Abstract
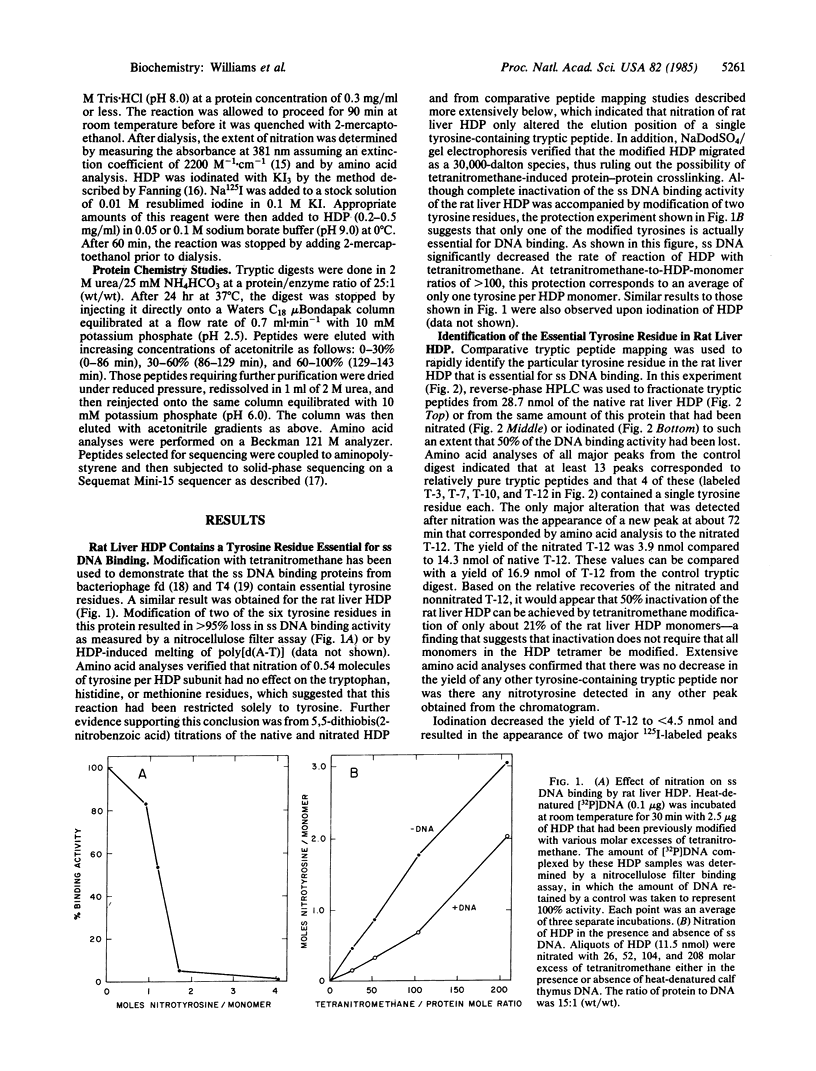
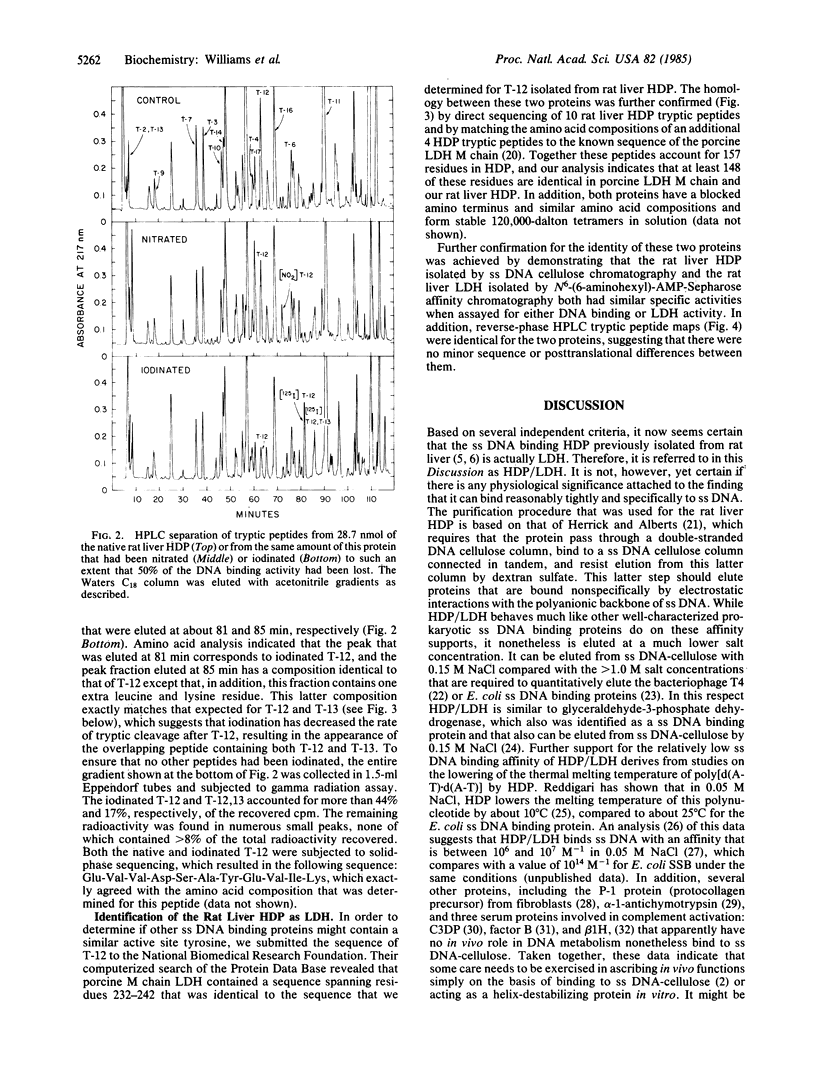
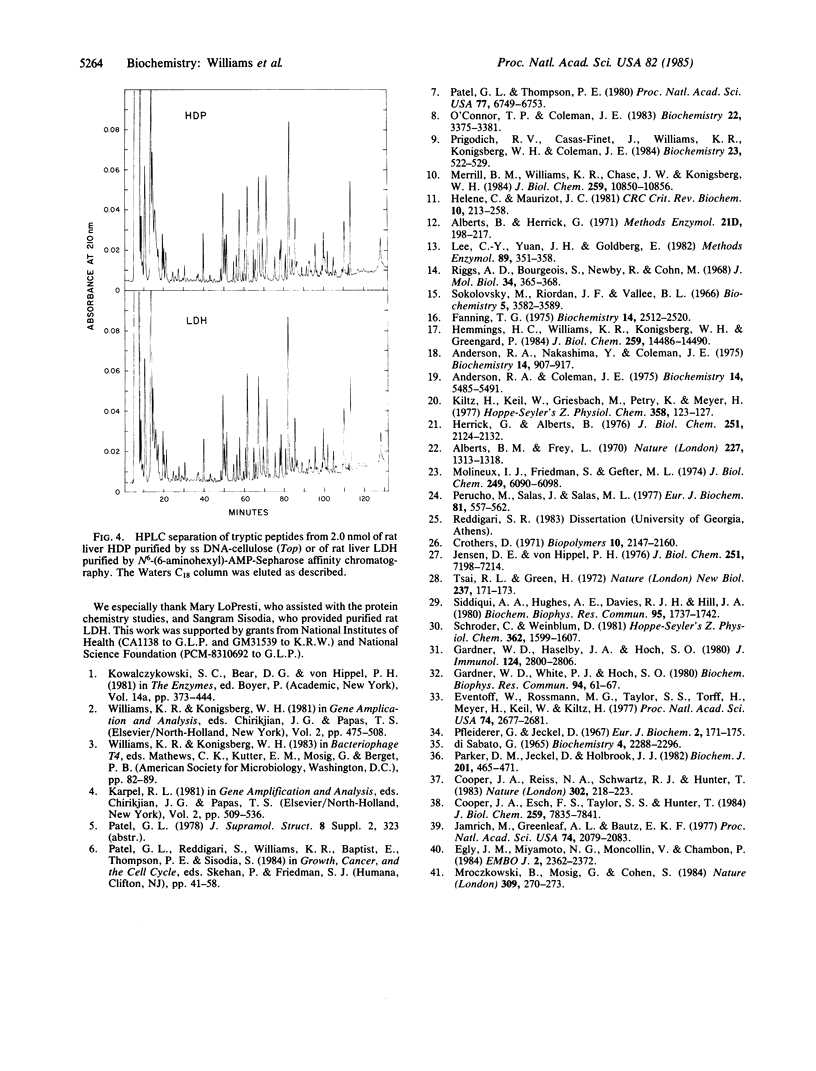
A rat liver DNA helix-destabilizing protein (HDP) that has previously been proposed to play a role in transcription has been identified as M chain lactate dehydrogenase (LDH-5; L-lactate:NAD+ oxidoreductase, EC 1.1.1.27). Tryptic peptides accounting for 157 amino acids in the rat liver HDP have been characterized and then matched to the published sequence for the M chain of porcine LDH. Based on amino acid compositions and direct solid-phase protein sequencing, at least 148 of the 157 residues that were compared are identical in both proteins. In addition, both porcine LDH and the rat liver HDP have blocked amino termini and similar amino acid compositions and molecular weights. Rat liver HDP and LDH-5 that were purified to molecular homogeneity had similar specific activities in both single-stranded DNA (ss DNA) binding and LDH assays. HPLC tryptic peptide maps were also identical for both the rat liver HDP and LDH proteins. Since preincubation of HDP in NADH prevents its binding to ss DNA, both NADH and ss DNA may be binding at the same site. Further support for this latter idea derives from chemical-modification studies which demonstrate that tyrosine-238, which is located near the coenzyme binding site of LDH, seems to be essential for the ability of HDP to bind ss DNA. These results indicate caution in ascribing in vivo roles solely on the basis of binding to ss DNA. Alternatively, they suggest that a single protein may play more than one biological role.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M., Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970 Sep 26;227(5265):1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- Anderson R. A., Coleman J. E. Physiochemical properties of DNA binding proteins: gene 32 protein of T4 and Escherichia coli unwinding protein. Biochemistry. 1975 Dec 16;14(25):5485–5491. doi: 10.1021/bi00696a017. [DOI] [PubMed] [Google Scholar]
- Anderson R. A., Nakashima Y., Coleman J. E. Chemical modifications of functional residues of fd gene 5 DNA-binding protein. Biochemistry. 1975 Mar 11;14(5):907–917. doi: 10.1021/bi00676a006. [DOI] [PubMed] [Google Scholar]
- Cooper J. A., Esch F. S., Taylor S. S., Hunter T. Phosphorylation sites in enolase and lactate dehydrogenase utilized by tyrosine protein kinases in vivo and in vitro. J Biol Chem. 1984 Jun 25;259(12):7835–7841. [PubMed] [Google Scholar]
- Cooper J. A., Reiss N. A., Schwartz R. J., Hunter T. Three glycolytic enzymes are phosphorylated at tyrosine in cells transformed by Rous sarcoma virus. Nature. 1983 Mar 17;302(5905):218–223. doi: 10.1038/302218a0. [DOI] [PubMed] [Google Scholar]
- Crothers D. M. Statistical thermodynamics of nucleic acid melting transitions with coupled binding equilibria. Biopolymers. 1971 Nov;10(11):2147–2160. doi: 10.1002/bip.360101110. [DOI] [PubMed] [Google Scholar]
- Eventoff W., Rossmann M. G., Taylor S. S., Torff H. J., Meyer H., Keil W., Kiltz H. H. Structural adaptations of lactate dehydrogenase isozymes. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2677–2681. doi: 10.1073/pnas.74.7.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G. Iodination of Escherichia coli lac repressor. Effect of tyrosine modification on repressor activity. Biochemistry. 1975 Jun 3;14(11):2512–2520. doi: 10.1021/bi00682a034. [DOI] [PubMed] [Google Scholar]
- Gardner W. D., Haselby J. A., Hoch S. O. Identification of a major serum DNA-binding protein as factor B of the alternative complement pathway. J Immunol. 1980 Jun;124(6):2800–2806. [PubMed] [Google Scholar]
- Gardner W. D., White P. J., Hoch S. O. Identification of a major human serum DNA-binding protein as beta 1H of the alternative pathway of complement activation. Biochem Biophys Res Commun. 1980 May 14;94(1):61–67. doi: 10.1016/s0006-291x(80)80187-2. [DOI] [PubMed] [Google Scholar]
- Helene C., Maurizot J. C. Interactions of oligopeptides with nucleic acids. CRC Crit Rev Biochem. 1981;10(3):213–258. doi: 10.3109/10409238109113600. [DOI] [PubMed] [Google Scholar]
- Hemmings H. C., Jr, Williams K. R., Konigsberg W. H., Greengard P. DARPP-32, a dopamine- and adenosine 3':5'-monophosphate-regulated neuronal phosphoprotein. I. Amino acid sequence around the phosphorylated threonine. J Biol Chem. 1984 Dec 10;259(23):14486–14490. [PubMed] [Google Scholar]
- Herrick G., Alberts B. Purification and physical characterization of nucleic acid helix-unwinding proteins from calf thymus. J Biol Chem. 1976 Apr 10;251(7):2124–2132. [PubMed] [Google Scholar]
- Jamrich M., Greenleaf A. L., Bautz E. K. Localization of RNA polymerase in polytene chromosomes of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1977 May;74(5):2079–2083. doi: 10.1073/pnas.74.5.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen D. E., von Hippel P. H. DNA "melting" proteins. I. Effects of bovine pancreatic ribonuclease binding on the conformation and stability of DNA. J Biol Chem. 1976 Nov 25;251(22):7198–7214. [PubMed] [Google Scholar]
- Kiltz H. H., Keil W., Griesbach M., Petry K., Meyer H. The primary structure of porcine lactate dehydrogenase: isoenzymes M4 and H4. Hoppe Seylers Z Physiol Chem. 1977 Jan;358(1):123–127. doi: 10.1515/bchm2.1977.358.1.123. [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Yuan J. H., Goldberg E. Lactate dehydrogenase isozymes from mouse. Methods Enzymol. 1982;89(Pt 500):351–358. doi: 10.1016/s0076-6879(82)89063-0. [DOI] [PubMed] [Google Scholar]
- Merrill B. M., Williams K. R., Chase J. W., Konigsberg W. H. Photochemical cross-linking of the Escherichia coli single-stranded DNA-binding protein to oligodeoxynucleotides. Identification of phenylalanine 60 as the site of cross-linking. J Biol Chem. 1984 Sep 10;259(17):10850–10856. [PubMed] [Google Scholar]
- Molineux I. J., Friedman S., Gefter M. L. Purification and properties of the Escherichia coli deoxyribonucleic acid-unwinding protein. Effects on deoxyribonucleic acid synthesis in vitro. J Biol Chem. 1974 Oct 10;249(19):6090–6098. [PubMed] [Google Scholar]
- Mroczkowski B., Mosig G., Cohen S. ATP-stimulated interaction between epidermal growth factor receptor and supercoiled DNA. Nature. 1984 May 17;309(5965):270–273. doi: 10.1038/309270a0. [DOI] [PubMed] [Google Scholar]
- Parker D. M., Jeckel D., Holbrook J. J. Slow structural changes shown by the 3-nitrotyrosine-237 residue in pig heart [Tyr(3NO2)237] lactate dehydrogenase. Biochem J. 1982 Mar 1;201(3):465–471. doi: 10.1042/bj2010465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G. L., Thompson P. E. Immunoreactive helix-destabilizing protein localized in transcriptionally active regions of Drosophila polytene chromosomes. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6749–6753. doi: 10.1073/pnas.77.11.6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perucho M., Salas J., Salas M. L. Identification of the mammalian DNA-binding protein P8 as glyceraldehyde-3-phosphate dehydrogenase. Eur J Biochem. 1977 Dec;81(3):557–562. doi: 10.1111/j.1432-1033.1977.tb11982.x. [DOI] [PubMed] [Google Scholar]
- Pfleiderer G., Jeckel D. Kupplungsreaktion an Lactatdehydrogenase nach reversibler Maskierung der SH-Gruppen. Umsetzung mit p-Diazobenzolsulfonsäure zur Markierung essentieller Histidin- oder Tyrosinreste. Eur J Biochem. 1967 Sep;2(2):171–175. doi: 10.1111/j.1432-1033.1967.tb00122.x. [DOI] [PubMed] [Google Scholar]
- Prigodich R. V., Casas-Finet J., Williams K. R., Konigsberg W., Coleman J. E. 1H NMR (500 MHz) of gene 32 protein--oligonucleotide complexes. Biochemistry. 1984 Jan 31;23(3):522–529. doi: 10.1021/bi00298a019. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Newby R. F., Cohn M. DNA binding of the lac repressor. J Mol Biol. 1968 Jul 14;34(2):365–368. doi: 10.1016/0022-2836(68)90261-1. [DOI] [PubMed] [Google Scholar]
- Schröder C., Weinblum D. Equilibrium binding constants of C3DP, a nucleic acid-binding protein isolated from human serum. Hoppe Seylers Z Physiol Chem. 1981 Dec;362(12):1599–1607. doi: 10.1515/bchm2.1981.362.2.1599. [DOI] [PubMed] [Google Scholar]
- Siddiqui A. A., Hughes A. E., Davies R. J., Hill J. A. The isolation and identification of alpha 1-antichymotrypsin as a DNA-binding protein from human serum. Biochem Biophys Res Commun. 1980 Aug 29;95(4):1737–1742. doi: 10.1016/s0006-291x(80)80099-4. [DOI] [PubMed] [Google Scholar]
- Sokolovsky M., Riordan J. F., Vallee B. L. Tetranitromethane. A reagent for the nitration of tyrosyl residues in proteins. Biochemistry. 1966 Nov;5(11):3582–3589. doi: 10.1021/bi00875a029. [DOI] [PubMed] [Google Scholar]
- Tsai R. L., Green H. Study of intracellular collagen precursors using DNA-cellulose chromatography. Nat New Biol. 1972 Jun 7;237(75):171–173. doi: 10.1038/newbio237171a0. [DOI] [PubMed] [Google Scholar]



