value |
35.24*** |
|
Cilp |
Cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
Down-regulated |
9.73 |
9.76 |
11.3*** |
|
Rxrg |
Retinoid X receptor gamma |
Down-regulated |
7.05 |
12.29 |
5.25** |
|
Dll1 |
Delta-like 1 |
Down-regulated |
2.21 |
3.16 |
1.78* |
|
Spp1 |
Secreted phospoprotein 1 |
Down-regulated |
5.23(ns) |
2.9 |
2.59* |
|
Vstm2a |
V-set and transmembrane domain containing 2A |
Up-regulated |
8.56 |
3.09 |
14.17** |
|
Figf |
c-fos induced growth factor |
Up-regulated |
3.52 |
3.17 |
4.06** |
|
Fgf10 |
Fibroblast growth factor 10 |
Up-regulated |
2.48 |
2.14 |
3.28 |
| Sfrp2 | Secreted frizzled-related protein 2 | Up-regulated | 2.62 | 2.09 | 3.38* |
#No numerical value for fold change as no transcripts detected in the mutant.
***p ≤ 0.001, **p ≤ 0.01, *p ≤ 0.05 indicates significance of differential expression from qRT-PCR.
p ≤ 0.05 for all microarray and RNA-seq differences except Spp1 in the Microarray (ns).
Biological Interpretation of differentially expressed genes: Down-regulated genes are associated with development and differentiation, cytoskeletal architecture and cell signalling
To reveal any enrichment of functionally related genes among the DE data sets, two web based tools, DAVID and GOstat, were used to analyse Gene Ontology (GO) term associations [28,29] (Tables 3 and 4). A significant enrichment (normalised by the number of genes in the genome with that associated GO term) indicates specific biological processes that are affected when mechanical stimuli are reduced. The strength of the enrichment is indicated by the calculated p-value. Independent analysis of data sets from microarray and RNA-seq showed the same enriched groups so analysis of the combined DE gene sets is presented (Table 1). The individual GO terms found to be enriched have been grouped for the purpose of interpretation as indicated in Tables 3 and 4. Analysis of the down-regulated gene set (Table 3) indicated that genes associated with Development and Differentiation are most highly enriched (total of 155/680 with p-values down to 2.73 × 10-15). Categories within this group are involved in signal transduction, including genes that encode signalling molecules, receptors, and transcription factors (TFs), for example signalling ligands Fgf4,Fgf5, Fgf6, Fgf8 from the fibroblast growth factor signalling pathway; receptors Fzd10 and Rxrg from the Wnt and Retinoic acid pathways respectively. 26 of these genes encode TFs including; Barx2, Scx, Hes6, Pitx2, Pitx3 and Tead4. The down-regulation of such signalling pathway component genes also underlies the enrichment of ontology groups related to Cell Signalling (Table 3: p-value down to 4.78 × 10-5).
Table 3.
Enrichment of gene groups down-regulated in muscle-less humeri based on GO terms under subontologies biological process and cellular component
| Gene ontology term | Gene count in study | Enrichment 1 | |
|---|---|---|---|
|
Development/Differentiation |
155 |
2.73 × 10-15 to 0.0084 |
|
| |
GO:0048856: Anatomical structure development |
134 |
2.73 × 10-15 |
| GO:0032502: Developmental process |
155 |
1.52 × 10-14 |
|
| GO:0048513: Organ development |
107 |
4.23 × 10-13 |
|
| GO:0007275: Multicellular organismal development |
142 |
4.92 × 10-13 |
|
| GO:0048731: System development |
122 |
5.57 × 10-13 |
|
| GO:0009888: Tissue development |
58 |
8.37 × 10-13 |
|
| GO:0030154: Cell differentiation |
94 |
8.86 × 10-10 |
|
| GO:0048869: Cellular developmental process |
96 |
1.67 × 10-9 |
|
| GO:0048468: Cell development |
45 |
2.66 × 10-7 |
|
| GO:0009887: Organ morphogenesis |
38 |
6.12 × 10-5 |
|
| GO:0001756: Somitogenesis |
8 |
2.61 × 10-4 |
|
| GO:0035282: Segmentation |
9 |
2.66 × 10-4 |
|
| GO:0009952: Anterior/posterior pattern formation |
13 |
0.0033 |
|
| GO:0009790: Embryonic development |
34 |
0.0074 |
|
| GO:0043009: Chordate embryonic development |
24 |
0.0075 |
|
| GO:0009792: Embryonic development ending in birth |
24 |
0.0084 |
|
|
Cytoskeleton |
30 |
5.30 × 10-19 to 0.0995 |
|
| Biological process |
GO:0030029: Actin filament-based process |
25 |
1.45 × 10-9 |
| GO:0030036: Actin cytoskeleton organization |
23 |
1.08 × 10-8 |
|
| GO:0007010: Cytoskeleton organization |
31 |
1.50 × 10-7 |
|
| GO:0007015: Actin filament organization |
5 |
0.0995 |
|
| GO:0015629: Actin cytoskeleton |
39 |
5.30 × 10-19 |
|
| Cellular component |
GO:0005856: Cytoskeleton |
86 |
2.20 × 10-14 |
| GO:0016459: Myosin complex |
21 |
1.26 × 10-15 |
|
| GO:0032982: Myosin filament |
11 |
2.32 × 10-12 |
|
| GO:0044430: Cytoskeletal part |
51 |
1.16 × 10-6 |
|
| GO:0005884: Actin filament |
5 |
0.0134 |
|
|
Cell signalling |
31 |
4.78 × 10-5 to 0.0899 |
|
| GO:0007154: Cell communication |
32 |
4.78 × 10-5 |
|
| GO:0007267: Cell-cell signalling |
23 |
1.33 × 10-4 |
|
| GO:0007169: Transmembrane protein tyrosine kinase signalling |
17 |
3.85 × 10-4 |
|
| GO:0007167: Enzyme linked receptor protein signalling pathway |
20 |
0.0011 |
|
| GO:0008543: Fibroblast growth factor receptor signalling pathway |
5 |
0.0139 |
|
| GO:0045168: Cell-cell signalling involved in cell fate specification |
4 |
0.0271 |
|
| GO:0040036: Regulation of fibroblast growth factor signalling |
3 |
0.0451 |
|
| GO:0043409: Negative regulation of MAPKKK cascade |
3 |
0.0614 |
|
| GO:0007219: Notch signalling pathway | 5 | 0.0899 | |
All GO terms are associated with the subontology “Biological Process” except where indicated under Cytoskeleton, where Cellular Component was also used.
1P-value of enrichment of GO terms using DAVID analysis software; similar results were found with GOstat.
Table 4.
Enrichment of gene groups up-regulated in muscle-less humeri based on GO terms under subontologies biological process and cellular component
| Gene ontology: biological process | Gene count in study | Enrichment 1 | |
|---|---|---|---|
|
Adhesion/Extracellular matrix |
32 |
1.99 × 10-10 to 0.0842 |
|
| GO:0007155: Cell adhesion |
41 |
1.99E-10 |
|
| GO:0022610: Biological adhesion |
41 |
2.12E-10 |
|
| GO:0007156: Homophilic cell adhesion |
15 |
4.95 × 10-7 |
|
| GO:0016337: Cell-cell adhesion |
19 |
9.38 × 10-6 |
|
| GO:0043062: Extracellular structure organization |
9 |
0.0223 |
|
| GO:0030198: Extracellular matrix organisation |
6 |
0.0842 |
|
|
Cell signalling |
33 |
3.45 × 10-9 to 0.0148 |
|
| GO:0007267: Cell-cell signalling |
27 |
3.45 × 10-9 |
|
| GO:0007154: Cell communication |
33 |
2.05 × 10-9 |
|
| GO:0007223: Wnt receptor signalling pathway, calcium modulating pathway |
7 |
0.0092 |
|
| GO:0016055: Wnt receptor signalling pathway |
4 |
0.0104 |
|
| GO:0008589: Regulation of smoothened signalling pathway |
9 |
0.0105 |
|
| GO:0010648: Negative regulation of smoothened signalling pathway |
4 |
0.0135 |
|
| GO:0048010: Vascular endothelial growth factor receptor signalling pathway |
10 |
0.0288 |
|
| GO:0007169: Transmembrane receptor protein tyrosine kinase signalling pathway |
3 |
0.0402 |
|
| GO:0007167: Enzyme linked receptor protein signalling pathway |
12 |
0.0051 |
|
| GO:0007165: Signal transduction |
80 |
0.0148 |
|
|
Development/Differentiation |
[34] |
4.38 × 10-6 to 0.0083 |
|
| GO:0048731: System development |
78 |
4.38 × 10-6 |
|
| GO:0048856: Anatomical structure development |
81 |
8.42 × 10-6 |
|
| GO:0007275: Multicellular organismal development |
87 |
6.80 × 10-5 |
|
| GO:0048468: Cell development |
31 |
8.17 × 10-5 |
|
| GO:0032502: Developmental process |
91 |
1.82 × 10-4 |
|
| GO:0048869: Cellular developmental process |
59 |
5.41 × 10-4 |
|
| GO:0030154: Cell differentiation |
57 |
5.74 × 10-4 |
|
| GO:0048513: Organ development |
56 |
0.0036 |
|
| GO:0045595: Regulation of cell differentiation |
18 |
0.0083 |
|
|
Gene ontology: cellular component |
Gene count in study |
Enrichment
1
|
|
|
Extracellular region |
35 |
2.34 × 10-10 to 0.022 |
|
| GO:0005576: Extracellular region |
81 |
2.34 × 10-10 |
|
| GO:0044421: Extracellular region part |
43 |
2.70 × 10-7 |
|
| GO:0031012: Extracellular matrix |
22 |
1.16 × 10-5 |
|
| GO:0005578: Proteinaceous extracellular matrix |
21 |
2.14 × 10-5 |
|
| GO:0005615: Extracellular space |
24 |
0.0020 |
|
| GO:0044420: Extracellular matrix part |
7 |
0.0202 |
|
|
Membrane association |
227 |
2.46 × 10-16 to 1.01 × 10-6 |
|
| GO:0005886: Plasma membrane |
134 |
2.46 × 10-16 |
|
| GO:0031224: Intrinsic to membrane |
191 |
1.07 × 10-8 |
|
| GO:0044425: Membrane part |
215 |
2.08 × 10-8 |
|
| GO:0016020: Membrane |
226 |
7.80 × 10-8 |
|
| GO:0031224: Intrinsic to membrane | 178 | 1.01 × 10-6 | |
1P-value of enrichment of GO terms using DAVID analysis software; similar results were found with GOstat.
110 genes associated with the Cytoskeleton were down-regulated in Pax3 Spd/Spd humeri, including those encoding microfilament, microtubule and intermediate filament components (Figure 4); 33 are directly associated with microfilaments (e.g. Acta1, Ablim2, Pdlim3), 13 with microtubules (e.g. Rassf5, Tubb2b, Tppp) and 4 with intermediate filaments (Des, Krt33b, Sync, Tchh); other DE genes associated with the cytoskeleton encode proteins that interact with myosin, or the extracellular matrix (ECM), including integrin and cadherin encoding genes (e.g. Lamb3, Cdh4, Itga4 and Pdgfa (not shown)).
Figure 4.
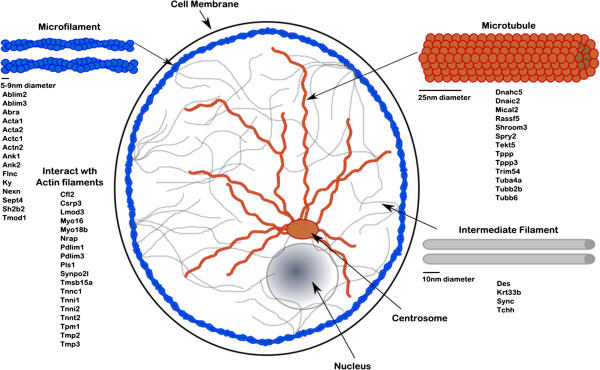
Visual representation of DE genes associated with cytoskeletal components.
Up-regulated genes are associated with cell adhesion, cell signalling and development and differentiation
Genes up-regulated in muscle-less humeri revealed by microarray and RNA-seq were similarly analysed for enrichment of genes associated with particular biological processes or cellular components, using associated GO terms (Table 4). For terms within the subontology biological process, the strongest enrichment was for cell adhesion and ECM associated genes (total of 50/447 genes, with p-values down to 1.99 × 10-10). The genes identified in this category include ECM glycoproteins (Lsamp, Svep1), ECM structural constituents (Col8a1, Col8a2, Frem3), cell-adhesion molecules (Cntn1, Cntn4, Cntn5) and calcium-dependent cell adhesion proteins (Pcdh8, Pcdhb2, Pcdhga1). This grouping in addition included genes involved in signalling pathways which overlap with the next most enriched terms; cell signalling (GO:0007154) and cell-cell communication (GO:0007267). The signalling pathway components identified in this category include: Hedgehog (Hhip, Ptch1), fibroblast growth factor (Fgf10, Fgf2), transforming growth factor (Frem2, Bmp3), Notch (Ctn1, Nrg1) and Wnt signalling (Dkk2, Cpz Rspo2, Rspo3, Sfrp2, Wnt2, Wnt2b, Wnt4 and Wnt16) and others, including receptors (Epha3, Epha4, Epha5, Grin2a, Grm4, Grm7, Grm8, Gfra2, and Pdgfra). Other signals identified as up-regulated included c-fos induced gowth factor (Figf), hepatocyte growth factor (Hgf) and Insulin-like growth factor (Igf1). The gene lists in the next most enriched set, Development and Differentiation, similarly show large overlap due again to the presence of the signalling pathway genes mentioned above, and also transcription factors Foxc2, Foxo3, Lmx1a, Lmx1b. z.
Under the subontology Cellular Component there was also striking enrichment of extracellular (total 81/447; with p-values down to 2.34 x10-10) and membrane associated gene products (total 227/447 p-values down to 2.46 × 10-16), including cell adhesion molecules (Cadm2, Cntn1), receptors (Fat2, Fat4, Grin2a, Grm4, Gfra2, Pdgfra, Robo2, Sorcs1, Sstr4), cell surface molecules (Cd83, Cd96), cadherins (Cdh20, Cdh8), trans-membrane proteins (Tmem26, Tmem28), voltage gated channels (Kcna1, Kcna2, Kcnc2) and cell adhesion and extracellular components: Alcam, Cntn4, Epha4, Col8a1, Col8a2, Pappa, Pcdh8.
Signalling pathway analysis of differentially expressed genes
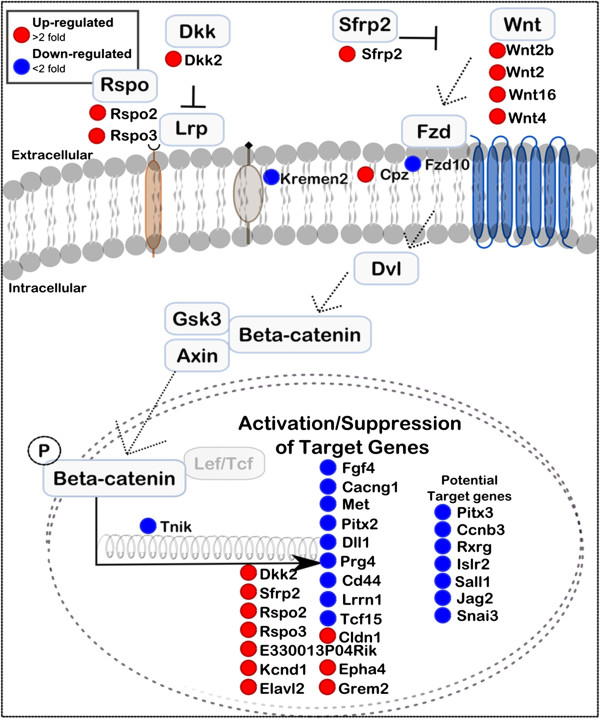
Given the strong enrichment of genes associated with Signalling Pathways and Development and Differentiation functions (Tables 3 and 4), we sorted DE genes according to participation in major developmental regulatory pathways (Figure 5 and Tables 5 and 6). By far the most strongly impacted cell communication pathway is Wnt signalling with 34 DE genes encoding signalling molecules, receptors, pathway antagonists, known targets or potential targets of the pathway (Figure 5, Table 5). From the diagrammatic representation of pathway components shown in Figure 5, it is apparent that the encoded products of DE genes act either at the cell surface in Wnt signal generation/modulation/interpretation or are targets of the pathway. The genes listed include known targets of the pathway and seven potential target genes (not verified), included here due to their similarity to known targets; for example Sall1is included because the orthologous gene Sall4 is a known direct target of the pathway [36]. In general, genes encoding cell surface components of the pathway are up-regulated, including signalling ligands and agonists (Wnt2, Wnt2b, Wnt4, Wnt16, Rspo2 and Rspo3), and extracellular antagonists (Dkk2, Sfrp2), while down-regulated genes identified are more commonly targets of the pathway (e.g. Fgf4, Cacng1, Pitx2, Dll1, Prg4, Lrrn1, Met, and Cd44). Interestingly nine known Wnt target genes are up-regulated, including Dkk2, Rspo2, Rspo3, Cldn1, Grem2, Kcnd1, Epha4 and Sfrp2, which encode membrane associated proteins, some of which regulate the Wnt pathway (Dkk2, Sfrp2, Rspo2, Rspo3).
Figure 5.
Visual representation of Wnt signalling pathway components, showing altered expression in developing muscle-less humeri. Blue circles indicate down-regulated genes red are up-regulated genes (Fold change ≥2, p ≤ 0.05, genes and details listed in Table 7).
Table 5.
Differentially expressed Wnt signalling pathway genes
|
Down-regulated Wnt Pathway genes | ||||||||
|---|---|---|---|---|---|---|---|---|
|
Fold change |
Gene symbol | Gene title |
Role in pathway |
Reference for role in pathway | ||||
| RNA-seq | Microarray | Ligand | Receptor | Intra-cellular | Target | |||
| # |
41.05 |
Fgf4 |
Fibroblast growth factor 4 |
|
|
|
T |
[37] |
| # |
|
Snai3 |
Snail homolog 3 |
|
|
|
PT |
Snai1
[38] |
| 90.87 |
32.06 |
Cacng1 |
Calcium channel, voltage-dependent, gamma subunit 1 |
|
|
|
T |
[39] |
| 42.86 |
18.77 |
Pitx3 |
Paired-like homeodomain transcription factor 3 |
|
|
|
PT |
Pitx2
[40] |
| 37.43 |
17.22 |
Kremen2 |
Kringle containing transmembrane protein 2 |
|
Co-Rec |
|
[41] |
|
| 29.52 |
|
Ccnb3 |
Cyclin B3 |
|
|
|
PT |
CyclinD1
[42] |
| 12.29 |
7.05 |
Rxrg |
Retinoid X receptor gamma |
|
|
|
PT |
Rarg
[43] |
| 5.95 |
5.20 |
Islr2 |
Immunoglobulin superfamily containing leucine-rich repeat 2 |
|
|
PT |
Islr
[44] |
|
| 4.87 |
|
Pitx2 |
Paired-like homeodomain transcription factor 2 |
|
|
|
T |
[40] |
| 3.58 |
|
Fzd10 |
Frizzled homolog 10 |
|
R |
|
|
[45] |
| 3.16 |
2.21 |
Dll1 |
Delta-like 1 |
|
|
|
T |
[46] |
| 3.05 |
|
Prg4 |
Lubricin |
|
|
|
T |
[30] |
| 2.81 |
2.57 |
Lrrn1 |
Leucine rich repeat protein 1, neuronal |
|
|
|
T |
[47] |
| 2.52 |
|
Sall1 |
Sal-like 1 |
|
|
|
PT |
Sall4:
[36] |
| 2.47 |
|
Tnik |
TRAF2 and NCK interacting kinase |
|
|
TF-act |
[48] |
|
| 2.32 |
|
Tcf15 |
Transcription factor 15 |
|
|
|
T |
|
| 2.32 |
|
Met |
Met proto-oncogene |
|
|
|
T |
[49] |
| 2.18 |
2.28 |
Cd44 |
CD44 antigen |
|
|
|
T |
[50] |
| 2.09 |
|
Jag2 |
Jagged2 |
|
|
|
PT |
Jag1
[51] |
| Up-regulated Wnt pathway genes | ||||||||
| 4.36 |
4.91 |
Grem2 |
Gremlin |
|
|
|
T |
[52] |
| 3.96 |
|
Wnt2 |
Wingless-related MMTV integration site 2 |
L |
|
|
|
|
| 3.50 |
|
Wnt2b |
Wingless-related MMTV integration site 2b |
L |
|
|
|
|
| 3.07 |
|
Wnt16 |
Wingless-related MMTV integration site 16 |
L |
|
|
|
|
| 2.85 |
2.90 |
Kcnd2 |
Potassium voltage-gated channel, shal-related family, member 2 |
|
T |
[47] |
||
| 2.73 |
|
Cldn1 |
Claudin-1 |
|
|
|
T |
[53] |
| 2.44 |
|
Dkk2 |
Dickkopf |
Ant |
|
T |
[47,54] |
|
| 2.43 |
3.08 |
Rspo2 |
R-spondin 2 |
L |
|
|
T |
[47,55,56] |
| 2.37 |
|
Epha4 |
Eph receptor A4 |
|
|
|
T |
[47] |
| 2.11 |
|
Elavl2 |
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 |
|
T |
[47] |
||
| 2.10 |
|
Cpz |
Carboxypeptidase Z |
Co-act |
|
|
[33] |
|
| 2.09 |
2.62 |
Sfrp2 |
Secreted frizzled related protein 2 |
Ant |
|
T |
[57] |
|
| 2.03 |
|
Wnt4 |
Wingless-related MMTV integration site 4 |
L |
|
|
|
|
| 2.02 |
|
Rspo3 |
R-spondin 3 |
L |
|
|
T |
[47,58] |
| 2.41 | E330013P04Rik | RIKEN cDNA E330013P04 gene | T | [47] | ||||
#No numerical value for fold change as no transcripts detected in the Mutant.
T -Target, PT -Potential Target, R -Receptor, Co-Rec -Co-Receptor, TF-act -Transcription Factor activator, L -Ligand, Ant -Antagonist, Co-act -Co-activator.
Table 6.
Differentially expressed genes associated with other signalling pathways
|
Down-regulated |
|
Up-regulated |
|
||
|---|---|---|---|---|---|
|
Bone morphogenetic protein signalling pathway | |||||
|
Fold change |
Gene symbol | Gene title |
Role in pathway |
||
| RNA-seq | Microarray | Reference | |||
| 87.94 |
50.88 |
Hfe2 |
Hemochromatosis type 2 |
Co-Rec |
[59] |
| 5.25 |
|
Fgf8 |
Fibroblast growth factor 8 |
T |
[60] |
| 2.44 |
|
Scx |
Scleraxis |
T |
[61] |
| 2.17 |
|
Egr2 |
Early growth response 2 |
T |
[62] |
| 2.03 |
|
Bmp3 |
Bone morphogenetic protein 3 |
L |
|
| Hedgehog signalling pathway | |||||
| 14.55 |
4.59 |
Hhatl |
Hedgehog acyltransferase-like |
L-act |
[63] |
| 2.70 |
|
Ptchd1 |
Patched domain containing 1 |
R |
|
| 2.27 |
|
Hhip |
Hedgehog interacting protein |
Ant, T |
[64] |
| 2.14 |
|
Ptch1 |
Patched 1 |
T |
[65] |
| Fibroblast growth factor signalling pathway | |||||
| # |
|
Fgf4 |
Fibroblast growth factor 4 |
L |
|
| # |
|
Fgf6 |
Fibroblast growth factor 5 |
L |
|
| 5.25 |
|
Fgf8 |
Fibroblast growth factor 8 |
L |
|
| 5.10 |
|
Fgf5 |
Fibroblast growth factor 5 |
L |
|
| 4.54 |
3.23 |
Fgfr4 |
Fibroblast growth factor receptor 4 |
R |
|
| 3.24 |
|
Spry1 |
Sprouty homolog 1 |
Ant |
[66] |
| 3.22 |
|
Spry4 |
Sprouty homolog 4 |
Ant |
|
| 2.75 |
|
Spry2 |
Sprouty homolog 2 |
Ant |
[66] |
| 2.39 |
|
Fgf2 |
Fibroblast growth factor 2 |
L |
|
| 2.134 |
2.48 |
Fgf10 |
Fibroblast growth factor 10 |
L |
|
| Hippo signalling pathway | |||||
| 98.29 |
49.05 |
Vgll2 |
Vestigial like 2 homolog (Drosophila) |
Co-TF |
[67] |
| 3.86 |
4.38 |
Tead4 |
TEA domain family member 4 |
Co-TF |
[68] |
| 2.96 |
|
Fat4 |
Fat tumor suppressor homolog 4 |
R |
[69] |
| 2.43 |
|
Fat2 |
Fat tumor suppressor homolog 2 |
R |
[69] |
| Notch signalling pathway | |||||
| 5.64 |
|
Dner |
Delta/notch-like EGF-related receptor |
R |
[70] |
| 3.16 |
2.21 |
Dll1 |
Delta-like 1 (Drosophila) |
L |
[71] |
| 2.69 |
|
Hes6 |
Hairy and enhancer of split 6 |
T |
[72] |
| 2.54 |
|
Dtx4 |
Deltex homolog 4 |
T |
[72] |
| 2.09 |
|
Jag2 |
Jagged 2 |
R |
[71] |
| 3.25 |
2.87 |
Nrg1 |
Neuregulin 1 |
T |
[73] |
| 2.36 |
2.09 |
Foxc2 |
Forkhead box C2 |
Upstream-activator |
[74] |
| 2.27 | 2.29 | Cntn1 | Contactin 1 | PL | [75] |
#No numerical value for fold change as no transcripts detected in the Mutant.
Co-Rec -Co-Receptor, T -Target, L -Ligand, L-act- Ligand activator, R -Receptor, Ant -Antagonist, Co-TF- -Co-Transcription Factor, PL –Potential Ligand.
Table 6 lists DE genes associated with other developmentally relevant signalling pathways, including the BMP, Hedgehog, Fibroblast growth factor, Hippo and Notch signalling pathways.
Spatial alteration of gene expression patterns
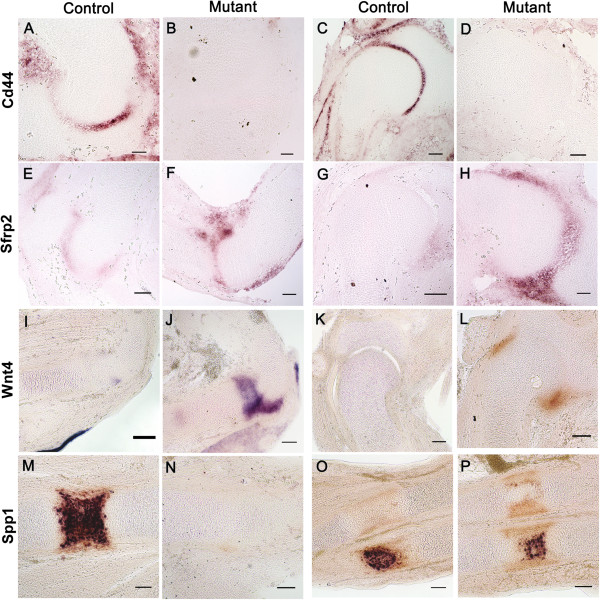
While Microarray and RNA-seq analysis provides data on quantitative changes in gene expression levels across the whole developing rudiment, it does not reveal alterations in the spatial distribution of transcripts or give clues to the specific developmental events affected. We therefore performed in situ hybridisation on control and muscle-less mutant (Pax3 Spd/Spd ) limb sections at TS23 for a selected subset of genes. Three of the genes selected encode components of Wnt signalling pathways known to be important in skeletal development: Cd44, Sfrp2 and Wnt4. Spp1 encodes an ECM protein Osteopontin which is a prominent component of mineralised matrices of bone and teeth [76]. Cd44 is a cell-surface glycoprotein involved in cell-cell interactions, cell adhesion and migration. It is a receptor for hyaluronic acid and can interact with other extracellular proteins, such as osteopontin, collagens and matrix metalloprteinases (MMPs) reviewed in [77]. It is a target gene of the Wnt signalling pathway [50]. Cd44 gene expression is down-regulated 2.28 fold in muscle-less humeri RNA (Table 5). The in situ hybridisation analysis reflects this down regulation dramatically with Cd44 transcripts hardly detectable in either shoulder or elbow joints of muscle-less mutant embryos, compared to the clear joint line restricted expression seen in controls (Figure 6A-D). Sfrp2 encodes a secreted protein that acts as a modulator of the Wnt signalling pathway, in particular during normal skeletal patterning in developing limbs [78]. Normal expression at TS23 (Figure 6E, G) can be detected in the elbow and shoulder joints. The level of up-regulation from microarray and RNA-seq analysis is 2.62 (p-value 0.01) and 2.09 (p-value 1.13E-06) fold respectively (Table 5). This up-regulation was reflected in the intensity of expression seen in mutant sections following in situ hybridisation (Figure 6F, H). In addition to the increased level of expression a change in the spatial pattern is also evident. In both the elbow and the shoulder joints expression is expanded, particularly on the ventral aspect of the joint and the staining is unevenly distributed (Figure 6F, H). A similar expression increase was seen in the phalangeal and carpal joints of the handplate, compared to control (not shown). Wnt4 is a signalling ligand of the Wnt signalling pathway. Expression of this gene has previously indicated its role in joint formation during limb development [79-81]. Control Wnt4 expression is seen at a low level in the ventral portion of the elbow joint (Figure 6I) at TS23; there is also distinct expression in the epidermis, as previously detected [82]. In the mutant there is a high level of expression in the elbow joint region; expression is spread across the whole joint line from ventral to dorsal, more extensive on the dorsal side (Figure 6J).
Figure 6.
Comparison of spatial distribution of differentially expressed genes in TS23 control (A,C,E,G,I,K,M) and TS23 mutant (B,D,F,H,J,L,N,P) elbow joint, shoulder joint, humerus, radius and ulna.Cd44(A-D) expression in the elbow joint (A-B), and the shoulder joint (C-D) (arrows in B and D indicate the position of joint-line cells based on cell morphology), Sfrp2(E-H) expression in the elbow joint (E-F) (arrow indicates abnormal expression on the ventral side of the joint), and the shoulder joint (G-H), Wnt4 expression in the elbow joint (I-J), and the shoulder joint (K-L), Spp1(K-N) expression in the humerus hypertrophic zone (hz) and perichondrium (p) (M-N) (the arrowhead in L indicates enlarged hypertropic chondrocytes), and in the ulna (u) and radius (r) (O-P). e epidermis. All scale bars are 100 μm.
The Spp1 gene is normally expressed in the hypertrophic zone and adjacent perichondrium [83]. Despite the appearance of hypertrophic chondrocytes at the mid-diaphysis of immobile Spd embryos (Figure 6N arrowhead), no Spp1 gene expression is detected in these cells. There is weak but detectable staining in the perichondrium at the site of the hypertrophic region, but again apparently lower than in control tissue (Figure 6M, N). Expression is also detected in the hypertrophic zone of the ulna (Figure 6O) and this is reduced but not absent in the muscle-less mutant ulna (Figure 6P), reflecting the reduced phenotypic effect seen in this rudiment [9]. Expression is seen only in the perichondrium of the radius in both the control and mutant (Figure 6O, P).
Discussion
Here we describe the spectrum of genes expressed in the developing humerus at TS23, early in the process of ossification and when territories of differentiating cells are being defined in the developing joint region. We also use microarray and RNA-sequencing to identify genes that are differentially regulated when mechanical stimulation of the developing skeletal rudiment is altered, giving an insight into the genes that respond to mechanical stimuli generated by muscle contractions. We reveal that the genes altered are highly enriched for genes that regulate development and differentiation, are involved in cytoskeletal rearrangement and components of extracellular matrix including cell adhesion and signalling molecules. Components of multiple signalling pathways important during development are affected, in particular 34 components of the Wnt signalling pathway. Although it is clear that appropriate mechanical stimulation from in utero muscle contractions is required for normal development of bone and cartilage, we know very little about the molecular mechanisms that incorporate mechanical cues with classical biochemical signalling pathways. The differentially regulated genes identified here, particularly those associated with signalling pathways and cytoskeletal changes represent a valuable focus for dissecting integrated regulation by biochemical and mechanical signals. These data represent an important resource that can be utilised to understand the molecular basis of mechanoregulation.
The transcriptome of a developing skeletal rudiment
Utilising RNA-sequencing technology to reveal the transcriptome in the normal developing humerus and associated joints at TS23 provides an insight to the processes that are occurring during this stage of skeletal development when chondrocytes are undergoing hypertrophy, the cartilaginous rudiment is beginning to ossify at the mid diaphysis reviewed in [84] and specific zones within the joint are differentiating reviewed in [85]. This adds a valuable resource to a growing set of data that can be combined to explore skeletal development. Previous transcriptome profiling studies have examined cartilage condensation in the tibia-fibula from E11.5 to E13.5 [86], ossification of metatarsals [87], in different zones of the growth plate in 2 week old mice [88] and in Runx2-/- mutant mice [89]. Earlier stages of limb development, although not specific to skeletal development, are informed by whole limb bud profiles between E9.5 and E11.5 [32]. Another important resource that has recently become available examines the spatial expression patterns of 18,000 coding genes and over 400 microRNAs within the whole mouse embryo at the same developmental stage examined here [90]. This digital transcriptome atlas is a powerful tool (Eurexpress; http://www.eurexpress.org/ee/intro.html) that can be used to examine spatial analysis of specific genes, exploring possible functional associations. Combining these resources gives information on quantitative and spatial expression of individual genes providing the basis to explore regulatory networks active during the development of skeletal rudiments.
Several of the findings of the transcriptome analysis are as expected; the most highly expressed genes (8 with ≥100,000 read counts) include 5 collagen encoding genes (Figure 2B); collagens have been shown to be the most abundant structural proteins in cartilage and show characteristic distribution patterns as skeletal rudiments develop [31,91]. The Insulin-like growth factor genes Igf2 and Igf1 and their associated receptors Ifg2r and Igf1r are also highly expressed (4,194 to 221,194 read counts); these are reported to play a prominent regulatory role in skeletal development (reviewed in [92]). Similarly, aggrecan (Acan) and osteopontin (Spp1), both involved in skeletal development, are highly expressed (Figure 2B). Although much is known about the regulatory network that controls early chondrogenesis and joint formation (Kronenberg [25]; Provot and Schipani [26]), open-ended whole transcriptome studies are required to add new information. Centred on regulatory signalling pathways known to be involved in skeletal development; HH, FGF, TGFβ (including BMP) and Wnt; a complete list of components of these pathways expressed in the humerus as TS23 was extracted (Figure 2B) indicating the potential role players in each of these pathways at this specific stage. The importance of Indian hedgehog (Ihh) expression in the early prehypertrophic and hypertrophic chondrocytes of cartilage condensations is well established [89,93,94]. The full spectrum of possible interacting molecules in HH signalling (Figure 2B) include the receptors Ptch1, Ptch2, Smo and transcription factors Gli1, Gli2, and Gli3. In addition to Ihh, Desert hedgehog (Dhh) expression was also detected (read count 70) and, examining the data presented by Cameron et al. confirms that Dhh is up-regulated (≥3 fold) in E13.5 fibual and tibual cartilage [86]. Dhh has not previously been functionally linked to skeletogenesis but this opens the possibility of regulatory contribution, perhaps co-operating with Ihh. No expression of Shh was detected. Similarly, novel components of the FGF, TGBβ and Wnt pathways were identified (Figure 2B).
The Wnt signalling pathway plays a central role during embryonic development and is known to be an important regulator of bone formation and bone remodelling reviewed in [95]. It also plays a pivotal role in joint formation and maintenance, shown through gain and loss of function experiments [80,96]. The key intracellular mediator of canonical Wnt signalling, β-catenin (Ctnnb1; 28,608 read counts) is the most highly expressed Wnt signalling component in the TS23 humerus and associated joints. The most highly expressed Wnt ligand is Wnt5a, previously associated with expression in joints and perichondrium [79] and proliferating chondrocytes [97]. Other highly expressed ligands include Wnt9a, Wnt5b, Wnt11, and Wnt4. Wnt5b and Wnt11 expression has been shown in the pre-hypertrophic chondrocytes and Wnt4, Wnt9 and Wnt16 in the developing joints [79-81,98]. High expression of Wnt9a could be due to its role in the temporal and spatial regulation of Ihh[96].
Numerous extracellular modulators of the pathway were detected; all five secreted frizzled related protein (Sfrp) genes, Dickoff (Dkk) 1, 2 and 3 genes and four R-spondins (Rspos), indicating a huge potential for pathway modulation. The most highly expressed antagonist modulators of the pathway were Dkk3 and Sfrp2 both of with are detected in joint cells at E13.5 and E15.5 [82]. The most highly expressed R-spondin agonist of the pathway was Rspo3, previously detected in phalanges [55].
Identification of Mechanoresponsive genes
The identification of differentially expressed genes between humeri from control and muscle-less embryonic limbs allows an investigation of the biological processes and the developmental regulatory signalling pathways that are affected by the removal of mechanical stimulation on skeletogenesis in vivo. We previously reported that muscleless (Splotch) mutants display abnormal ossification in the humerus, altered humeral morphogenesis and altered elbow and shoulder joint formation and these effects were first observed at TS23 [9]. This was chosen as the point of analysis for differential gene expression because, although limb muscles begin to contract from approximately E12.5 [99] it is uncertain how much stimulation is transmitted to the skeletal rudiments when the developing tendons are at early stages of morphogenesis [100]; the sole indication that the force is functionally transmitted is the mutant phenotype seen at TS23. Although analysis at TS23 may miss some of the earliest effects, it is relatively early in the response and the earliest time at which it is certain that the system is disturbed. Alteration in expression pattern of some selected candidate genes and pathways was previously revealed [8,10], but here we carry out the first genome wide study identifying a total of 1,132 independent genes as differentially expressed: with approximately 60% down-regulated and 40% up-regulated. The finding of more genes being down-regulated than up-regulated and to a greater extent is consistent with the proposal that mechanical stimuli support the correct differentiation of cells, as observed in the ossification phenotype [9], and for the maintenance of tissue patterning, as seen in the developing joint [16]. GO annotation analysis identified specific biological processes that are affected when mechanical stimuli are removed. This type of analysis has been used previously to interpret biological processes associated with developing skeletal tissue [87,89,101]. Analysis of the down-regulated DE gene set identified genes associated with development and differentiation as the most highly enriched categories, including developmental regulatory signalling pathway molecules and transcription factors. Similarly, analysis of up-regulated DE gene sets indicated genes associated with cell signalling and development and differentiation.
DE genes were also highly enriched for genes associated with the cytoskeleton. The cytoskeleton controls cell shape, organelle transport, cell motility and division, and connects the extracellular matrix to internal cell processes reviewed in [102]. It maintains the mechanical integrity of cells and has been implicated in relaying mechanical signals to downstream biochemical responses [7,103]. This was seen in the embryonic lung where cytoskeletal network inhibitors resulted in altered tissue morphogenesis and conversely when cytoskeletal tension was activated lung development was accelerated reviewed in [104], indicating the dynamic role the cytoskeleton has in morphogenesis.
In chondrocytes the actin microfilaments are predominantly located at the periphery of the cytoplasm [105], tubulin microtubules are uniformly distributed throughout the cytoplasm [106] as are intermediate filaments, connecting the nuclear membrane with the cell periphery [107]. In this study 84 genes annotated as cytoskeletal were down-regulated when mechanical stimulation was removed. These include 33 genes directly associated with actin microfilaments, 13 with microtubules and 4 with intermediate filaments (Figure 4). The most highly affected group, the Filamentous-actin cytoskeleton, has been shown to be involved in articular cartilage chondrocyte mechanotransduction, converting a mechanical stimulus into a biochemical response [103,108-110]. Other studies have confirmed the involvement of the actin cytoskeleton in cartilage chondrocyte mechanotransduction via manipulation of the actin accessory proteins [108,111], but there are few reports on the affect of mechanical stimulation on microtubule and intermediate filaments [112]. Among the DE genes is an actin-binding protein, cofilin2 (Cfl2); cofilin was previously shown to be increased following cyclic mechanical loading of chondrocytes [111].
The identification of cytoskeletal genes down-regulated following the removal of mechanical stimulation indicates that the cytoskeleton is affected, but is this because the mechanical integrity of the cell is altered or because mechanotransduction from the ECM is affected, or perhaps a combination of both? The finding that ECM and cell adhesion associated genes are also affected further supports changes in mechanotransduction pathways. The cell-adhesion associated integrins (Itga4, Itga7, Itgb6) and cadherins (Cdh4, Cdh15, Cdhr3) are down-regulated and these proteins potentially function to physically couple cells to the ECM and play a role in mechanical signal transduction [103,104]. Articular chondrocytes have been shown to express both integrin [113] and non-integrin [114] ECM receptors. Another actin-associated protein identified to be down-regulated is actinin-α2 (Actn2); this protein also couples the cytoskeleton to the ECM and may be involved in transducing mechanical stimulation.
Secreted phosphoprotein 1 (Spp1), previously known as Osteopontin (Opn), is one of the abundant non-collagenous proteins in bone matrix produced by osteoblasts and osteoclasts reviewed in [115]. Spp1 binds to hydroxyapatite and is a potent inhibitor of the mineralisation process, inhibiting the growth of bone matrix crystals [116]. Spp1 is expressed early in bone development, however it was concluded not to be required for normal development of bones as null mice (OPN-/-) have no apparent effect on the structure or distribution of cells within bone tissue [117]. However, Spp1 expression has been shown to be regulated by mechanical stimulation both in vitro and in vivo[35,118-120]. We found Spp1 to be down-regulated in the developing humerus at TS23 in muscle-less embryos and in situ hybridisation showed a dramatic absence of detectable Spp1 expression in hypertrophic chondrocytes whereas it is still detectable in the perichondrium (Figure 6N), indicating a specific effect on expression in hypertrophic chondrocytes and not a delay in the onset of normal expression. It was previously shown that OPN-/- mice did not suffer bone loss in response to mechanical unloading [120], suggesting that mice lacking Spp1 could not sense the changes in mechanical stress, thus indicating its potential role in the signal transduction of mechanical stimulation. It has been suggested that mechanotransduction through Spp1 is dependent on microfilament integrity, as mechanically stimulated increases in Spp1 expression was blocked by disruption with cytochalasin-D in osteoblasts [119]. This again highlights the link between an ECM component and the cytoskeleton in a mechanoresponse implicating these components in signal transduction, either directly through the cytoskeleton or through cell adhesion complexes via the cytoskeleton.
An example of a non-integrin ECM component that is down-regulated in the absence of mechanical stimulation is Cd44, a target gene of the Wnt signalling pathway [50], encoding a single-pass membrane glycoprotein that binds proteoglycan and hyaluronan (HA) to produce a pericellular matrix surrounding chondrocytes reviewed in [121]. Cd44 has been implicated in joint cavitation through interaction with HA [122]. We previously showed loss of expression of the Cd44 gene in the interzone of the forming knee joints of immobilised chick embryos [16], one of a number of gene expression patterns reflecting a general loss of organisation of differentiating tissue territories, and here we show a similar effect on Cd44 expression in the elbow and shoulder joint of muscle-less mouse limbs, where we previously showed a similar loss of tissue organisation [9]. The very restricted expression of Cd44 in the interzone of forming joints in control embryos at TS23 is barely detectable in muscle-less mutants (Figure 6A-D). As well as the gene being sensitive to mechanical stimulation, as an integral part of the ECM and a regulator of joint formation, the gene product may also be an important mediator of mechanical stimuli.
The link between Cd44 and the Wnt signalling pathway highlights perhaps the most striking finding of this analysis; the altered expression of 34 genes implicated in the Wnt signalling pathway (Figure 5). Canonical Wnt signalling has been shown to be involved in maintaining joint integrity [80,96] and is disturbed in the joints of muscle-less mouse embryos [8]. Wnt signalling might also be involved in co-ordinating ossification and joint development; both processes altered in muscle-less embryos [5]. Non canonical signalling has also been implicated in planar cell polarity during growth plate regulation [123,124]. There are also previous indications that the Wnt pathway is responsive to mechanical stimulation in mesenchymal stem cells [125] in mature bone in vivo[126] and in response to injury of articular cartilage [127]. Here, genes encoding four Wnt ligands are up-regulated in muscle-less embryos and in the case of Wnt4 we show specific up-regulation in the elbow and shoulder joint region (Figure 6I-L). Two of the up-regulated genes (Wnt2 and Wnt2b) have not previously been associated with skeletal development. The Wnt signalling antagonist Sfrp2 is also up-regulated specifically in the joint region (Figure 6E-H). The majority of known Wnt target genes affected are down-regulated (Figure 5), perhaps due to increased expression of negative regulators Sfrp2 and Dkk2 and down-regulation of the Fzd10 receptor; however ligands and agonists Rspo2 and Rspo3 are up-regulated as are some target genes indicating effects at multiple levels of regulation of the pathway. It is interesting that a number of the up-regulated targets feedback as negative regulators of the pathway (for example: Dkk2, Sfrp2). It is now important to functionally test the mechanisms linking mechanical stimulation with Wnt signalling. This work provides sets of candidate genes to use in functional assays to excavate this important link. Understanding how mechanical stimuli influence the Wnt signalling pathway would be a major step forward in understanding how mechanical cues work together with classical molecular positional information to guide spatially appropriate tissue differentiation and provide indications of how conditions can be effectively recreated in vitro to guide stem cell differentiation.
In situ hybridisation analysis showed altered gene expression of 3 Wnt pathway genes in the developing shoulder and elbow joints; one down regulated (Cd44) and two upregulated (Sfrp2 and Wnt4) (Figure 6). Changes in the spatial restriction of Sfrp2 and Wnt4 expression were also seen. This does not represent a delay in normal expression in the mutant because the altered pattern is not reminiscent of earlier stages [82] and the changes are consistent with altered patterning of the tissue territories in the forming joint and the fusion phenotype seen in the mutant, with cartilage forming across the joint at later stages [8,9].
In this study differential expression in developing skeletal rudiments is documented in the absence of limb muscle; this will include genes that respond to lack of mechanical stimulation but perhaps also as a paracrine response to adjacent muscle cells. We know that phenotypic effects on ossification and joint formation are due to the lack of mechanical stimulation rather than physical absence of muscle cells because phenotypic analysis of a range of mouse mutants where muscle is immobile [8], reduced [9] or absent [8,128] have similar effects and we see similar effects in immobilised chick embryos [15,16]. Therefore, although some of the genes identified here may respond to lack of adjacent muscle tissue, many must underlie the phenotypic effects seen in response to lack of mechanical stimulation. This is further supported by the overlap of some of the genes identified here and in skeletal cells in culture or adult tissues, in response to mechanical stimulation [18,19].
Another important limitation in this work is the possibility that a proportion of the down-regulated genes may be due to contamination of the dissected control humeri with adjacent mesenchyme/muscle, since this is being compared to tissue from muscle-less embryos. Although care was taken with the dissections, it is impossible to be sure that all muscle tissue was eliminated from the control. The down-regulated gene set also showed enrichment for muscle associated genes (Additional file 1: Table S4), consistent with possible contamination of the dissected control humeri by neighbouring muscle. To inform this we also sequenced the transcriptome of mesenchyme adjacent to the humerus of control embryos at TS23 and compared it to the transcriptome of control humeri. We then cross referenced this to the down-regulated gene set in control versus muscle-less humeri (Additional file 1: Table S2), noting any genes enriched more than 3 fold in mesenchyme compared to control humeri; these are indicated in column 2 of Additional file 1: Table S2. It is possible that these genes are involved in both cartilage and muscle development so no genes have been removed from the data set, however, DE genes also showing higher expression in mesenchyme compared to control humeri must be treated with caution with respect to a skeletal specific response to mechanical stimulation. Such genes have not been prioritised in any of our subsequent exploration of candidate mechanosensitive genes.
The developing humerus at TS23 constitutes different cell and tissue populations at different stages of differentiation including the joint region, the perichondrium and the organised zones within the cartilage rudiment. Therefore the experimental design employed here will capture genes associated with different cells types at different stages of differentiation. It will now be important to sort out which cells and tissues have altered expression of specific genes. This can be addressed for a sub set of genes by in situ hybridisation, with an initial analysis of 4 genes presented in Figure 6. It can be addressed in a high throughput manner by isolating specific cell populations using laser microdissection from tissue sections (laser capture), purification of RNA and quantitative RT-PCR gene expression profiling, comparing control and mutant tissue from,, for example the hypertrophic, prehypertrophic or the elbow joint region alone.
We used both RNA-sequencing (Illumina) and Microarray (Agilent) technologies in parallel to determine differential expression. Microarray technology has been utilised to determine expression of chondrogenic and osteogenic genes from developing whole tissues [32,87,89], and from in vitro differentiation procedures [19,34,129-131]. The use of RNA-seq technology to describe the transcriptome is more recent [132-134]. Previous direct comparisons between microarray and RNA sequencing-based approaches to reveal alterations in gene expression between tissues reported that RNA-seq identified more DE genes [23,24,135]. We also found that RNA-seq is more sensitive in reproducibly detecting alterations in gene expression, detecting more genes altered at lower quantitative levels (Additional file 1: Table S1; 5 > 2 fold). This was further emphasised by reducing the stringency of the statistical analysis to p ≤ 0.08, which increased the number of genes detected by microarray specifically (not shown). An example of the importance of the increased sensitivity and reproducibility of RNA-seq is shown by the Spp1 gene which did not show statistical significance by microarray but has been verified by qRT-PCR and in situ hybridisation (Table 2 and Figure 6). The larger dynamic range [24] and higher reproducibility across replicates [135] has also been found in other studies.
Conclusion
This study examines the set of genes active at a key stage of skeletal development (TS23) and reveals the genes that are differentially regulated in the developing humerus when skeletal muscle is absent. Since we previously showed that the lack of muscle contractions leads to common phenotypic defects in both ossification and joint formation in several chick and mouse models, this provides an insight into the genome wide alterations in gene transcription that take place when the mechanical environment is altered. Given the importance of appropriate mechanical stimulation generated by embryo movement on skeletal development we postulated that mechanical stimuli must integrate with biochemical cell signalling pathways known to be essential for normal development. We show that multiple signalling pathways are affected, with components of the Wnt signalling pathway most strongly disturbed including 4 Wnt ligands and both down-regulation and up-regulation of target genes. Down-regulated genes include Cd44, Dll1 and Fgf4 which are involved in further cellular interactions during joint formation or feed into other important cell communication events. Among the up-regulated Wnt targets are several genes that feed back into the Wnt pathway itself as antagonists (Sfrp2 and Dkk2) or agonists (Rspo2, Rspo3). This finding, together with alteration of cytoskeletal components, indicates the biological processes involved in integrating biophysical stimuli during cell differentiation and patterning. Understanding the mechanistic basis for how developing cells interpret and respond to biophysical cues is a major challenge, relevant to all developing systems, and will impact our ability to control differentiation of progenitor cells for regenerative therapies. This work is an early step in unravelling the mechanistic basis of biophysical regulation of skeletal development and provides a focus for future studies.
Methods
RNA preparation
Heterozygous Splotch-delayed (Pax3 Spd/+ ) [12] mice were purchased from Jackson Laboratories (Jax®). All animal work was carried out under the guidelines of Trinity College Dublin Bioresources Unit and Bioethics Committee. The generation of homozygous Pax3 Spd/Spd mutant embryos was achieved by crossing heterozygous Pax3 Spd/+ males and females. Embryonic material was collected from timed pregnancies on the afternoon of the 14th day (E14.5). Individual embryos were dissected and the developmental stage according to Theiler criteria [21], and the phenotype were recorded. All embryos were genotyped following PCR amplification as described in [136]. The humeri, including the associated joint regions, were finely dissected from control and mutant embryos at stage TS23 (Figure 1A). Tissue was mechanically homogenised and total RNA extracted (SV Total RNA Isolation System: Promega, UK). Pooling of rudiment tissue from multiple embryos of the same genotype (2–4) was performed. RNA integrity was assessed on a 2100 Bioanalyser (Agilent Technologies); RNA samples with RIN (RNA integrity number) values of 8.2-9.6 were used for Microarray and RNA-seq analysis.
Microarray
Four independent pooled sets of samples were used for microarray (n = 4 biological replicates) analysis. All microarrays were processed at IMGM® Laboratories (Martinsried, Germany). 100 ng of total RNA per sample was reverse transcribed into cDNA and then converted into labelled cRNA by in vitro transcription incorporating cyanine-3-CTP (Low input quick-amp labelling kit one-colour, Agilent Technologies). Genome wide expression profiling was carried out using the Agilent Mouse GE v2 Microarrays (4x44K format) (G4846A, Agilent Technologies) which contains 39,485 coding and non-coding sequences of the mouse genome (Figure 1B). A one-colour based hybridisation protocol was performed at 65°C for 17 hours on separate mouse GE v2 microarray platforms. Microarrays were then washed with increased stringency using Gene Expression Wash Buffers (Agilent Technologies) followed by drying with Acetonitrile (Sigma). Fluorescent signal intensities were detected with Scan Control A.8.4.1 software (Agilent Technologies) in the Agilent DNA microarray scanner and extracted from the images using Feature Extraction 10.7.3.1 software (Agilent Technologies). The software tools Feature Extraction 10.7.3.1, GeneSpring GX 11.5.1 and Spotfire Decision Site 9.1.2 (TIBCO) were used for quality control and statistical data analysis. Quantile normalisation was applied to each data set in order to impose the same distribution of probe signal intensities for each array [137], thus adjusting them to a uniform level that can allow for comparable downstream analysis. Welch’s approximate t-test (“unpaired unequal variance”, parametric) was applied to compare the control and mutant groups. A corrected p-value was calculated based on the algorithm of Benjamini and Hochberg [27], based on control of the False Discovery Rate (FDR). A fold change of ≥ 2 and FDR-adjusted p-value of ≤0.05 were used as criteria to indicate differential expression between the two groups.
RNA-sequencing: alignment and differential expression analysis
Three independent pooled sets of samples were used for RNA-seq (n = 3 biological replicates) analysis. The DNase-treated RNA (3 μg) was used to prepare RNA-Seq libraries with the TruSeq RNA Sample Prep kit. A total of six cDNA libraries were constructed, representing triplicate biological replicates for each group. 40 bp single end reads were obtained from an Illumina GAII in FASTQ format, one sample per sequencing lane. The Tophat aligner (http://tophat.cbcb.umd.edu/) was used to align the reads to the mouse reference genome (mm9). After alignment the read counts for each gene were extracted using htseq-count (http://www-huber.embl.de/users/anders/HTSeq/) based on an mm9 Refseq gff file. Differential expression in our two groups was evaluated using DESeq version 1.4.1, implemented in R 2.14.1. DESeq uses a negative binomial distribution to model genic read counts following normalisation based on size factors and variance. As for the microarray analysis, p-values were adjusted by the procedure of Benjamini and Hochberg to control the type I error rate, and a cut off of p ≤ 0.05, and a fold change of ≥ 2 were used as a threshold to define differential expression.
Quantitative real-time reverse transcription-polymerase chain reaction
Quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) was used to verify the relative gene expression changes in nine genes indicated to be differentially expressed (DE) by microarray and RNA seq analysis; Fgf4, Cilp, Rxrg, Dll1, Spp1 Vstm2a, Figf, Fgf10 and Sfrp2 (Table 7). All primers were designed using Primer Express Software®, version 3.0, under default settings for TaqMan® quantification and purchased through Sigma (Sigma-Aldrich, UK). Primers sets were designed with a primer Tm range of 58°-60°, an optimal length of 20 bp and an amplicon range of 50–150 bp. Total RNA was reverse transcribed (100 ng) into cDNA using iScript™ cDNA systhesis kit (BioRad) as per manufacturer’s instructions. SYBR green gene expression quantification was performed using QuantiTect SYBR green kit. 5 μl of cDNA preparation was diluted 1:5 with RNase free water, 10 μl of 2x QuantiTect SYBR green PCR master mix, 0.5 ul (10 μM) of each primer and 4ul RNase free water). Samples were assayed in triplicate in one run (40 cycles), which was composed of three stages, 95°C for 10 min, 95°C for 15 s for each cycle (denaturation) and 60°C for 1 min (annealing and extension). Real-time PCR was performed using an ABI 7500 Sequence Detection system (Applied Biosystems). qRT-PCR data was analysed using relative quantification and the Ct method as described previously [138], with the Gapdh gene as the endogenous control. The level of gene expression was calculated by subtracting the averaged Ct values (Ct is the threshold cycle) for Gapdh from those of the gene of interest. The relative expression was calculated as the difference (ΔΔCt) between the Ct of the test sample (mutant) and that of the control sample. The relative expression of genes of interest were calculated and expressed as 2-ΔΔCt. Relative quantification values are presented as fold changes plus/minus the standard error of the mean relative to the control group, which was normalised to one.
Table 7.
Primer sequences for qRT-PCR analysis of differentially expressed genes
| Gene | Primer sequence | Amplicon length | |
|---|---|---|---|
| Fgf4 |
Fwd |
CCGACGAGTGTAAATTCAAAGAAA |
97 |
| Rv |
TTCTTACTGAGGGCCATGAACA |
||
| Cilp |
Fwd |
AAAAAGACGGCTTTCCAAATCA |
78 |
| Rv |
GGCATAGATAGGCCCATTGC |
||
| Rxrg |
Fwd |
CGTTGAGTGGGCCAAACG |
75 |
| Rv |
CCTGCCCGGAGTAGAATGAC |
||
| Dll1 |
Fwd |
GACCGCCGCTTCCTAATAGG |
74 |
| Rv |
GCCCAGATGTTCAGCTTAATTCC |
||
| Spp1 |
Fwd |
CCCTCGATGTCATCCCTGTT |
69 |
| Rv |
TGCCCTTTCCGTTGTTGTC |
||
| Vstm2a |
Fwd |
GTGGAGCTCTTACCCGACAGA |
73 |
| Rv |
CATTGCCTTGGACTTTCACTGTAC |
||
| Figf |
Fwd |
GGTTGCCTGAAACAGAGTAGTAGGT |
71 |
| Rv |
AGCATTGCCCTTGGACTTTG |
||
| Fgf10 |
Fwd |
GGGCTGCTGTTGCTGCTT |
94 |
| Rv |
GGCCTCCTGTGACACCATGT |
||
| Sfrp2 | Fwd | CAGAGAGAGTTCAAGCGCATCTC | 68 |
Gene ontology annotation analysis
Gene Ontology (GO) terms were utilised to reveal significant enrichment of groups of genes among the DE datasets from the microarray and the RNA-seq analysis using the Database for Annotation, Visualisation and Integrated Discovery, DAVID (http://david.abcc.ncifcrf.gov/) [28], and GOstat (http://gostat.wehi.edu.au/) software. Analysis of GO terms associated with biological process, molecular function and cellular component was performed on all data-sets independently and combined to identify significantly (p ≤ 0.05) enriched gene sets. The strength of the enrichment of any GO term-associated gene set is reflected in the calculated p-values, comparing the proportion of genes in the data-set and the proportion of genes in the genome bearing that annotation.
In situ Hybridisation
Expression probes were prepared from cDNA clones obtained from a mouse expressed sequence tag (EST) library (IMAGE, Source Biosciences); details given in Table 8. Antisense and sense digoxigenin-labelled RNA was transcribed in vitro from 1 μg of linearised plasmid using T7, T3 and SP6 promoter sites (according to insert orientation and vector), all components for in vitro transcription were purchased from Roche, Germany. DNA template was degraded by incubation of probes with RNase free DNase (Roche) and probes purified on G25 columns (Amersham Biosciences, USA) according to manufacturer’s instructions. Probe concentrations were determined by spectophotometry and probes stored at -20°C. Embryonic limbs at TS23 were fixed (4% Paraformaldehyde (PFA)), dehydrated (graded series of Methanol/Phosphate Buffered Saline with 0.1% Triton (PBT) washes) and stored at -20°C prior to sectioning. Limbs were rehydrated in a reverse series of Methanol/PBT washes. Sectioning was performed with a vibrating microtome (VT1000S, Leica; embedded in 4% low melting point agarose (Invitrogen)/PBS (80 μm)). Hybridisation of sections was largely carried out as described previously [128].
Table 8.
Details of cDNA clones used as expression probes
| Gene | Extent of probe on genbank sequence | Source |
|---|---|---|
| Sfrp2 |
Nucleotide 82 to 852 on gene bank sequence U88567 |
A. Rattner |
| Cd44 |
Nucleotide 222 to 3020 on gene bank sequence NM_00103915.1 |
IMAGE Library |
| Spp1 |
Nucleotide 27 to 1472 on gene bank sequence NM_001204201 |
IMAGE Library |
| Wnt4 | Nucleotide 639 to 1101 on NM_009523.1 | A. McMahon |
Availability of supporting data
The data sets supporting the results of this article are available in the EMBL-EBI ArrayExpress repository (http://www.ebi.ac.uk/arrayexpress/). The differential expressed data set from the Microarray [E-MTAB-1744], the differential expressed data set from the RNA-sequencing [E-MTAB-1746] and for the transcriptome [E-MTAB-1745]. Lists of differentially expressed genes are available in Additional file 1.
Abbreviations
TS: Theiler stage; RNA-sseq: RNA-sequencing; PCR: Polymerase chain reaction; RNA: Ribonucleic acid; RIN: RNA integrity number; cDNA: complementary deoxyribonucleic acid; FDR: False discovery rate; qRT-PCR: quantitative real time PCR; DE: Differential expression; Tm: Melting temperature; bp: Base pairs; ng: Nanogram; μM: Micro molar; min: Minute; Ct: Cycle threshold; GO: Gene ontology; EST: Expressed sequence tag; PFA: Paraformaldehyde; PBS: Phosphate buffer saline.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RR carried out all the experiments and analysis of data, collected all samples and drafted the manuscript. NN and PP were involved in preliminary work and conception of the present study. EK, PC and DM provided an RNA sequencing service at TrinSeq generating the primary gene lists. DK contributed to discussions of the findings and drafting of the manuscript. PM conceived and designed the study, participated in data analysis and oversaw drafting of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplementary data Tables S1-S4.
Contributor Information
Rebecca A Rolfe, Email: rolfera@tcd.ie.
Niamh C Nowlan, Email: n.nowlan@imperial.ac.uk.
Elaine M Kenny, Email: elaneyk@gmail.com.
Paul Cormican, Email: cormicp@tcd.ie.
Derek W Morris, Email: derek.morris@tcd.ie.
Patrick J Prendergast, Email: pprender@tcd.ie.
Daniel Kelly, Email: kellyd9@tcd.ie.
Paula Murphy, Email: paula.murphy@tcd.ie.
Acknowledgements
This research was funded by a TCD-UCD Innovation Academy PhD student scholarship Bursary (2010–2014) and the Wellcome Trust (083539/Z/07/Z). Next generation sequencing was performed in TrinSeq (http://www.medicine.tcd.ie/sequencing), a core facility funded by Science Foundation Ireland (SFI) under Grant No. [SFI/07/RFP/GEN/F327/EC07] with support from the Trinity Centre for High Performance Computing.
References
- Chen JH, Liu C, You L, Simmons CA. Boning up on Wolff’s Law: mechanical regulation of the cells that make and maintain bone. J Biomech. 2009;15(1):108–118. doi: 10.1016/j.jbiomech.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Rodriguez JI, Garcia-Alix A, Palacios J, Paniagua R. Changes in the long bones due to fetal immobility caused by neuromuscular disease. A radiographic and histological study. J Bone Joint Surg Am. 1988;15(7):1052–1060. [PubMed] [Google Scholar]
- Rodriguez JI, Palacios J, Garcia-Alix A, Pastor I, Paniagua R. Effects of immobilization on fetal bone development. A morphometric study in newborns with congenital neuromuscular diseases with intrauterine onset. Calcif Tissue Int. 1988;15(6):335–339. doi: 10.1007/BF02553275. [DOI] [PubMed] [Google Scholar]
- Nowlan NC, Sharpe J, Roddy KA, Prendergast PJ, Murphy P. Mechanobiology of embryonic skeletal development: Insights from animal models. Birth Defects Res C Embryo Today. 2010;15(3):203–213. doi: 10.1002/bdrc.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe R, Roddy K, Murphy P. Mechanical regulation of skeletal development. Curr Osteoporos Rep. 2013;15(2):107–116. doi: 10.1007/s11914-013-0137-4. [DOI] [PubMed] [Google Scholar]
- Mammoto T, Mammoto A, Torisawa YS, Tat T, Gibbs A, Derda R, Mannix R, de Bruijn M, Yung CW, Huh D. et al. Mechanochemical control of mesenchymal condensation and embryonic tooth organ formation. Dev Cell. 2011;15(4):758–769. doi: 10.1016/j.devcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto A, Mammoto T, Ingber DE. Mechanosensitive mechanisms in transcriptional regulation. J Cell Sci. 2012;15(Pt 13):3061–3073. doi: 10.1242/jcs.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn J, Shwartz Y, Blitz E, Krief S, Sharir A, Breitel DA, Rattenbach R, Relaix F, Maire P, Rountree RB. et al. Muscle contraction is necessary to maintain joint progenitor cell fate. Dev Cell. 2009;15(5):734–743. doi: 10.1016/j.devcel.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Nowlan NC, Bourdon C, Dumas G, Tajbakhsh S, Prendergast PJ, Murphy P. Developing bones are differentially affected by compromised skeletal muscle formation. Bone. 2010;15(5):1275–1285. doi: 10.1016/j.bone.2009.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowlan NC, Dumas G, Tajbakhsh S, Prendergast PJ, Murphy P. Biophysical stimuli induced by passive movements compensate for lack of skeletal muscle during embryonic skeletogenesis. Biomech Model Mechanobiol. 2012;15(1–2):207–219. doi: 10.1007/s10237-011-0304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz T, Kothary R, Surani MA, Halata Z, Grim M. The Splotch mutation interferes with muscle development in the limbs. Anat Embryol (Berl) 1993;15(2):153–160. doi: 10.1007/BF00171747. [DOI] [PubMed] [Google Scholar]
- Vogan KJ, Epstein DJ, Trasler DG, Gros P. The splotch-delayed (Spd) mouse mutant carries a point mutation within the paired box of the Pax-3 gene. Genomics. 1993;15(2):364–369. doi: 10.1006/geno.1993.1333. [DOI] [PubMed] [Google Scholar]
- Sharir A, Stern T, Rot C, Shahar R, Zelzer E. Muscle force regulates bone shaping for optimal load-bearing capacity during embryogenesis. Development. 2011;15(15):3247–3259. doi: 10.1242/dev.063768. [DOI] [PubMed] [Google Scholar]
- Roddy KA, Kelly GM, van Es MH, Murphy P, Prendergast PJ. Dynamic patterns of mechanical stimulation co-localise with growth and cell proliferation during morphogenesis in the avian embryonic knee joint. J Biomech. 2011;15(1):143–149. doi: 10.1016/j.jbiomech.2010.08.039. [DOI] [PubMed] [Google Scholar]
- Nowlan NC, Prendergast PJ, Murphy P. Identification of mechanosensitive genes during embryonic bone formation. PLoS Comput Biol. 2008;15(12):e1000250. doi: 10.1371/journal.pcbi.1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roddy KA, Prendergast PJ, Murphy P. Mechanical influences on morphogenesis of the knee joint revealed through morphological, molecular and computational analysis of immobilised embryos. PLoS One. 2011;15(2):e17526. doi: 10.1371/journal.pone.0017526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh E, Church VL, Osborne AC, Lamb KJ, Archer CW, Francis-West PH, Pitsillides AA. Differential regulation of GDF-5 and FGF-2/4 by immobilisation in ovo exposes distinct roles in joint formation. Dev Dyn. 2006;15(3):826–834. doi: 10.1002/dvdy.20679. [DOI] [PubMed] [Google Scholar]
- Patel MJ, Liu W, Sykes MC, Ward NE, Risin SA, Risin D, Jo H. Identification of mechanosensitive genes in osteoblasts by comparative microarray studies using the rotating wall vessel and the random positioning machine. J Cell Biochem. 2007;15(3):587–599. doi: 10.1002/jcb.21218. [DOI] [PubMed] [Google Scholar]
- Bougault C, Aubert-Foucher E, Paumier A, Perrier-Groult E, Huot L, Hot D, Duterque-Coquillaud M, Mallein-Gerin F. Dynamic compression of chondrocyte-agarose constructs reveals new candidate mechanosensitive genes. PLoS One. 2012;15(5):e36964. doi: 10.1371/journal.pone.0036964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironen RK, Karjalainen HM, Elo MA, Kaarniranta K, Torronen K, Takigawa M, Helminen HJ, Lammi MJ. cDNA array reveals mechanosensitive genes in chondrocytic cells under hydrostatic pressure. Biochim Biophys Acta. 2002;15(1–3):45–54. doi: 10.1016/s0167-4889(02)00247-1. [DOI] [PubMed] [Google Scholar]
- Theiler K. The House Mouse. Berlin, Heidelberg, New York: Springer; 1972. [Google Scholar]
- Malone JH, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011;15:34. doi: 10.1186/1741-7007-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiry P, Lee LJ, Frey BJ, Rupar CA, Siu VM, Blencowe BJ, Hegele RA. Transcriptional profiling of endocrine cerebro-osteodysplasia using microarray and next-generation sequencing. PLoS One. 2011;15(9):e25400. doi: 10.1371/journal.pone.0025400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomly D, Walter NA, Hunter JE, Darakjian P, Kawane S, Buck KJ, Searles RP, Mooney M, McWeeney SK, Hitzemann R. Evaluating gene expression in C57BL/6 J and DBA/2 J mouse striatum using RNA-Seq and microarrays. PLoS One. 2011;15(3):e17820. doi: 10.1371/journal.pone.0017820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;15(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- Provot S, Schipani E. Molecular mechanisms of endochondral bone development. Biochem Biophys Res Commun. 2005;15(3):658–665. doi: 10.1016/j.bbrc.2004.11.068. [DOI] [PubMed] [Google Scholar]
- Benjamini YHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;15(1):289–300. [Google Scholar]
- da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;15(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;15(9):1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Koyama E, Shibukawa Y, Nagayama M, Sugito H, Young B, Yuasa T, Okabe T, Ochiai T, Kamiya N, Rountree RB. et al. A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev Biol. 2008;15(1):62–73. doi: 10.1016/j.ydbio.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner T, Stove J. Collagens–major component of the physiological cartilage matrix, major target of cartilage degeneration, major tool in cartilage repair. Adv Drug Deliv Rev. 2003;15(12):1569–1593. doi: 10.1016/j.addr.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Taher L, Collette NM, Murugesh D, Maxwell E, Ovcharenko I, Loots GG. Global gene expression analysis of murine limb development. PLoS One. 2011;15(12):e28358. doi: 10.1371/journal.pone.0028358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Shao YY, Ballock RT. Carboxypeptidase Z (CPZ) links thyroid hormone and Wnt signaling pathways in growth plate chondrocytes. J Bone Miner Res. 2009;15(2):265–273. doi: 10.1359/jbmr.081014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Qing H, He Y, Wang J, Zhu Z, Wang H. Gene expression patterns of osteocyte-like MLO-Y4 cells in response to cyclic compressive force stimulation. Cell Biol Int. 2010;15(5):425–432. doi: 10.1042/CBI20090061. [DOI] [PubMed] [Google Scholar]
- Klein-Nulend J, Roelofsen J, Semeins CM, Bronckers AL, Burger EH. Mechanical stimulation of osteopontin mRNA expression and synthesis in bone cell cultures. J Cell Physiol. 1997;15(2):174–181. doi: 10.1002/(SICI)1097-4652(199702)170:2<174::AID-JCP9>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Bohm J, Sustmann C, Wilhelm C, Kohlhase J. SALL4 is directly activated by TCF/LEF in the canonical Wnt signaling pathway. Biochem Biophys Res Commun. 2006;15(3):898–907. doi: 10.1016/j.bbrc.2006.07.124. [DOI] [PubMed] [Google Scholar]
- Kratochwil K, Galceran J, Tontsch S, Roth W, Grosschedl R. FGF4, a direct target of LEF1 and Wnt signaling, can rescue the arrest of tooth organogenesis in Lef1(-/-) mice. Genes Dev. 2002;15(24):3173–3185. doi: 10.1101/gad.1035602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D, Koole W, Fuerer C, Fish M, Eroglu E, Nusse R. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell. 2008;15(5):508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska MB, Misztal K, Michowski W, Szczot M, Purta E, Lesniak W, Klejman ME, Dabrowski M, Filipkowski RK, Nagalski A. et al. LEF1/beta-catenin complex regulates transcription of the Cav3.1 calcium channel gene (Cacna1g) in thalamic neurons of the adult brain. J Neurosci. 2010;15(14):4957–4969. doi: 10.1523/JNEUROSCI.1425-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J. et al. Identification of a Wnt/Dvl/beta-Catenin – > Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;15(5):673–685. doi: 10.1016/S0092-8674(02)01084-X. [DOI] [PubMed] [Google Scholar]
- Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C. et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;15(6889):664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, Ben-Ze’ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci USA. 1999;15(10):5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew LL, Takemaru K, Bates R, Moon RT. Direct regulation of the Xenopus engrailed-2 promoter by the Wnt signaling pathway, and a molecular screen for Wnt-responsive genes, confirm a role for Wnt signaling during neural patterning in Xenopus. Mech Dev. 1999;15(1–2):21–32. doi: 10.1016/s0925-4773(99)00136-7. [DOI] [PubMed] [Google Scholar]
- Tice DA, Szeto W, Soloviev I, Rubinfeld B, Fong SE, Dugger DL, Winer J, Williams PM, Wieand D, Smith V. et al. Synergistic induction of tumor antigens by Wnt-1 signaling and retinoic acid revealed by gene expression profiling. J Biol Chem. 2002;15(16):14329–14335. doi: 10.1074/jbc.M200334200. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Wada N, Nishimatsu S, Nohno T. Involvement of frizzled-10 in Wnt-7a signaling during chick limb development. Dev Growth Differ. 2000;15(6):561–569. doi: 10.1046/j.1440-169x.2000.00545.x. [DOI] [PubMed] [Google Scholar]
- Hofmann M, Schuster-Gossler K, Watabe-Rudolph M, Aulehla A, Herrmann BG, Gossler A. WNT signaling, in synergy with T/TBX6, controls Notch signaling by regulating Dll1 expression in the presomitic mesoderm of mouse embryos. Genes Dev. 2004;15(22):2712–2717. doi: 10.1101/gad.1248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenpusch-Theil K, Magnani D, Amaniti EM, Han L, Armstrong D, Theil T. Transcriptional analysis of Gli3 mutants identifies Wnt target genes in the developing hippocampus. Cereb Cortex. 2012;15(12):2878–2893. doi: 10.1093/cercor/bhr365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T, Li VS, Ng SS, Taouatas N, Vries RG, Mohammed S, Heck AJ, Clevers H. The kinase TNIK is an essential activator of Wnt target genes. EMBO J. 2009;15(21):3329–3340. doi: 10.1038/emboj.2009.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon EM, van der Neut R, van de Wetering M, Clevers H, Pals ST. Wnt signaling regulates expression of the receptor tyrosine kinase met in colorectal cancer. Cancer Res. 2002;15(18):5126–5128. [PubMed] [Google Scholar]
- Wielenga VJ, Smits R, Korinek V, Smit L, Kielman M, Fodde R, Clevers H, Pals ST. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am J Pathol. 1999;15(2):515–523. doi: 10.1016/S0002-9440(10)65297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrach S, Ambler CA, Lo Celso C, Hozumi K, Watt FM. Jagged 1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development. 2006;15(22):4427–4438. doi: 10.1242/dev.02644. [DOI] [PubMed] [Google Scholar]
- Im J, Kim H, Kim S, Jho EH. Wnt/beta-catenin signaling regulates expression of PRDC, an antagonist of the BMP-4 signaling pathway. Biochem Biophys Res Commun. 2007;15(1):296–301. doi: 10.1016/j.bbrc.2006.12.205. [DOI] [PubMed] [Google Scholar]
- Miwa N, Furuse M, Tsukita S, Niikawa N, Nakamura Y, Furukawa Y. Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol Res. 2001;15(11–12):469–476. doi: 10.3727/096504001108747477. [DOI] [PubMed] [Google Scholar]
- Li X, Liu P, Liu W, Maye P, Zhang J, Zhang Y, Hurley M, Guo C, Boskey A, Sun L. et al. Dkk2 has a role in terminal osteoblast differentiation and mineralized matrix formation. Nat Genet. 2005;15(9):945–952. doi: 10.1038/ng1614. [DOI] [PubMed] [Google Scholar]
- Nam JS, Park E, Turcotte TJ, Palencia S, Zhan X, Lee J, Yun K, Funk WD, Yoon JK. Mouse R-spondin2 is required for apical ectodermal ridge maintenance in the hindlimb. Dev Biol. 2007;15(1):124–135. doi: 10.1016/j.ydbio.2007.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abed E, Chan TF, Delalandre A, Martel-Pelletier J, Pelletier JP, Lajeunesse D. R-spondins are newly recognized players in osteoarthritis that regulate Wnt signaling in osteoblasts. Arthritis Rheum. 2011;15(12):3865–3875. doi: 10.1002/art.30625. [DOI] [PubMed] [Google Scholar]
- Lescher B, Haenig B, Kispert A. sFRP-2 is a target of the Wnt-4 signaling pathway in the developing metanephric kidney. Dev Dyn. 1998;15(4):440–451. doi: 10.1002/(SICI)1097-0177(199812)213:4<440::AID-AJA9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Nam JS, Turcotte TJ, Smith PF, Choi S, Yoon JK. Mouse cristin/R-spondin family proteins are novel ligands for the Frizzled 8 and LRP6 receptors and activate beta-catenin-dependent gene expression. J Biol Chem. 2006;15(19):13247–13257. doi: 10.1074/jbc.M508324200. [DOI] [PubMed] [Google Scholar]
- Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ. et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;15(5):531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- Heymer J, Ruther U. Syndactyly of Ft/+ mice correlates with an imbalance in bmp4 and fgf8 expression. Mech Dev. 1999;15(2):173–181. doi: 10.1016/S0925-4773(99)00186-0. [DOI] [PubMed] [Google Scholar]
- Blitz E, Viukov S, Sharir A, Shwartz Y, Galloway JL, Pryce BA, Johnson RL, Tabin CJ, Schweitzer R, Zelzer E. Bone ridge patterning during musculoskeletal assembly is mediated through SCX regulation of Bmp4 at the tendon-skeleton junction. Dev Cell. 2009;15(6):861–873. doi: 10.1016/j.devcel.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc N, Luppen CA, Ho VV, Nagpal S, Hacia JG, Smith E, Frenkel B. Gene expression profiling of glucocorticoid-inhibited osteoblasts. J Mol Endocrinol. 2004;15(1):175–193. doi: 10.1677/jme.0.0330175. [DOI] [PubMed] [Google Scholar]
- Abe Y, Kita Y, Niikura T. Mammalian Gup1, a homolog of Saccharomyces cerevisiae glycerol uptake/transporter 1, acts as a negative regulator for N-terminal palmitoylation of Sonic hedgehog. FEBS J. 2008;15(2):318–331. doi: 10.1111/j.1742-4658.2007.06202.x. [DOI] [PubMed] [Google Scholar]
- Chuang PT, McMahon AP. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;15(6720):617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Furuta Y, Rush MG, Henderson R, Winnier G, Hogan BL. Involvement of Sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Development. 1997;15(1):53–63. doi: 10.1242/dev.124.1.53. [DOI] [PubMed] [Google Scholar]
- Gross I, Bassit B, Benezra M, Licht JD. Mammalian sprouty proteins inhibit cell growth and differentiation by preventing ras activation. J Biol Chem. 2001;15(49):46460–46468. doi: 10.1074/jbc.M108234200. [DOI] [PubMed] [Google Scholar]
- Johnson CW, Hernandez-Lagunas L, Feng W, Melvin VS, Williams T, Artinger KB. Vgll2a is required for neural crest cell survival during zebrafish craniofacial development. Dev Biol. 2011;15(1):269–281. doi: 10.1016/j.ydbio.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chan SW, Zhang X, Walsh M, Lim CJ, Hong W, Song H. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 2010;15(3):290–300. doi: 10.1101/gad.1865310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. Function and cancer genomics of FAT family genes (review) Int J Oncol. 2012;15(6):1913–1918. doi: 10.3892/ijo.2012.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Tohgo A, Ono K, Kaneko M, Fujishima K, Hirano T, Kengaku M. DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat Neurosci. 2005;15(7):873–880. doi: 10.1038/nn1492. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Chiba S, Hosoya N, Kumano K, Saito T, Kurokawa M, Kanda Y, Hamada Y, Hirai H. Binding of Delta1, Jagged1, and Jagged2 to Notch2 rapidly induces cleavage, nuclear translocation, and hyperphosphorylation of Notch2. Mol Cell Biol. 2000;15(18):6913–6922. doi: 10.1128/MCB.20.18.6913-6922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fior R, Henrique D. A novel hes5/hes6 circuitry of negative regulation controls Notch activity during neurogenesis. Dev Biol. 2005;15(2):318–333. doi: 10.1016/j.ydbio.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Zhang K, Wong P, Zhang L, Jacobs B, Borden EC, Aster JC, Bedogni B. A Notch1-neuregulin1 autocrine signaling loop contributes to melanoma growth. Oncogene. 2012;15(43):4609–4618. doi: 10.1038/onc.2011.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume T, Jiang H, Topczewska JM, Hogan BL. The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 2001;15(18):2470–2482. doi: 10.1101/gad.907301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzoca A, Corsi P, Polizzi A, Pinto MF, Xenaki D, Furley AJ, Gennarini G. F3/Contactin acts as a modulator of neurogenesis during cerebral cortex development. Dev Biol. 2012;15(1):133–151. doi: 10.1016/j.ydbio.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Sodek J, Ganss B, McKee MD. Osteopontin. Crit Rev Oral Biol Med. 2000;15(3):279–303. doi: 10.1177/10454411000110030101. [DOI] [PubMed] [Google Scholar]
- Woods A, Wang G, Beier F. Regulation of chondrocyte differentiation by the actin cytoskeleton and adhesive interactions. J Cell Physiol. 2007;15(1):1–8. doi: 10.1002/jcp.21110. [DOI] [PubMed] [Google Scholar]
- Ikegawa M, Han H, Okamoto A, Matsui R, Tanaka M, Omi N, Miyamae M, Toguchida J, Tashiro K. Syndactyly and preaxial synpolydactyly in the single Sfrp2 deleted mutant mice. Dev Dyn. 2008;15(9):2506–2517. doi: 10.1002/dvdy.21655. [DOI] [PubMed] [Google Scholar]
- Church V, Nohno T, Linker C, Marcelle C, Francis-West P. Wnt regulation of chondrocyte differentiation. J Cell Sci. 2002;15(Pt 24):4809–4818. doi: 10.1242/jcs.00152. [DOI] [PubMed] [Google Scholar]
- Guo X, Day TF, Jiang X, Garrett-Beal L, Topol L, Yang Y. Wnt/beta-catenin signaling is sufficient and necessary for synovial joint formation. Genes Dev. 2004;15(19):2404–2417. doi: 10.1101/gad.1230704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Tabin CJ. Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development. 2000;15(14):3141–3159. doi: 10.1242/dev.127.14.3141. [DOI] [PubMed] [Google Scholar]
- Witte F, Dokas J, Neuendorf F, Mundlos S, Stricker S. Comprehensive expression analysis of all Wnt genes and their major secreted antagonists during mouse limb development and cartilage differentiation. Gene Expr Patterns. 2009;15(4):215–223. doi: 10.1016/j.gep.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Kim IS, Otto F, Zabel B, Mundlos S. Regulation of chondrocyte differentiation by Cbfa1. Mech Dev. 1999;15(2):159–170. doi: 10.1016/S0925-4773(98)00210-X. [DOI] [PubMed] [Google Scholar]
- Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;15:629–648. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- Pitsillides AA, Ashhurst DE. A critical evaluation of specific aspects of joint development. Dev Dyn. 2008;15(9):2284–2294. doi: 10.1002/dvdy.21654. [DOI] [PubMed] [Google Scholar]
- Cameron TL, Belluoccio D, Farlie PG, Brachvogel B, Bateman JF. Global comparative transcriptome analysis of cartilage formation in vivo. BMC Dev Biol. 2009;15:20. doi: 10.1186/1471-213X-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugars RV, Karner E, Petersson U, Ganss B, Wendel M. Transcriptome analysis of fetal metatarsal long bones by microarray, as a model for endochondral bone formation. Biochim Biophys Acta. 2006;15(10):1031–1039. doi: 10.1016/j.bbamcr.2006.08.027. [DOI] [PubMed] [Google Scholar]
- Belluoccio D, Bernardo BC, Rowley L, Bateman JF. A microarray approach for comparative expression profiling of the discrete maturation zones of mouse growth plate cartilage. Biochim Biophys Acta. 2008;15(5):330–340. doi: 10.1016/j.bbagrm.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Hecht J, Seitz V, Urban M, Wagner F, Robinson PN, Stiege A, Dieterich C, Kornak U, Wilkening U, Brieske N. et al. Detection of novel skeletogenesis target genes by comprehensive analysis of a Runx2(-/-) mouse model. Gene Expr Patterns. 2007;15(1–2):102–112. doi: 10.1016/j.modgep.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I. et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;15(1):e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen BR. Role of cartilage collagens in formation of the skeleton. Ann N Y Acad Sci. 1996;15:124–130. doi: 10.1111/j.1749-6632.1996.tb56250.x. [DOI] [PubMed] [Google Scholar]
- Yakar S, Courtland HW, Clemmons D. IGF-1 and bone: New discoveries from mouse models. J Bone Miner Res. 2010;15(12):2543–2552. doi: 10.1002/jbmr.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina E, Schneider S, Rosowski M, Lauster R, Vortkamp A. Expression of Fgf and Tgfbeta signaling related genes during embryonic endochondral ossification. Gene Expr Patterns. 2005;15(1):102–109. doi: 10.1016/j.modgep.2005.04.012. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;15(16):2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macsai CE, Foster BK, Xian CJ. Roles of Wnt signalling in bone growth, remodelling, skeletal disorders and fracture repair. J Cell Physiol. 2008;15(3):578–587. doi: 10.1002/jcp.21342. [DOI] [PubMed] [Google Scholar]
- Spater D, Hill TP, O’Sullivan RJ, Gruber M, Conner DA, Hartmann C. Wnt9a signaling is required for joint integrity and regulation of Ihh during chondrogenesis. Development. 2006;15(15):3039–3049. doi: 10.1242/dev.02471. [DOI] [PubMed] [Google Scholar]
- Lu C, Wan Y, Cao J, Zhu X, Yu J, Zhou R, Yao Y, Zhang L, Zhao H, Li H, Zhao J, He L, Ma G, Yang X, Yao Z, Guo X. Wnt-mediated reciprocal regulation between cartilage and bone development during endochondral ossification. Bone. 2013;15(2):566–574. doi: 10.1016/j.bone.2012.12.016. [DOI] [PubMed] [Google Scholar]
- Hartmann C, Tabin CJ. Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell. 2001;15(3):341–351. doi: 10.1016/S0092-8674(01)00222-7. [DOI] [PubMed] [Google Scholar]
- Suzue T. Movements of mouse fetuses in early stages of neural development studied in vitro. Neurosci Lett. 1996;15(2):131–134. doi: 10.1016/S0304-3940(96)13141-4. [DOI] [PubMed] [Google Scholar]
- Marturano JE, Arena JD, Schiller ZA, Georgakoudi I, Kuo CK. Characterization of mechanical and biochemical properties of developing embryonic tendon. Proc Natl Acad Sci USA. 2013;15(16):6370–6375. doi: 10.1073/pnas.1300135110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn P, Cox M, Chen D, Serra R. Molecular profiling of the developing mouse axial skeleton: a role for Tgfbr2 in the development of the intervertebral disc. BMC Dev Biol. 2010;15:29. doi: 10.1186/1471-213X-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenghat FJ, Ingber DE. Mechanotransduction: all signals point to cytoskeleton, matrix, and integrins. Sci STKE. 2002;15(119):e6. doi: 10.1126/stke.2002.119.pe6. [DOI] [PubMed] [Google Scholar]
- Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;15(5111):1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- Ingber DE. Mechanical control of tissue morphogenesis during embryological development. Int J Dev Biol. 2006;15(2–3):255–266. doi: 10.1387/ijdb.052044di. [DOI] [PubMed] [Google Scholar]
- Idowu BD, Knight MM, Bader DL, Lee DA. Confocal analysis of cytoskeletal organisation within isolated chondrocyte sub-populations cultured in agarose. Histochem J. 2000;15(3):165–174. doi: 10.1023/A:1004095207330. [DOI] [PubMed] [Google Scholar]
- Jortikka MO, Parkkinen JJ, Inkinen RI, Karner J, Jarvelainen HT, Nelimarkka LO, Tammi MI, Lammi MJ. The role of microtubules in the regulation of proteoglycan synthesis in chondrocytes under hydrostatic pressure. Arch Biochem Biophys. 2000;15(2):172–180. doi: 10.1006/abbi.1999.1543. [DOI] [PubMed] [Google Scholar]
- Blain EJ, Gilbert SJ, Hayes AJ, Duance VC. Disassembly of the vimentin cytoskeleton disrupts articular cartilage chondrocyte homeostasis. Matrix Biol. 2006;15(7):398–408. doi: 10.1016/j.matbio.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Knight MM, Toyoda T, Lee DA, Bader DL. Mechanical compression and hydrostatic pressure induce reversible changes in actin cytoskeletal organisation in chondrocytes in agarose. J Biomech. 2006;15(8):1547–1551. doi: 10.1016/j.jbiomech.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Guilak F. Compression-induced changes in the shape and volume of the chondrocyte nucleus. J Biomech. 1995;15(12):1529–1541. doi: 10.1016/0021-9290(95)00100-X. [DOI] [PubMed] [Google Scholar]
- Parkkinen JJ, Lammi MJ, Inkinen R, Jortikka M, Tammi M, Virtanen I, Helminen HJ. Influence of short-term hydrostatic pressure on organization of stress fibers in cultured chondrocytes. J Orthop Res. 1995;15(4):495–502. doi: 10.1002/jor.1100130404. [DOI] [PubMed] [Google Scholar]
- Campbell JJ, Blain EJ, Chowdhury TT, Knight MM. Loading alters actin dynamics and up-regulates cofilin gene expression in chondrocytes. Biochem Biophys Res Commun. 2007;15(2):329–334. doi: 10.1016/j.bbrc.2007.06.185. [DOI] [PubMed] [Google Scholar]
- Steward AJ, Wagner DR, Kelly DJ. The pericellular environment regulates cytoskeletal development and the differentiation of mesenchymal stem cells and determines their response to hydrostatic pressure. Eur Cell Mater. 2013;15:167–178. doi: 10.22203/ecm.v025a12. [DOI] [PubMed] [Google Scholar]
- Woods VL Jr, Schreck PJ, Gesink DS, Pacheco HO, Amiel D, Akeson WH, Lotz M. Integrin expression by human articular chondrocytes. Arthritis Rheum. 1994;15(4):537–544. doi: 10.1002/art.1780370414. [DOI] [PubMed] [Google Scholar]
- Knudson W, Aguiar DJ, Hua Q, Knudson CB. CD44-anchored hyaluronan-rich pericellular matrices: an ultrastructural and biochemical analysis. Exp Cell Res. 1996;15(2):216–228. doi: 10.1006/excr.1996.0320. [DOI] [PubMed] [Google Scholar]
- Standal T, Borset M, Sundan A. Role of osteopontin in adhesion, migration, cell survival and bone remodeling. Exp Oncol. 2004;15(3):179–184. [PubMed] [Google Scholar]
- Steitz SA, Speer MY, McKee MD, Liaw L, Almeida M, Yang H, Giachelli CM. Osteopontin inhibits mineral deposition and promotes regression of ectopic calcification. Am J Pathol. 2002;15(6):2035–2046. doi: 10.1016/S0002-9440(10)64482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittling SR, Matsumoto HN, McKee MD, Nanci A, An XR, Novick KE, Kowalski AJ, Noda M, Denhardt DT. Mice lacking osteopontin show normal development and bone structure but display altered osteoclast formation in vitro. J Bone Miner Res. 1998;15(7):1101–1111. doi: 10.1359/jbmr.1998.13.7.1101. [DOI] [PubMed] [Google Scholar]
- Terai K, Takano-Yamamoto T, Ohba Y, Hiura K, Sugimoto M, Sato M, Kawahata H, Inaguma N, Kitamura Y, Nomura S. Role of osteopontin in bone remodeling caused by mechanical stress. J Bone Miner Res. 1999;15(6):839–849. doi: 10.1359/jbmr.1999.14.6.839. [DOI] [PubMed] [Google Scholar]
- Toma CD, Ashkar S, Gray ML, Schaffer JL, Gerstenfeld LC. Signal transduction of mechanical stimuli is dependent on microfilament integrity: identification of osteopontin as a mechanically induced gene in osteoblasts. J Bone Miner Res. 1997;15(10):1626–1636. doi: 10.1359/jbmr.1997.12.10.1626. [DOI] [PubMed] [Google Scholar]
- Ishijima M, Tsuji K, Rittling SR, Yamashita T, Kurosawa H, Denhardt DT, Nifuji A, Noda M. Resistance to unloading-induced three-dimensional bone loss in osteopontin-deficient mice. J Bone Miner Res. 2002;15(4):661–667. doi: 10.1359/jbmr.2002.17.4.661. [DOI] [PubMed] [Google Scholar]
- Knudson W, Loeser RF. CD44 and integrin matrix receptors participate in cartilage homeostasis. Cell Mol Life Sci. 2002;15(1):36–44. doi: 10.1007/s00018-002-8403-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowthwaite GP, Edwards JC, Pitsillides AA. An essential role for the interaction between hyaluronan and hyaluronan binding proteins during joint development. J Histochem Cytochem. 1998;15(5):641–651. doi: 10.1177/002215549804600509. [DOI] [PubMed] [Google Scholar]
- Li Y, Dudley AT. Noncanonical frizzled signaling regulates cell polarity of growth plate chondrocytes. Development. 2009;15(7):1083–1092. doi: 10.1242/dev.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. Wnt signaling in development and disease. Cell Biosci. 2012;15(1):14. doi: 10.1186/2045-3701-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudenschild AK, Hsieh AH, Kapila S, Lotz JC. Pressure and distortion regulate human mesenchymal stem cell gene expression. Ann Biomed Eng. 2009;15(3):492–502. doi: 10.1007/s10439-008-9629-2. [DOI] [PubMed] [Google Scholar]
- Robling AG, Niziolek PJ, Baldridge LA, Condon KW, Allen MR, Alam I, Mantila SM, Gluhak-Heinrich J, Bellido TM, Harris SE. et al. Mechanical stimulation of bone in vivo reduces osteocyte expression of Sost/sclerostin. J Biol Chem. 2008;15(9):5866–5875. doi: 10.1074/jbc.M705092200. [DOI] [PubMed] [Google Scholar]
- Dell’Accio F, De Bari C, El Tawil NM, Barone F, Mitsiadis TA, O’Dowd J, Pitzalis C. Activation of WNT and BMP signaling in adult human articular cartilage following mechanical injury. Arthritis Res Ther. 2006;15(5):R139. doi: 10.1186/ar2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowlan NC, Murphy P, Prendergast PJ. A dynamic pattern of mechanical stimulation promotes ossification in avian embryonic long bones. J Biomech. 2008;15(2):249–258. doi: 10.1016/j.jbiomech.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Bernstein P, Sticht C, Jacobi A, Liebers C, Manthey S, Stiehler M. Expression pattern differences between osteoarthritic chondrocytes and mesenchymal stem cells during chondrogenic differentiation. Osteoarthritis Cartilage. 2010;15(12):1596–1607. doi: 10.1016/j.joca.2010.09.007. [DOI] [PubMed] [Google Scholar]
- James CG, Appleton CT, Ulici V, Underhill TM, Beier F. Microarray analyses of gene expression during chondrocyte differentiation identifies novel regulators of hypertrophy. Mol Biol Cell. 2005;15(11):5316–5333. doi: 10.1091/mbc.E05-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallheden T, Karlsson C, Brunner A, Van Der Lee J, Hagg R, Tommasini R, Lindahl A. Gene expression during redifferentiation of human articular chondrocytes. Osteoarthritis Cartilage. 2004;15(7):525–535. doi: 10.1016/j.joca.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;15(4):493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Landry JR. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;15(3):249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;15(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;15(9):1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller-Peck CR, Mullen RJ. Altered cell proliferation in the spinal cord of mouse neural tube mutants curly tail and Pax3 splotch-delayed. Brain Res Dev Brain Res. 1997;15(2):177–188. doi: 10.1016/S0165-3806(97)00095-3. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;15(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;15(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data Tables S1-S4.