Abstract
Objectives: A number of studies conducted to assess the association between Clara cell 10-kDa protein (CC10) +38A/G polymorphism and susceptibility to asthma have yielded inconsistent and inconclusive results. In the present study, the possible association was assessed by a meta-analysis.
Methods: Relevant articles were identified for the period ranging from Jan 1998 up to March 2013. Pooled odds ratios (OR) with 95% confidence intervals (CI) were appropriately derived from fixed effects or random-effects models.
Results: Ten case-control studies with a total of 1529 asthma cases and 2399 controls were included in this meta-analysis. The association between CC10 +38A/G polymorphism and asthma risk was determined in dominant model, recessive model, additive model, and codominant model. In dominant model, CC10 +38A/G polymorphism seemed to be associated with elevated asthma risk (OR = 1.62; 95% CI, 1.23-2.12; P = 0.0005). Subgroup analyses by ethnicity also found significant associations between this polymorphism and asthma risk in Asians and Caucasians. Results from other genetic models further identified this possible association.
Conclusion: This meta-analysis suggests that CC10 +38A/G polymorphism confers asthma risk.
Key Words: Asthma, CC10, Meta-analysis, Polymorphism
INTRODUCTION
Asthma is an inflammatory disorder of the airways characterized by reversible airway obstruction and bronchial hyper-responsiveness. Although environmental factors are important determinants of asthma, numerous studies have revealed that asthma has a strong genetic component.1
Clara cell 10-kDa protein (CC10, also known as CC16, SCGB1A1, uteroglobin) is a 10-kDa protein produced by non-ciliated Clara cells and is one of the most abundant proteins in the fluids lining the airways.2 The CC10 gene is located on chromosome 11q12-13, a region that has been associated with asthma and atopy in several genome-wide linkage studies.3,4 A single nucleotide polymorphism, the +38A/G polymorphism (rs3741240), has been identified in this gene.5 Some papers have reported that this polymorphism was associated with asthma risk, whereas others found no such association.5-14 It was possible that most of these studies only included a modest sample size, and thus each of them might not achieve a reliable conclusion. Meta-analysis is a useful method for investigating associations between genetic factors and diseases, because a quantitative approach is used to combine the results from different studies on the same topic, thereby providing more reliable conclusions.15 Here, we use meta-analysis to clarify the association of CC10 +38A/G polymorphism with asthma. To our knowledge, this is the first genetic meta-analysis of the association between CC10 +38A/G polymorphism and the risk of asthma.
METHODS
Search for publications: We performed a literature search using the MEDLINE and China National Knowledge Infrastructure (CNKI) databases to identify articles that examined associations between CC10 +38A/G polymorphism and asthma. The following key words and subject terms were searched: Clara cell 10-kDa protein, CC10, CC16, SCGB1A1, uteroglobin, asthma, genetic, and polymorphism. Reference lists of articles retained for review were inspected for relevant publications. No publication date or language restrictions were imposed.
Inclusion and exclusion criteria: The following inclusion criteria were used: (i) the study should have evaluated the association between the +38A/G polymorphism and asthma risk; (ii) the study should have had a case-control design; (iii) sufficient data should have been provided in order to calculate odds ratios (OR) and 95% confidence intervals (CI). Studies were excluded if any of the following conditions applied: (i) only abstracts or reviews were available, without sufficient data; (ii) genotype frequencies were not reported; (iii) studies were repeated or publications overlapped.
Data extraction: The following information was extracted from each study: author, year of publication, ethnicity of the study population, numbers of cases and controls, and CC10 +38A/G genotype numbers.
Statistical analysis: Meta-analyses were performed using dominant model (AA + AG vs GG). Heterogeneity among studies was assessed using the chi-square based Q and I2 statistics. Heterogeneity was considered to be significant for I2 > 50%. Fixed-effects or random-effects models were used to pool the data according to heterogeneity. Other comparative genetic models were also used to assess the association between the polymorphism and the risk of asthma (AA vs AG + GG, AA vs GG, AG vs GG, and A vs G). For the subgroup analysis by ethnicity, the study populations were stratified into two groups: Asian and Caucasian. Galbraith plot was used to spot the outliers which were the sources of heterogeneity. Funnel plot and Egger’s test were used to detect publication bias.16 Analyses were performed using Revman 5.1 and STATA 11.0 software.
RESULTS
Characteristics of studies: A total of 10 relevant articles on CC10 +38A/G polymorphism in asthma met the study inclusion criteria (Table-I), and were included in the meta-analysis.5-14 Finally, 1529 genotyped asthma cases and 2399 genotyped non-asthmatic controls were included in the meta-analysis. Five case-control studies were performed in Asian populations; while five were performed in Caucasian populations. The characteristics of each case-control study and the genotype and allele distributions in each case-control study are presented in Table-I.
Table-I.
Characteristics of the individual studies included in the meta-analysis
| Cases/ |
Asthma genotypes
|
|
Asthma genotypes
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Author (Ref) | Year | Ethnicity | Controls (n) | AA | AG | GG | AA | AG | GG | |
| Laing (5) | 1998 | Caucasian | 137/68 | 26 | 67 | 44 | 5 | 26 | 37 | |
| Mansur (6) | 2002 | Caucasian | 127/67 | 14 | 57 | 56 | 8 | 31 | 28 | |
| Sengler (7) | 2003 | Caucasian | 188/118 | 15 | 91 | 82 | 12 | 50 | 56 | |
| Gui (8) | 2003 | Asian | 50/50 | 16 | 21 | 13 | 8 | 16 | 26 | |
| Sharma (9) | 2004 | Asian | 259/251 | 72 | 119 | 68 | 74 | 115 | 62 | |
| Candelaria (10) | 2005 | Caucasian | 127/801 | 23 | 66 | 38 | 81 | 386 | 334 | |
| Zhang (11) | 2009 | Caucasian | 42/561 | 5 | 21 | 16 | 77 | 249 | 235 | |
| Ku (12) | 2011 | Asian | 202/217 | 63 | 78 | 61 | 33 | 82 | 102 | |
| Chen (13) | 2012 | Asian | 277/211 | 177* | 100 | 86* | 125 | |||
| Zhang YL (14) | 2012 | Asian | 120/55 | 19 | 47 | 54 | 5 | 17 | 33 | |
* number of AA genotype and AG genotype. NA, not available
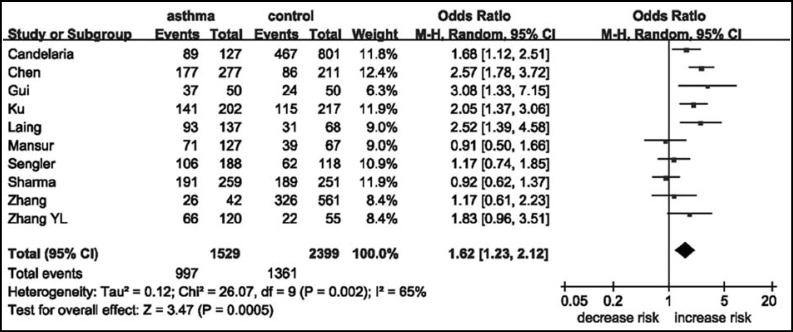
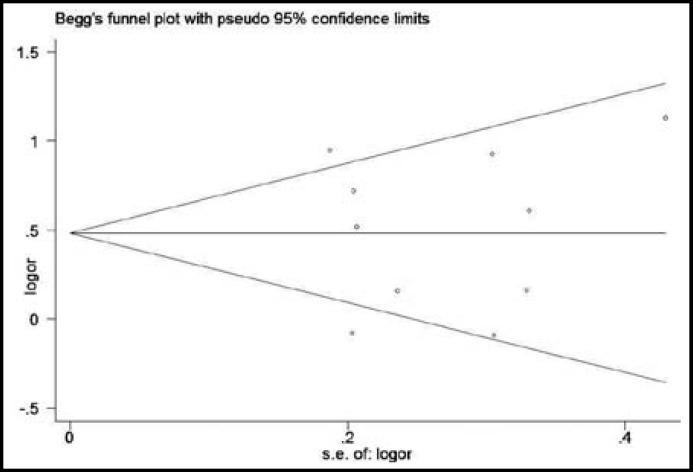
Results of meta-analyses: As shown in Fig.1, the heterogeneity of AA + AG vs GG was assessed for all 10 studies, and the chi-square value was 26.07 with 9 degrees of freedom and P = 0.002 in a random-effects model. Therefore, a random-effects model was used for synthesis of the data. The overall OR was 1.62 (95% CI, 1.23-2.12), and the Z-test value for overall effect was 3.47 (P = 0.0005) for the AA + AG vs GG. Subgroup analysis by ethnicity was performed. For ethnicity, the populations were stratified into two groups: Asian (908 cases and 784 controls), Caucasian (621 cases and 1615 controls). There were significant associations with asthma risk in these populations: Asian (OR = 1.87; 95% CI, 1.20-2.91; P = 0.005) and Caucasian (OR = 1.41; 95% CI, 1.02-1.94; P = 0.04). Publication bias was assessed graphically by using funnel plots and assessed statistically by using Egger’s test. The shape of the Begg’s funnel plots appeared symmetrical for the AA + AG vs GG model (Fig.2). Egger’s test was performed to provide statistical evidence of funnel plot asymmetry (P = 0.99). Summary results from the other comparisons are presented in Table-II.
Fig.1.
Meta-analysis of the correlation between the CC10 +38A/G polymorphism and asthma risk using a dominant model
Fig.2.
Begg’s funnel plot for evaluation of publication bias in the selection of studies on the association between asthma risk and the CC10 +38A/G polymorphism (dominant model).
Table-II.
Summary of comparisons based on different genotypes
|
Sample size
|
No. of |
Test of association
|
||||||
|---|---|---|---|---|---|---|---|---|
| Study | case | control | studies | OR (95% CI) | Z | P value | ||
| AA + AG vs. GG | Overall | 1529 | 2399 | 10 | 1.62 (1.23 – 2.12) | 3.47 | 0.0005 | |
| Asian | 908 | 784 | 5 | 1.87 (1.20 – 2.91) | 2.78 | 0.005 | ||
| Caucasian | 621 | 1615 | 5 | 1.41 (1.02 – 1.94) | 2.08 | 0.04 | ||
| AA vs. AG + GG | Overall | 1252 | 2188 | 9 | 1.48 (1.02 – 2.15) | 2.09 | 0.04 | |
| Asian | 631 | 573 | 4 | 1.42 (0.92 – 3.26) | 1.69 | 0.09 | ||
| Caucasian | 621 | 1615 | 5 | 1.31 (0.79 – 2.18) | 1.05 | 0.29 | ||
| AA vs. GG | Overall | 685 | 1216 | 9 | 1.77 (1.11 – 2.82) | 2.41 | 0.02 | |
| Asian | 366 | 343 | 4 | 2.14 (0.96 – 4.81) | 1.85 | 0.06 | ||
| Caucasian | 319 | 873 | 5 | 1.53 (0.82 – 2.86) | 1.33 | 0.18 | ||
| AG vs. GG | Overall | 999 | 1885 | 9 | 1.37 (1.14 – 1.64) | 3.43 | 0.0006 | |
| Asian | 461 | 453 | 4 | 1.36 (1.04 – 1.78) | 2.26 | 0.02 | ||
| Caucasian | 538 | 1432 | 5 | 1.37 (1.08 – 1.75) | 2.58 | 0.01 | ||
| A vs. G | Overall | 2504 | 4376 | 9 | 1.40 (1.10 – 1.78) | 2.72 | 0.006 | |
| Asian | 1262 | 1146 | 4 | 1.60 (1.00 – 2.57) | 1.97 | 0.05 | ||
| Caucasian | 1242 | 3230 | 5 | 1.27 (0.96 – 1.68) | 1.70 | 0.09 | ||
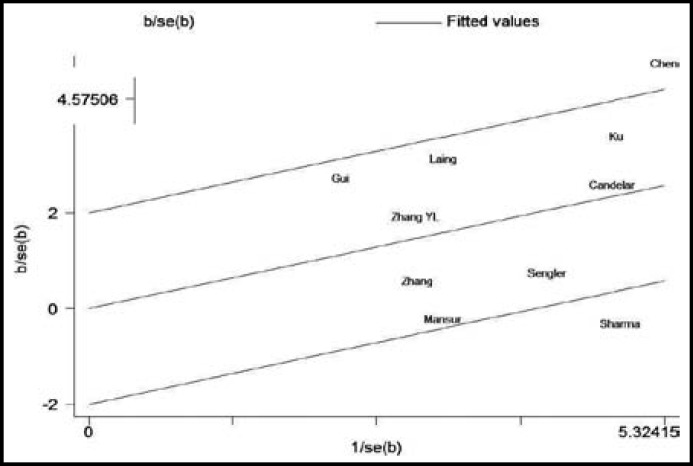
Heterogeneity analysis: The between-study heterogeneity was obvious in dominant model (I2 = 65%). Galbraith plots spotted Sharma’s study and Chen’s study as the outliers and the major source of the heterogeneity (Fig.3). The heterogeneity remarkably decreased in dominant model after excluding Sharma’s study and Chen’s study (I2 = 43%). Besides, a significant association was also observed after excluding Sharma’s study and Chen’s study (OR = 1.63; 95% CI, 1.26-2.11; P = 0.0002).
Fig.3.
Galbraith plots of association between CC10 +38A/G polymorphism and asthma risk
DISCUSSION
CC10 with primary expression in the uterus and non-ciliated bronchiolar cells, has an anti-inflammatory effect on the urogenital and respiratory tracts.17,18 Deficiency of CC10 aggravates pulmonary allergic inflammation through augmentation of the TH2 response, and reconstitution of CC10 in CC10-deficient mice is able to reverse the altered phenotypes.19 These results suggested that CC10 played an important role in the development of asthma. Although asthma can have many causes, genetic factors are considered strong determinants, and therefore, researchers have searched for the genes responsible. Several studies have investigated the role of CC10 +38A/G polymorphism in asthma susceptibility both in Asians and Caucasians, with contrasting results. Meta-analysis has been recognized as an important tool to more precisely define the effect of selected genetic polymorphisms on risk of disease. Therefore, we performed the meta-analysis specifically to assess this association.
The present meta-analysis found a significant association between the CC10 +38A/G polymorphism and asthma risk (OR = 1.62; 95% CI, 1.23-2.12; P = 0.0005). In addition, subgroup analysis by ethnicity showed that this polymorphism was significantly associated with increased asthma risk in both the Caucasians (OR = 1.41; 95% CI, 1.02-1.94; P = 0.04) and Asians (OR = 1.87; 95% CI, 1.20-2.91; P = 0.005). Moreover, other genetic models also found positive results. Thus, meta-analysis of available data suggests CC10 +38A allele contributes to increased asthma risk.
The importance of heterogeneity should be mentioned, as it is an important issue when interpreting the results of meta-analyses. In this present meta-analysis, we found obvious heterogeneity (I2 = 65%). Galbraith plot was used to spot the outliers as the possible major sources of heterogeneity. Two studies were spotted as the outliers. After excluding those two studies, the between-study heterogeneity decreased and there was no obvious heterogeneity among the left 8 studies (I2 = 43%), which suggested the heterogeneity might come from those two studies. Meta-analysis of the left 8 studies also showed CC10 +38A/G polymorphism was associated with increased risk of asthma (OR = 1.63; 95% CI,1.26-2.11; P = 0.0002).
Some possible limitations in this meta-analysis should be acknowledged. First, only published studies that were included in the selected electronic databases were identified; it is possible that some relevant published or unpublished studies that supported the null hypothesis may have been missed, which may have biased the results, and this may not have been detected by the statistical tests that were performed. Second, the effect of gene-gene and gene-environment interactions was not addressed in this meta-analysis status, because of limited available data. Third, all the data were used without adjustment by detailed individual information such as age, sex, and lifestyle in our meta-analysis. Finally, the number of included studies was moderate.
To sum up, this meta-analysis suggests that CC10 +38A/G polymorphism confers asthma risk. Further studies can assess the possible gene-environmental and gene-gene interactions in the association between this polymorphism and asthma risk.
Author Contribution:
Guangri Zhao, Xiaodan Lin, Ming Zhou and Jian Zhao: All the authors had substantial contributions to conception and design, acquisition of data, analysis and interpretation of data, drafting the manuscript and final approval of the version to be published.
Conflicts of interest: None.
References
- 1.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 201;242(1):10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mukherjee AB, Zhang Z, Chilton BS. Uteroglobin: a steroid-inducible immunomodulatory protein that founded the Secretoglobin superfamily. Endocr Rev. 2007;28(7):707–725. doi: 10.1210/er.2007-0018. [DOI] [PubMed] [Google Scholar]
- 3.Huang SK, Mathias RA, Ehrlich E, Plunkett B, Liu X, Cutting GR, et al. Evidence for asthma susceptibility genes on chromosome 11 in an African-American population. Hum Genet. 2003;113(1):71–75. doi: 10.1007/s00439-003-0934-4. [DOI] [PubMed] [Google Scholar]
- 4.Cookson W, Faux J, Sharp P, Hopkin J. Linkage between immunoglobulin E responses underlying asthma and rhinitis and chromosome 11q. Lancet. 1989;1(8650):1292–1295. doi: 10.1016/s0140-6736(89)92687-1. [DOI] [PubMed] [Google Scholar]
- 5.Laing IA, Goldblatt J, Eber E, Hayden CM, Rye PJ, Gibson NA, et al. A polymorphism of the CC16 gene is associated with an increased risk of asthma. J Med Genet. 1998;35(6):463–467. doi: 10.1136/jmg.35.6.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mansur AH, Fryer AA, Hepple M, Strange RC, Spiteri MA. An association study between the Clara cell secretory protein CC16 A38G polymorphism and asthma phenotypes. Clin Exp Allergy. 2002;32(7):994–999. doi: 10.1046/j.1365-2222.2002.01426.x. [DOI] [PubMed] [Google Scholar]
- 7.Sengler C, Heinzmann A, Jerkic SP, Haider A, Sommerfeld C, Niggemann B, et al. Clara cell protein 16 (CC16) gene polymorphism influences the degree of airway responsiveness in asthmatic children. J Allergy Clin Immunol. 2003;111(3):515–519. doi: 10.1067/mai.2003.180. [DOI] [PubMed] [Google Scholar]
- 8.Gui Q, Qian GS, Huang GJ, Li SP. Study on association between CC16 gene G38A mutation and asthma in the patients of Han population in Chongqing, China. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2003;20(5):542–543. [PubMed] [Google Scholar]
- 9.Sharma S, Ghosh B. Association of an intragenic microsatellite marker in the CC16 gene with asthma in the Indian population. J Hum Genet. 2004;49(12):677–683. doi: 10.1007/s10038-004-0206-8. [DOI] [PubMed] [Google Scholar]
- 10.Candelaria PV, Backer V, Laing IA, Porsbjerg C, Nepper-Christensen S, de Klerk N, et al. Association between asthma-related phenotypes and the CC16 A38G polymorphism in an unselected population of young adult Danes. Immunogenetics. 2005;57(1-2):25–32. doi: 10.1007/s00251-005-0778-2. [DOI] [PubMed] [Google Scholar]
- 11.Zhang G, Khoo SK, Laatikainen T, Pekkarinen P, Vartiainen E, von Hertzen L, et al. Opposite gene by environment interactions in Karelia for CD14 and CC16 single nucleotide polymorphisms and allergy. Allergy. 2009;64(9):1333–1341. doi: 10.1111/j.1398-9995.2009.02006.x. [DOI] [PubMed] [Google Scholar]
- 12.Ku MS, Sun HL, Lu KH, Sheu JN, Lee HS, Yang SF, et al. The CC16 A38G polymorphism is associated with the development of asthma in children with allergic rhinitis. Clin Exp Allergy. 2011;41(6):794–800. doi: 10.1111/j.1365-2222.2010.03679.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen LC, Tseng HM, Wu CJ, Kuo ML, Wu CJ, Gao PS, et al. Evaluation of a common variant of the gene encoding clara cell 10 kd protein (CC10) as a candidate determinant for asthma severity and steroid responsiveness among Chinese children. J Asthma. 2012;49(7):665–672. doi: 10.3109/02770903.2012.697954. [DOI] [PubMed] [Google Scholar]
- 14.Zhang YL, Wang XF, Hou YY, Bi D. Relationship between Clara cell protein 10 gene G38A polymorphism and pathogens of wheezing in children under 5 years old. J Appl Clin Pediatr. 2012;27(2):769–770. [Google Scholar]
- 15.Kavvoura FK, Ioannidis JP. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum Genet. 2008;123(1):1–14. doi: 10.1007/s00439-007-0445-9. [DOI] [PubMed] [Google Scholar]
- 16.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh G, Katyal SL, Brown WE, Kennedy AL, Singh U, Wong-Chong M-L. Clara cell 10 kDa protein (CC10): Comparison of structure and function to uterogloblin. Biochim Biophys Acta. 1990;1039(3):348–355. doi: 10.1016/0167-4838(90)90270-p. [DOI] [PubMed] [Google Scholar]
- 18.Bernard A, Roels H, Lauwerys R, Witters R, Gielens C, Soumillion A, et al. Human urinary protein 1: evidence for identity with the Clara cell protein and occurrence in respiratory tract and urogenital secretions. Clin Chim Acta. 1992;207(3):239–249. doi: 10.1016/0009-8981(92)90122-7. [DOI] [PubMed] [Google Scholar]
- 19.Chen L-C, Zhang Z, Myers AC, Huang S-K. Cutting edge: altered pulmonary eosinophilic inflammation in mice deficient for Clara cell secretory 10-kDa protein. J Immunol. 2001;167(6):3025–3028. doi: 10.4049/jimmunol.167.6.3025. [DOI] [PubMed] [Google Scholar]